Abstract
The use of non-steroidal anti-inflammatory drugs (NSAIDs) is associated with reduced risk of colorectal neoplasia. Previous studies have reported that polymorphisms in NSAID-metabolizing enzymes central to NSAID metabolism including UDP-glucuronosyltransferases (UGT) and cytochrome P450 (CYP) 2C9 may modify this protective effect. We investigated whether 35 functionally relevant polymorphisms within CYP2C9 and UGT genes were associated with colorectal cancer risk or modified the protective effect of NSAIDs on colorectal cancer susceptibility, using 1,584 colorectal cancer cases and 2,516 unaffected sibling controls from the Colon Cancer Family Registry. A three-SNP genotype in UGT1A6 (G-A-A; Ala7-Thr181-Arg184) and the Asp85 variant in UGT2B15 increased the risk of colorectal cancer (OR 3.87; 95% CI 1.04-14.45 and OR 1.34; 95% CI 1.10-1.63, respectively). We observed interactions between UGT1A3 Thr78Thr (A>G) and NSAID use (p-interaction=0.02), a three-SNP genotype within UGT2B4 and ibuprofen use (p-interaction=0.0018), as well as UGT2B15 Tyr85Asp (T>G) and aspirin use (p-interaction=0.01). The interaction with the UGT2B4 and the UGT2B15 polymorphisms were noteworthy at the 25% FDR level. This study highlights the need for further pharmacogenetic studies to identify individuals who might benefit from NSAID use as part of developing effective strategies for prevention of colorectal neoplasia.
Keywords: UGT, CYP2C9, colorectal cancer, ibuprofen, aspirin, non-steroidal anti-inflammatory drugs, NSAIDs
Introduction
Several studies, including randomized trials, have reported protective effects of non-steroidal anti-inflammatory drugs (NSAIDs) on colorectal carcinogenesis as well as on the etiology of other tumors, illustrating the chemopreventive potential of NSAIDs (Flossmann and Rothwell, 2007; Cross et al., 2008; Chia et al., 2012; Rothwell et al., 2010; 2012a). Many questions remain, however, unanswered, especially regarding dosage, treatment duration and long-term benefit of NSAID use (Potter, 2012). NSAIDs reduce inflammation through inhibition of prostaglandin synthesis (by blocking prostaglandin-endoperoxide synthases 1 (COX-1) and 2 (COX-2)) and thus affect pathways that are relevant for carcinogenesis such as proliferation, apoptosis and angiogenesis (Ulrich et al., 2006). In support, recent meta-analyses among >24,000 individuals showed a 37% reduction of colorectal cancer and a 19% reduction of cancer mortality overall among individuals using aspirin (Rothwell et al., 2012a). Interestingly, data on molecular tumor pathology of colorectal cancers suggest that the preventive effect of aspirin may be limited to a subset of tumors carrying mutations in PI3KCA (Ogino et al., 2013; Tougeron et al., 2013). Furthermore, the effectiveness of NSAIDs depends on the bioavailability of the active drug compound. Inter-individual differences in drug metabolism may modify therapeutic effects, which can be a direct consequence of polymorphisms within related genes (Ulrich et al., 2006; Cross et al., 2008). Polymorphisms in genes of drug-metabolizing enzymes have long been known to have functional impacts on pharmacokinetics, particularly on bioavailability (Weber and Hein, 1979; Ciotti et al., 1997; Takahashi et al., 1998). The metabolism of NSAIDs primarily involves glycine N-acyltransferase and uridine 5′ diphosphate glucuronosyl transferases (e.g. UGT1A6) and cytochrome P450 2C9 (CYP2C9) (Hutt et al., 1986; Leemann et al., 1993). Thus, interactions between polymorphisms in CYP2C9 or the UGT gene families and NSAID use may modify the risk of colorectal cancer.
Previous studies have investigated the effect of selected polymorphisms (CYP2C9*2, CYP2C9*3, UGT1A6*2) (Rettie et al., 1994; Haining et al., 1996; Sullivan-Klose et al., 1996; Ciotti et al., 1997; Steward et al., 1997; Takahashi et al., 1998; Gill et al., 1999; Bigler et al., 2001; Chan et al., 2005; Samowitz et al., 2006; Bae et al., 2011) that appear to have functional consequences on colorectal cancer risk. Their findings suggest interactions between polymorphisms in drug-metabolizing enzymes and NSAID use, highlighting the potential of pharmacogenetics to tailor chemoprevention. Variants in UGT genes modified the risk of colorectal adenoma dependent on the use of NSAIDs (Bigler et al., 2001; Chan et al., 2005). UGT enzymes are phase II drug metabolizing enzymes, which modify xenobiotic or endobiotic compounds through glucuronidation. UGT1A6 variant enzymes were reported to display lower activity, resulting in a prolonged exposure to the active drug and consequently reduced colorectal adenoma risk (Bigler et al., 2001; Samowitz et al., 2006; Chan et al., 2011). However, only little is known about other UGT polymorphisms and their interaction with NSAIDs in colorectal cancer susceptibility.
The UGT1A gene family consists of four common exons and at least 13 variable exons, resulting in many shared sequences and consequently shared polymorphisms within this gene family giving rise to nine functional UGT1A enzymes (Mackenzie et al., 2005). In addition, several UGT enzymes share substrate specificity (Kuehl et al., 2005). Therefore, in order to study both the effect of UGT1A and UGT2B polymorphisms on risk of colorectal cancer and their potential to modify the protective effect of NSAID use on colorectal cancer risk, pharmacogenetic investigations of the UGT loci need to be performed in a targeted and comprehensive manner. Due to the complex structure of the UGT loci, genetic variation within this region is insufficiently covered on standard genome-wide association platforms. Consequently, GWAS consortia cannot provide thorough information to improve our understanding of the impact of UGT gene polymorphisms on cancer risk and many other phenotypes. Thus, many of the UGT variants reported here are being studied for the first time for an association with colorectal cancer risk.
We conducted a matched case-sibling control study, based on 1,584 colorectal cancer cases and 2,516 healthy controls, investigating a selection of putatively functional single nucleotide polymorphisms (SNPs) in ten genes of phase I (CYP2C9) and phase II (UGT) drug metabolizing enzymes in relation to risk of colorectal cancer. We also investigated combined genotypes across UGT genes, as multiple ‘hits’ in this detoxification machinery may be required to have an impact on colorectal carcinogenesis. Finally, a focus of this study was interactions between targeted polymorphisms and NSAID use in colorectal cancer risk.
Materials and Methods
Study Population
The study population has been described previously (Newcomb et al., 2007). Briefly, colorectal cancer cases were recruited for the Colon Cancer Family Registry (CCFR) from six registry centers. The CCFR cases were patients and affected relatives diagnosed with primary invasive colorectal cancer between 1998 and 2002 who were interviewed within five years of diagnosis. Controls were siblings without a colorectal cancer diagnosis at the time of enrollment. Although eligibility requirements varied slightly across registry centers, participants typically were required to be between the ages of 20 and 74 (Newcomb et al., 2007). Standard questionnaires were used to collect epidemiologic data from CCFR participants regarding demographic characteristics, medical history, NSAID use, family history of cancer, smoking history, diet, physical activity, height, and weight. NSAID use was defined as regular use in the two years prior to study enrollment. Blood and tissue samples were collected according to standardized procedures. Individuals were excluded from this study if the case did not have at least one matched unaffected sibling as a control or if an individual's sex determined by genotyping did not match reported sex on the questionnaire. Only individuals self-reported as Caucasian and collected through population-based recruitment, were included in these analyses. Informed consent was obtained from all participants. The Institutional Review Boards at each CCFR site approved the study. Blood samples were collected according to standardized procedures as described earlier (Newcomb et al., 2007) and DNA was extracted from peripheral blood leukocytes and quantified using the PicoGreen kit (Invitrogen, Paisley, United Kingdom).
Selection of polymorphisms and genotyping
Polymorphisms were selected based on a candidate gene approach with the aim of assessing the association of the complex genetic variation within UGT1A and UGT2B gene families, as well as CYP2C9, on both risk and effect modification of NSAID use on colorectal cancer. We selected 35 polymorphisms with minor allele frequencies of at least 3% in ten genes to capture genetic variants in UGT genes with previously demonstrated functional impact or with known amino acid changes (Haining et al., 1996; Ciotti et al., 1997; Takahashi et al., 1998; Krishnaswamy et al., 2005a; Thomas et al., 2006).
TagSNPs were specifically chosen to capture common non-synonymous variation in the UGT1A locus. We used extensive resequencing data from 92 Caucasian individuals (Thomas et al., 2006) to determine the LD structure of the locus and identify nsSNPs with minor allele frequencies greater than 3%. Outside of the UGT1A locus, the candidate genes CYP2C9, UGT2B4, UGT2B7 and UGT2B15 were chosen based on earlier functional work supporting their role in NSAID metabolism (Gill et al., 1999; Kuehl et al., 2005; Kuehl et al., 2006). With regards to the UGT2B genes, UGT2B4 and UGT2B7 were included because of their demonstrated glucuronidation of ibuprofen (Kuehl et al., 2005) and salicylic acid (Kuehl et al., 2006). In these experiments, there was little evidence that UGT2B15 or UGT2B17 could form acyl and phenolic glucuronides of salicylic acid at detectable levels. Furthermore, there were no non-synonymous SNPs in UGT2B17 with minor allele frequencies >3%. We included UGT2B15 due to its demonstrated, albeit low, glucuronidation of ibuprofen and naproxen in vitro (Kuehl et al., 2006).
TaqMan-based assays were performed for the CYP2C9 polymorphisms R144C (rs1799853) and I359L (rs1057910), the UGT1A6 polymorphisms T181A (rs2070959) and R184S (rs1105879), the UGT2B4 polymorphism D458E (rs13119049), the UGT2B15 polymorphism D85Y (rs1902023), and the UGT2B7 polymorphism Y268H (rs7439366) at the Fred Hutchinson Cancer Research Center using the Applied Biosystems 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Genotypes were assigned using the Allelic Discrimination Software (Applied Biosystems SDS Software, version 2.3). The CYP2C9 and UGT1A6 assays contained 3.75 ng of DNA, 2.5 μl of 2X TaqMan Universal PCR Mix (No AmpErase UNG), 300 nmol/L forward primer, 300 nmol/L reverse primer, 100nmol/L FAM-labeled MGB probe, 100nmol/L VIC-labeled MGB probe and double-distilled water to a final volume of 5 μl. Cycling conditions were 50°C 2min, 95°C 10min, 40 × 95°C, 15sec; 60°C, 1 min. The UGT2B15 polymorphism D85Y (rs1902023) was genotyped as above but with 900nM primer and 200nM probe concentrations and the PCR was extended to 50 cycles. UGT2B4 and UGT2B7 genotyping reactions used TaqMan core reagents (cat#4304439), 5ng DNA, 200nM primers, 5mM (UGT2B4) and 4mM (UGT2B7) MgCl2, 50nM (UGT2B4) and 200nM (UGT2B7) probes, and 50 PCR cycles. All assays were validated on 30 CEPH family trios purchased from the Coriell Cell Repository (CEPH HAPMAPPT01).
Five polymorphisms (UGT1A5 A158G (rs12475068), UGT1A6 S7A (rs6759892), UGT1A9 -275T>A (rs6714486), UGT2B4 3′UTR (rs1131878) and UGT2B4 5′UTR (rs1966151)) were genotyped using the Illumina GoldenGate platform and the Veracode BeadXpress Reader (Illumina, Inc. San Diego, CA) as described previously (Kleinstein et al., 2013). Call rates for all five genotypes exceeded 98%. The concordance for blinded duplicates was 100%.
The promoter repeat polymorphism in UGT1A1 (rs34815109) was genotyped using the GeneScan assay, as previously described (Thomas et al., 2006). The exon 1 polymorphisms in UGT1A3 and UGT1A7 were genotyped using direct sequencing, as described (Thomas et al., 2006).
For quality control, all assays were validated using the 30 CEPH family trios prior to running study samples. Each assay batch contained negative and positive controls and 5% of the total number of samples were re-genotyped to confirm reproducibility. All genotypes were also reviewed independently by two technicians. Laboratory staff was blinded to case-control status for all assays.
Two polymorphisms (rs28898617 and rs6431625), both located in UGT1A3 were not in Hardy-Weinberg Equilibrium and thus were excluded from further analyses.
Statistical Analysis
The associations between polymorphisms and the risk of colorectal cancer were assessed with conditional logistic regression models estimating odds ratios (OR) and 95% confidence intervals (CI) (Horvath and Laird, 1998; Gauderman et al., 1999). All models used kinship as the matching variable and adjusted for age and sex. For each polymorphism, the risk of colorectal cancer, rectal cancer, and colon cancer (combining proximal and distal) was estimated using a co-dominant model unless any cell had fewer than 30 individuals, in which case a dominant model was used. A recessive model was not estimated as the sample sizes were too small for this analysis. A trend test was performed using the log-additive model to investigate dose-dependent associations with the variant allele. Combined analyses of all polymorphisms within each gene and haplotype analyses were also conducted. For the combined genotype analyses, genotype combinations within each gene were analyzed both individually and in a dominant analysis, where those with at least one variant allele in a genotype combination were compared with those carrying only the major alleles. For the haplotype analyses, haplotype frequencies in cases and controls were inferred with the SAS/Genetics software module. For haplotypes with fewer than 5 individuals per cell, haplotypes were combined in one category (HRare). The analyses were carried out using the expectation-maximization algorithm to generate maximum likelihood estimates of haplotype frequencies.
We also investigated polymorphism interactions with NSAID, aspirin or ibuprofen use between current users vs. never/former users. In this study population, current NSAID use was more strongly inversely associated with colorectal cancer risk than other aspects of NSAID use and thus we chose it as the primary variable for interaction analysis. The reference groups were comprised of individuals who were homozygous for the wild-type allele(s) and never/former users of NSAIDs. Because use of NSAIDs may be associated with other known risk factors for colorectal cancer, NSAID interactions were adjusted for smoking (continuous pack-years), body mass index (BMI; continuous), and physical activity (four-level ordinal variable based on average MET hours) in addition to age (continuous) and sex. We assessed the interaction between genotype and the use of both aspirin and ibuprofen on a multiplicative scale using the cross-product of NSAID exposure and genotype; the likelihood-ratio test was used to determine the statistical significance of the interaction.
All tests of statistical significance were two-sided at α=0.05. The false discovery rate (FDR) of Benjamini and Hochberg was used to correct for multiple testing (Benjamini and Hochberg, 1995; Benjamini et al., 2001). Most of the investigated polymorphisms were selected based on functional significance at an in vitro or in silico levels, thus we used the FDR at 25%. Analyses were conducted using SAS Version 9.3 for Windows (SAS Institute Inc., Cary, NC).
Results
Characteristics of the study population are presented in Table 1. We observed no statistically significant differences between cases and unaffected sibling controls by age; regular NSAID, ibuprofen, or aspirin use; physical activity; or BMI. Cases were more likely to be male (p<0.01). A summary of genotyped polymorphisms and respective minor allele frequencies is given in Supplementary Table 1. Allele frequencies were consistent with previous studies in Caucasian populations (Bigler et al., 2001; Chan et al., 2005; Dura et al., 2012).
Table 1. Selected characteristics of population-based Colon Cancer Family Registry kinships restricted to Caucasians (N=4100).
| Cases (N=1584)a N (%) |
Controls (N=2516)a N (%) |
|
|---|---|---|
| Age (y), mean (SD) | 53.5±10.8 | 54.0±11.7 |
| Gender | ||
| Female | 774 (48.9) | 1390 (55.3) |
| Male | 810 (51.1) | 1126 (44.7) |
| Family History of Cancerb | ||
| Yes | 506 (32.5) | 771 (31.1) |
| No | 1051 (67.5) | 1708 (68.9) |
| Center | ||
| Ontario | 296 (18.7) | 491 (19.5) |
| USC (University of Southern California) | 319 (20.1) | 444 (17.7) |
| Australia | 317 (20.0) | 554 (22.0) |
| Hawaii | 6 (0.4) | 7 (0.3) |
| Mayo Foundation | 266 (16.8) | 502 (20.0) |
| Seattle | 380 (24.0) | 518 (20.6) |
| Tumor site | ||
| Proximal | 526 (33.2) | - |
| Distal | 460 (29.0) | - |
| Rectal | 523 (33.0) | - |
| Regular Aspirin usec | ||
| No | 1320 (84.1) | 2080 (84.1) |
| Yes | 250 (15.9) | 394 (15.9) |
| Regular Ibuprofen usec | ||
| No | 1444 (92.5) | 2255 (90.9) |
| Yes | 117 (7.5) | 226 (9.1) |
| Regular NSAID usec | ||
| No | 1234 (78.2) | 1916 (76.9) |
| Yes | 343 (21.8) | 577 (23.1) |
| Physical activity | ||
| Inactive | 375 (23.7) | 585 (23.3) |
| Less active | 424 (26.8) | 679 (27.0) |
| Active | 374 (23.6) | 564 (22.4) |
| Very active | 338 (21.3) | 548 (21.8) |
| BMI [kg/m2]±SD | 27.4 ± 6.0 | 26.8 ± 5.5 |
| Cigarette smoking (pack-years)±SD | 12.9 ±19.5 | 11.7 ± 19.3 |
Numbers may not add to total because of missing data.
First degree relative
Regular Aspirin, Ibuprofen and NSAID use defined as regular use of least two pills per week for at least one month.
SNP risk estimates
Out of 18 analyzed polymorphisms, one showed a significant association with colorectal cancer risk (Supplementary Table 2). A non-synonymous polymorphism in the UGT2B15 gene (rs1902023, T>G; UGT2B15*2), which leads to a tyrosine to aspartic acid change at position 85, statistically significantly increased the risk of colorectal cancer (OR 1.34; 95% CI 1.10-1.63, p-value=0.02) in individuals with the TG genotype (Table 2). The risk of colorectal cancer for homozygous carriers of the variant allele was not statistically significant (OR 1.27; 95% CI 0.97-1.67). Similarly, in an additive analysis, no significant association was observed. No other polymorphisms were statistically significantly associated with risk of colorectal cancer.
Table 2. Association between UGT2B15 Tyr85Asp (rs1902023, UGT2B15*2) and risk of colorectal cancer.
| Cases | Controls | ORa,b | 95% CI | p-valuec | |
|---|---|---|---|---|---|
|
| |||||
| UGT2B15 (Tyr85Asp) | |||||
| T/T | 403 | 693 | 1.00 | ||
| T/G | 792 | 1161 | 1.34 | 1.10 – 1.63 | |
| G/G | 354 | 586 | 1.27 | 0.97 – 1.67 | 0.017 |
| TG or GG | 1146 | 1747 | 1.33 | 1.10 – 1.62 | 0.019 |
OR, odds ratio; CI, confidence interval.
All analyses were adjusted for age and sex.
Global p-value from log-likelihood ratio test.
As previous studies have shown that haplotypes or genotype combinations of UGT polymorphisms may have a larger impact on enzyme activity than a single nucleotide change, we also investigated haplotypes within each UGT gene and across the UGT1A locus, as well as combined genotypes within each genotyped gene for association with colorectal cancer risk (Supplementary Tables 3, 4 and 5). Although none of the haplotypes were statistically significantly associated, a three-SNP genotype within UGT1A6 was associated with risk of colorectal cancer (Table 3). Individuals who were homozygous for the major alleles of the polymorphisms rs2070959 (A>G; Thr181Ala) and rs1105879 (A>C; Arg184Ser) and, additionally, homozygous for the rs6759892 (T>G; Ser7Ala) minor G-allele of the UGT1A6 gene, i.e. the UGT1A6*3 allele (Bock et al., 2005), were at a 3.9-fold higher risk of colorectal cancer (OR 3.87; 95% CI 1.04-14.45) than individuals carrying only the major alleles of all three polymorphisms (UGT1A6*1). This association was noteworthy at the 25% FDR level.
Table 3. Association between 3-SNP-genotypes (Ser7Ala, Thr181Ala, Arg184Ser) in UGT1A6 and risk of colorectal cancer.
| UGT1A6a | Cases | Controls | ORb,c | 95% CI | p-valued | p-valuee |
|---|---|---|---|---|---|---|
| TT/AA/AA | 477 | 775 | 1.00 | - | - | 0.002f |
| TG/AG/AC | 508 | 806 | 1.11 | 0.89 - 1.37 | 0.36 | |
| GG/GG/CC | 159 | 239 | 1.12 | 0.80 - 1.57 | 0.49 | |
| TG/AA/AA | 115 | 187 | 1.04 | 0.72 - 1.49 | 0.84 | |
| GG/AG/AC | 65 | 97 | 1.26 | 0.77 - 2.06 | 0.36 | |
| TG/AA/AC | 43 | 43 | 1.72 | 0.94 - 3.16 | 0.08 | |
| GG/AG/CC | 16 | 35 | 0.51 | 0.24 - 1.22 | 0.13 | |
| GG/AA/AA | 9 | 11 | 3.87 | 1.04 - 14.45 | 0.04 | |
| GG/AA/AC | 5 | 5 | 3.28 | 0.76 - 14.11 | 0.11 | |
| TT/AG/AC | 1 | 1 | n.a. | n.a. | n.a. | |
| GG/AA/CC | 0 | 1 | n.a. | n.a. | n.a. |
Sequence of variants: rs1105879/ rs2070959/ rs6759892
OR, odds ratio; CI, confidence interval.
All analyses were adjusted for age and sex.
Wald test p-value.
Global p-value from log-likelihood ratio test.
Noteworthy at the 25% FDR level.
NSAID interactions
Several of the investigated variants showed a significant (p<0.05) interaction with the modification of colorectal cancer risk by NSAID use; however, some of these results were based on small cell sizes and should be considered tentative.
The UGT1A3 Thr78Thr (rs17868336; A>G) variant showed a statistically significant interaction with NSAID use (p-interaction=0.02), increasing the risk of non-NSAID users with the homozygous minor G-allele genotype by 50% (OR 1.57; 95% CI 1.06-2.34, Table 4.a) in comparison to non-users with the major A-allele.
Table 4. Association between NSAID use and colorectal cancer and rectal cancer risk stratified by UGT1A genotypes.
| a) Association between NSAID use and colorectal cancer risk stratified by UGT1A3 (Thr78Thr) genotypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| No NSAID use | NSAID use | |||||||
| CRCa Cases | Controls | ORb,c | 95% CI | CRC Cases | Controls | OR | 95% CI | |
|
| ||||||||
| UGT1A3 (Thr78Thr) | ||||||||
| A/A | 964 | 1527 | 1.00 | 277 | 451 | 0.95 | 0.78 - 1.16 | |
| A/G or G/G | 95 | 123 | 1.57 | 1.06 - 2.34 | 18 | 41 | 0.66 | 0.34 - 1.27 |
| p-interaction 0.02d | ||||||||
| b) Association between NSAID use and rectal cancer risk stratified by UGT1A6 (Ser7Ala) | ||||||||
|---|---|---|---|---|---|---|---|---|
| No NSAID use | NSAID use | |||||||
| RCe Cases | Controls | OR | 95% CI | RC Cases | Controls | OR | 95% CI | |
|
| ||||||||
| UGT1A6 (Ser7Ala) | ||||||||
| T/T | 99 | 187 | 1.00 | 37 | 55 | 1.33 | 0.75 - 2.37 | |
| T/G or G/G | 222 | 342 | 1.40 | 0.94 - 2.08 | 41 | 83 | 0.77 | 0.42 - 1.41 |
| p-interaction 0.03d | ||||||||
| c) Association between NSAID use and rectal cancer risk stratified by UGT1A6 (Arg184Ser) genotypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| No NSAID use | NSAID use | |||||||
| RC Cases | Controls | OR | 95% CI | RC Cases | Controls | OR | 95% CI | |
|
| ||||||||
| UGT1A6 (Arg184Ser) | ||||||||
| A/A | 146 | 266 | 1.00 | 46 | 67 | 1.23 | 0.76 - 1.99 | |
| A/C or C/C | 217 | 328 | 1.25 | 0.89 - 1.77 | 36 | 82 | 0.66 | 0.37 - 1.15 |
| p-interaction 0.02d | ||||||||
CRC, Colorectal cancer
OR, odds ratio; CI, confidence interval.
All analyses were adjusted for age, sex, BMI, pack-years and physical activity.
For multiplicative interaction term.
RC, Rectal cancer
A stratified analysis by tumor site showed a statistically significant interaction between two of the UGT1A6 polymorphisms (Ser7Ala and Arg184Ser) and NSAID use for risk of rectal cancer (p-interaction=0.03, Table 4.b, and p-interaction=0.02, Table 4.c respectively).
We also investigated whether haplotypes across the UGT1A locus, within any of the investigated UGT genes, where several polymorphisms were genotyped (i.e. UGT1A3, UGT1A6, UGT1A7, UGT2B4 and CYP2C9) or combined genotypes within any of the investigated genes interacted with NSAID use, or specifically, ibuprofen or aspirin in colorectal cancer predisposition. No interactions were observed with overall NSAID use; however, the use of ibuprofen was associated with a higher risk of colorectal cancer in individuals who were homozygous for the major alleles of all three investigated UGT2B4 polymorphisms (rs1966151, A>G, 5′UTR; rs13119049, A>T, Asp458Glu; rs1131878, A>G, 3′UTR; OR 2.31; 95% CI 1.07-4.97), whereas ibuprofen users who carried at least one variant allele at any of the three loci tended to be at lower risk (OR 0.73; 95% CI 0.48-1.12) of colorectal cancer than non-users with only major alleles (p-interaction=0.0018, Table 5). When we accounted for multiple testing based on FDR, this association was noteworthy at the 25% FDR level.
Table 5. Association between ibuprofen use and colorectal cancer risk stratified by combined UGT2B4 genotypes (5′UTR, Asp458Glu, 3′UTR).
| No Ibuprofen use | Ibuprofen use | |||||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ORa,b | 95% CI | Cases | Controls | ORa,b | 95% CI | |
|
| ||||||||
| UGT2B4 (5′UTR, Asp458Glu, 3′UTR) | ||||||||
| AA_AA_AA | 198 | 328 | 1.00 | - | 28 | 20 | 2.31 | 1.07 - 4.97 |
| any variant | 934 | 1408 | 1.12 | 0.86 - 1.50 | 67 | 157 | 0.73 | 0.48 - 1.12 |
| p-interaction 0.0018c,d | ||||||||
OR, odds ratio; CI, confidence interval.
All analyses were adjusted for age, sex, BMI, pack-years and physical activity.
p-value for multiplicative interaction term.
Noteworthy at the 25% FDR level.
Additionally, the coding variant Tyr85Asp in the UGT2B15 (UGT2B15*2) gene showed a statistically significant interaction with aspirin use (p-interaction=0.01). Individuals who were homozygous for the minor G-allele and used aspirin were at higher risk of colorectal cancer (OR 1.71; 95% CI 1.07-2.73), than non-users who carried only the major T-allele (Figure 1 and Table 6). This association was noteworthy at the 25% FDR level.
Figure 1.
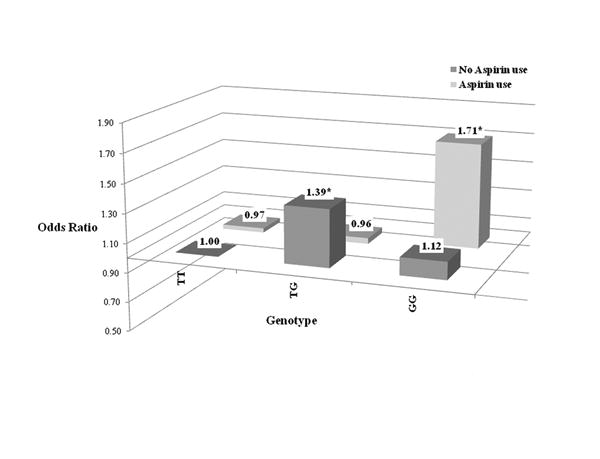
Table 6. Association between aspirin use and colorectal cancer risk stratified by UGT2B15*2 (Tyr85Asp) genotypes.
| No Aspirin use | Aspirin use | |||||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ORa,b | 95% CI | Cases | Controls | ORa,b | 95% CI | |
|
| ||||||||
| UGT2B15 (Tyr85Asp) | ||||||||
| T/T | 316 | 533 | 1.00 | 54 | 88 | 0.97 | 0.63 – 1.50 | |
| T/G | 604 | 866 | 1.39 | 1.11 – 1.74 | 112 | 191 | 0.96 | 0.68 – 1.34 |
| G/G | 259 | 465 | 1.12 | 0.82 – 1.53 | 58 | 61 | 1.71 | 1.07 – 2.73 |
| p-interaction 0.01c,d | ||||||||
OR, odds ratio; CI, confidence interval.
All analyses were adjusted for age, sex, BMI, pack-years and physical activity.
For multiplicative interaction term.
Noteworthy at the 25% FDR level.
Discussion
Colorectal cancer is a heterogeneous disease and targeted prevention strategies are required to reduce the public health burden of the disease (Colussi et al., 2013). In light of extensive data on the preventive effects of aspirin and other NSAIDs on risk of cancer, there is growing interest in determining whether genetic variation in NSAID-metabolizing enzymes can be used to predict the protective effect of NSAIDs on cancer development and on complications such as gastrointestinal bleedings (Derry and Loke, 2000; Ulrich et al., 2006; Cross et al., 2008; Cuzick et al., 2009; Chia et al., 2012; Kraus et al., 2013; Rothwell at al., 2010; 2012a; 2012b; 2013). Since metabolizing enzymes can modify, conjugate, and/or excrete endobiotic or xenobiotic compounds, including the detoxification of carcinogens and metabolism of chemopreventive or chemotherapeutic compounds, genetic variability in these enzymes may thus alter the toxicity or efficacy of xenobiotics and consequently alter cancer susceptibility.
In this large population-based study of sibling pairs in the CCFR, we investigated whether polymorphisms within genes of phase I (CYP2C9) and phase II (UGT) drug-metabolizing enzymes were associated with or modified the protective effect of NSAIDs on colorectal cancer risk. In addition to previously investigated polymorphisms, we targeted several previously unstudied polymorphisms in UGT genes in this study.
Variants in four genes (UGT1A3, UGT1A6, UGT2B4 and UGT2B15) were statistically significantly associated with colorectal or rectal cancer risk either in individual analysis, combined genotype analysis, or in combination with NSAID use. The interaction between the UGT2B15 Tyr85Asp polymorphism and aspirin use (which was also associated with risk overall) as well as between a three-SNP genotype in UGT2B4 and ibuprofen use were noteworthy at the 25% FDR level.
This study is the first to report the association of the UGT1A6*3 polymorphism, a three-SNP genotype (Ala7, Thr181, Arg184), with increased colorectal cancer risk; previous studies had focused on a two-SNP genotype within UGT1A6 (Thr181Ala, Arg184Ser, i.e. UGT1A6*8 also referred to as UGT1A6*2 in some studies) (Bigler et al., 2001; Chan et al., 2005; McGreavey et al., 2005; Samowitz et al., 2006). Nonetheless, these results require further validation, as the association was based on few individuals and thus stratification by tumor location was not possible. In vitro investigations of the UGT1A6*3 variant enzyme showed no significant change of the enzymatic activity compared to the wild type enzyme UGT1A6*1 (Krishnaswamy et al., 2005b). However, the family of UGT1A genes represents a complex locus consisting of four common exons and at least 13 variable exons with many shared sequences and polymorphisms among the UGT1A genes (Mackenzie et al., 2005). The complexity of the UGT1A region is further increased by its LD structure; many SNPs are linked to each other, thus making it difficult to identify the causal variation and the related gene or enzyme product. The Ser7Ala polymorphism is in complete LD with other polymorphisms in the UGT1A locus, including three polymorphisms located in the 5′UTR of the gene, associated with 50% decreased gene expression of UGT1A6, but not with lower protein levels or reduced enzyme activity (Krishnaswamy et al., 2005a; Thomas et al., 2006), thus the functional consequence of these polymorphisms remains to be elucidated. Previous studies have shown that the UGT1A6*8 allele (two-SNP genotype: Ala181 and Ser184) modified the protective effect of aspirin on colon or colorectal adenoma; however, no study has shown similar effects on the risk of colon carcinoma (Bigler et al., 2001; Chan et al., 2005; McGreavey et al., 2005; Samowitz et al., 2006; Thompson et al., 2009), concordant with our observations. It has been proposed earlier that the protective effect of NSAIDs may occur earlier during carcinogenesis and may thus be relevant for adenoma prevention rather than for prevention of carcinomas (McGreavey et al., 2005). However, recent meta-analyses (Rothwell et al., 2012a; 2012b) strongly suggest that the preventive effect of NSAIDs exists across all stages of colorectal neoplasia. Nevertheless, further investigation regarding dose, treatment duration and long-term benefit of NSAID use are required to optimize its use in cancer prevention (Cuzick et al., 2009; Potter, 2012).
In the present study we observed an association of a UGT2B15 variant with increased risk of colorectal cancer, which also interacted with aspirin use in colorectal cancer predisposition, as well as novel interactions between UGT2B4 polymorphisms and ibuprofen. Both the UGT2B4 and the UGT2B15 enzymes are primarily involved in the metabolism of sex hormones. Polymorphisms within these genes have been associated with increased breast and prostate cancer risk, respectively (MacLeod et al., 2000; Park et al., 2004; Low et al., 2010; Grant et al., 2013). While UGT2B4 has been shown to metabolize NSAIDs at quite high rates, UGT2B15-mediated glucuronidation was observed only at low rates (Kuehl et al., 2005; 2006). In vitro studies of the UGT2B15 variant enzyme displayed similar substrate specificity between the Tyr85 and the Asp85 enzymes; however, the Asp85 variant enzyme had a faster turn-around time (i.e. higher Vmax) (Levesque et al., 1997). This may lead to a reduced internal dose of the active drug compound accompanied by reduction of its preventive effect. The previously reported associations with cancer risk are probably due to changed sex steroid metabolism resulting in altered tissue exposure to hormones capable of stimulating cell proliferation (Yong et al., 2011). However, the strong interaction between the UGT2B15 polymorphism and aspirin use in relation to colorectal cancer risk, which remained significant after accounting for multiple testing, suggests a meaningful role of the enzyme in NSAID metabolism. Nevertheless, it seems that larger structural genetic variation such as copy number variations (CNV) have a stronger impact on cancer risk as previously reported in recent study. It was shown that a large CNV that leads to the deletion of the UGT2B17 gene, significantly decreased the risk of rectal cancer (Angstadt et al., 2013), but not colon cancer.
Concordantly with a recent meta-analysis which covered 13 studies, we did not observe an association between any of the CYP2C9 polymorphisms and risk of colorectal cancer (Liang et al., 2012). While an interaction of the polymorphisms with NSAID use on colorectal cancer risk was reported previously, in this study no interaction was observed (Bigler et al., 2001; Samowitz et al., 2006).
There are several strengths to this study. The case–unaffected sibling control design helps avoid false positives that can result from population stratification and increases the power to detect gene–NSAID interactions. The relatively large overall sample size in this study made it possible to examine combined genotypes, as well as interaction analysis for NSAID use. There are several potential limitations to our study. Current NSAID use was only modestly associated with reduced CRC risk in our study population, which may limit the power for detection of gene-NSAID interaction. The family-based study design likely reduced the power of the main effect analyses. Furthermore, despite the large sample size, interaction analysis with combined genotypes or specific NSAIDs resulted in small cell sizes, reducing the precision of estimates and the power. When we tested the interactions for multiple comparisons at the 25% FDR level, only the interaction of the UGT2B4 combined genotype with NSAID use and the UGT2B15*2 with aspirin use were noteworthy. On the other hand, we investigated specific hypotheses with respect to putative gene-NSAID interactions and functional polymorphisms; in this setting adjustment for multiple comparisons is less critical. Finally, our study was based on the investigations of SNPs, thus we did not cover larger structural variants in UGT genes, which may have a stronger impact on colorectal cancer risk.
In summary, our results suggest that variation in four genes (UGT1A3, UGT1A6, UGT2B4 and UGT2B15) modifies the risk of colorectal cancer either independent or in conjunction with NSAID use. Our results underscore the importance of pharmacogenetics as a tool to identify individuals who may benefit from NSAIDs as chemopreventives. In addition, our study results suggest that it appears to be more important to consider combinations of genotypes rather than individual polymorphisms to identify the interaction between genetic variability and the protective effect of NSAID use on colorectal cancer development, particularly for UGT families.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute, National Institutes of Health under RFA # CA-95-011 and through cooperative agreements with members of the Colon Cancer Family Registry (CCFR) and Principal Investigators. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFRs, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CFR. CFR centers providing data for the analysis include: Australasian Colorectal Cancer Family Registry (U01 CA097735); Familial Colorectal Neoplasia Collaborative Group (U01 CA074799); Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800); Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); Seattle Colorectal Cancer Family Registry (U01 CA074794);University of Hawaii Colorectal Cancer Family Registry (U01 CA074806); and University of California, Irvine Informatics Center (U01 CA078296). We thank Allyson Templeton for providing us with data on CCFR enrollment and follow-up and Yesilda Balavarca for data analysis. We also thank Sushma Thomas, Christine Rimorin, Jeanne DaGloria and Ling-Yu Kuan of the FHCRC Molecular Epidemiology Laboratory for genotyping assistance.
Supported by: This study received support from the National Cancer Institute grants R01 CA129063, T32 CA09168, R01 CA112516, and R01 CA114467-05
References
- Angstadt AY, Berg A, Zhu J, Miller P, Hartman TJ, Lesko SM, Muscat JE, Lazarus P, Gallagher CJ. The effect of copy number variation in the phase II detoxification genes UGT2B17 and UGT2B28 on colorectal cancer risk. Cancer. 2013;119:2477–2485. doi: 10.1002/cncr.28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JW, Choi CI, Jang CG, Lee SY. Effects of CYP2C9*1/*13 on the pharmacokinetics and pharmacodynamics of meloxicam. Br J Clin Pharmacol. 2011;71:550–555. doi: 10.1111/j.1365-2125.2010.03853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Royal Stat Soc, Series B, Methodological. 1995;57:289–300. [Google Scholar]
- Bigler J, Whitton J, Lampe JW, Fosdick L, Bostick RM, Potter JD. CYP2C9 and UGT1A6 genotypes modulate the protective effect of aspirin on colon adenoma risk. Cancer Res. 2001;61:3566–3569. [PubMed] [Google Scholar]
- Bock KW, Burchell B, Guillemette C, Mackenzie PI, Nebert DW, Court M, Owens I. UGT Alleles Nomenclature Home Page. UGT Nomenclature Commitee. 2005 13/12/09. [Google Scholar]
- Chan AT, Hsu M, Zauber A, Hawk E, Bertagnolli MM. The Influence of UGT 1A6 Variants and Aspirin Use in a Randomized Trial of Celecoxib for Prevention of Colorectal Adenoma. Cancer Prev Res. 2011;5:61–72. doi: 10.1158/1940-6207.CAPR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, Tranah GJ, Giovannucci EL, Hunter DJ, Fuchs CS. Genetic variants in the UGT1A6 enzyme, aspirin use, and the risk of colorectal adenoma. J Natl Cancer Inst. 2005;97:457–460. doi: 10.1093/jnci/dji066. [DOI] [PubMed] [Google Scholar]
- Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer--reinterpreting paradigms. Nat Rev Clin Oncol. 2012;9:561–570. doi: 10.1038/nrclinonc.2012.137. [DOI] [PubMed] [Google Scholar]
- Ciotti M, Marrone A, Potter C, Owens IS. Genetic polymorphism in the human UGT1A6 (planar phenol) UDP-glucuronosyltransferase: pharmacological implications. Pharmacogenetics. 1997;7:485–495. doi: 10.1097/00008571-199712000-00007. [DOI] [PubMed] [Google Scholar]
- Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JT, Poole EM, Ulrich CM. A review of gene-drug interactions for nonsteroidal anti-inflammatory drug use in preventing colorectal neoplasia. Pharmacogenomics J. 2008;8:237–247. doi: 10.1038/sj.tpj.6500487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Otto F, Baron JA, Brown PH, Burn J, Greenwald P, Jankowski J, La Vecchia C, Meyskens F, Senn HJ, Thun M. Aspirin and non-steroidal anti-inflammatory drugs for cancer prevention: an international consensus statement. Lancet Oncol. 2009;10:501–507. doi: 10.1016/S1470-2045(09)70035-X. [DOI] [PubMed] [Google Scholar]
- Derry S, Loke YK. Risk of gastrointestinal haemorrhage with long term use of aspirin: meta-analysis. British Medical Journal. 2000;321:1183–1187. doi: 10.1136/bmj.321.7270.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura P, Salomon J, Te Morsche RH, Roelofs HM, Kristinsson JO, Wobbes T, Witteman BJ, Tan AC, Drenth JP, Peters WH. High enzyme activity UGT1A1 or low activity UGT1A8 and UGT2B4 genotypes increase esophageal cancer risk. Int J Oncol. 2012;40:1789–1796. doi: 10.3892/ijo.2012.1385. [DOI] [PubMed] [Google Scholar]
- Flossmann E, Rothwell PM. Effect of aspirin on long-term risk of colorectal cancer: consistent evidence from randomised and observational studies. Lancet. 2007;369:1603–1613. doi: 10.1016/S0140-6736(07)60747-8. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ, Witte JS, Thomas DC. Family-based association studies. J Natl Cancer Inst Monogr. 1999;26:31–37. doi: 10.1093/oxfordjournals.jncimonographs.a024223. [DOI] [PubMed] [Google Scholar]
- Gill HJ, Tjia JF, Kitteringham NR, Pirmohamed M, Back DJ, Park BK. The effect of genetic polymorphisms in CYP2C9 on sulphamethoxazole N-hydroxylation. Pharmacogenetics. 1999;9:43–53. doi: 10.1097/00008571-199902000-00007. [DOI] [PubMed] [Google Scholar]
- Grant DJ, Hoyo C, Oliver SD, Gerber L, Shuler K, Calloway E, Gaines AR, McPhail M, Livingston JN, Richardson RM, Schildkraut JM, Freedland SJ. Association of uridine diphosphate-glucuronosyltransferase 2B gene variants with serum glucuronide levels and prostate cancer risk. Genet Test Mol Biomarkers. 2013;17:3–9. doi: 10.1089/gtmb.2012.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining RL, Hunter AP, Veronese ME, Trager WF, Rettie AE. Allelic variants of human cytochrome P450 2C9: baculovirus-mediated expression, purification, structural characterization, substrate stereoselectivity, and prochiral selectivity of the wild-type and I359L mutant forms. Arch Biochem Biophys. 1996;333:447–458. doi: 10.1006/abbi.1996.0414. [DOI] [PubMed] [Google Scholar]
- Horvath S, Laird NM. A discordant-sibship test for disequilibrium and linkage: no need for parental data. Am J Hum Genet. 1998;63:1886–1897. doi: 10.1086/302137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt AJ, Caldwell J, Smith RL. The metabolism of aspirin in man: a population study. Xenobiotica. 1986;16:239–249. doi: 10.3109/00498258609043527. [DOI] [PubMed] [Google Scholar]
- Kleinstein SE, Heath L, Makar KW, Poole EM, Seufert BL, Slattery ML, Xiao L, Duggan DJ, Hsu L, Curtin K, Koepl L, Muehling J, Taverna D, Caan BJ, Carlson CS, Potter JD, Ulrich CM. Genetic variation in the lipoxygenase pathway and risk of colorectal neoplasia. Genes Chromosomes Cancer. 2013;52:437–449. doi: 10.1002/gcc.22042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus S, Hummler S, Toriola AT, Poole EM, Scherer D, Kotzmann J, Makar KW, Kazanov D, Galazan L, Naumov I, Coghill AE, Duggan D, Gigic B, Arber N, Ulrich CM. Impact of genetic polymorphisms on adenoma recurrence and toxicity in a COX2 inhibitor (celecoxib) trial: results from a pilot study. Pharmacogenet Genomics. 2013;23:428–437. doi: 10.1097/FPC.0b013e3283631784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Hao Q, Al-Rohaimi A, Hesse LM, von Moltke LL, Greenblatt DJ, Court MH. UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: I. Identification of polymorphisms in the 5′-regulatory and exon 1 regions, and association with human liver UGT1A6 gene expression and glucuronidation. J Pharmacol Exp Ther. 2005a;313:1331–1339. doi: 10.1124/jpet.104.081950. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy S, Hao Q, Al-Rohaimi A, Hesse LM, von Moltke LL, Greenblatt DJ, Court MH. UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: II. Functional impact of the three most common nonsynonymous UGT1A6 polymorphisms (S7A, T181A, and R184S) J Pharmacol Exp Ther. 2005b;313:1340–1346. doi: 10.1124/jpet.104.081968. [DOI] [PubMed] [Google Scholar]
- Kuehl GE, Bigler J, Potter JD, Lampe JW. Glucuronidation of the aspirin metabolite salicylic acid by expressed UDP-glucuronosyltransferases and human liver microsomes. Drug Metab Dispos. 2006;34:199–202. doi: 10.1124/dmd.105.005652. [DOI] [PubMed] [Google Scholar]
- Kuehl GE, Lampe JW, Potter JD, Bigler J. Glucuronidation of nonsteroidal anti-inflammatory drugs: identifying the enzymes responsible in human liver microsomes. Drug Metab Dispos. 2005;33:1027–1035. doi: 10.1124/dmd.104.002527. [DOI] [PubMed] [Google Scholar]
- Leemann TD, Transon C, Bonnabry P, Dayer P. A major role for cytochrome P450TB (CYP2C subfamily) in the actions of non-steroidal antiinflammatory drugs. Drugs Exp Clin Res. 1993;19:189–195. [PubMed] [Google Scholar]
- Levesque E, Beaulieu M, Green MD, Tephly TR, Belanger A, Hum DW. Isolation and characterization of UGT2B15(Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 1997;7:317–325. doi: 10.1097/00008571-199708000-00007. [DOI] [PubMed] [Google Scholar]
- Liang S, Hu J, Cao W, Cai S. Meta-analysis of cytochrome P-450 2C9 polymorphism and colorectal cancer risk. PLoS ONE. 2012;7:e49134. doi: 10.1371/journal.pone.0049134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low YL, Li Y, Humphreys K, Thalamuthu A, Darabi H, Wedren S, Bonnard C, Czene K, Iles MM, Heikkinen T, Aittomaki K, Blomqvist C, Nevanlinna H, Hall P, Liu ET, Liu J. Multi-variant pathway association analysis reveals the importance of genetic determinants of estrogen metabolism in breast and endometrial cancer susceptibility. PLoS Genet. 2010;6:e1001012. doi: 10.1371/journal.pgen.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- MacLeod SL, Nowell S, Plaxco J, Lang NP. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann Surg Oncol. 2000;7:777–782. doi: 10.1007/s10434-000-0777-3. [DOI] [PubMed] [Google Scholar]
- McGreavey LE, Turner F, Smith G, Boylan K, Timothy Bishop D, Forman D, Roland Wolf C, Barrett JH. No evidence that polymorphisms in CYP2C8, CYP2C9, UGT1A6, PPARdelta and PPARgamma act as modifiers of the protective effect of regular NSAID use on the risk of colorectal carcinoma. Pharmacogenet Genomics. 2005;15:713–721. doi: 10.1097/01.fpc.0000174786.85238.63. [DOI] [PubMed] [Google Scholar]
- Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, Hall D, Hopper JL, Jass J, Le Marchand L, Limburg P, Lindor N, Potter JD, Templeton AS, Thibodeau S, Seminara D. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- Ogino S, Lochhead P, Giovannucci E, Meyerhardt JA, Fuchs CS, Chan AT. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene. 2013 doi: 10.1038/onc.2013.244. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen L, Shade K, Lazarus P, Seigne J, Patterson S, Helal M, Pow-Sang J. Asp85tyr polymorphism in the udp-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J Urol. 2004;171:2484–2488. doi: 10.1097/01.ju.0000117748.44313.43. [DOI] [PubMed] [Google Scholar]
- Potter JD. Aspirin and cancer prevention and treatment: are we there yet? Cancer Epidemiol Biomarkers Prev. 2012;21:1439–1440. doi: 10.1158/1055-9965.EPI-12-0837. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- Rothwell PM. Aspirin in prevention of sporadic colorectal cancer: current clinical evidence and overall balance of risks and benefits. Recent Results Cancer Res. 2013;191:121–142. doi: 10.1007/978-3-642-30331-9_7. [DOI] [PubMed] [Google Scholar]
- Rothwell PM, Price JF, Fowkes FG, Zanchetti A, Roncaglioni MC, Tognoni G, Lee R, Belch JF, Wilson M, Mehta Z, Meade TW. Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet. 2012a;379:1602–1612. doi: 10.1016/S0140-6736(11)61720-0. [DOI] [PubMed] [Google Scholar]
- Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, Meade TW. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet. 2012b;379:1591–1601. doi: 10.1016/S0140-6736(12)60209-8. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Wolff RK, Curtin K, Sweeney C, Ma KN, Andersen K, Levin TR, Slattery ML. Interactions between CYP2C9 and UGT1A6 polymorphisms and nonsteroidal anti-inflammatory drugs in colorectal cancer prevention. Clin Gastroenterol Hepatol. 2006;4:894–901. doi: 10.1016/j.cgh.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Steward DJ, Haining RL, Henne KR, Davis G, Rushmore TH, Trager WF, Rettie AE. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997;7:361–367. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kashima T, Nomoto S, Iwade K, Tainaka H, Shimizu T, Nomizo Y, Muramoto N, Kimura S, Echizen H. Comparisons between in-vitro and in-vivo metabolism of (S)-warfarin: catalytic activities of cDNA-expressed CYP2C9, its Leu359 variant and their mixture versus unbound clearance in patients with the corresponding CYP2C9 genotypes. Pharmacogenetics. 1998;8:365–373. doi: 10.1097/00008571-199810000-00001. [DOI] [PubMed] [Google Scholar]
- Thomas SS, Li SS, Lampe JW, Potter JD, Bigler J. Genetic variability, haplotypes, and htSNPs for exons 1 at the human UGT1A locus. Hum Mutat. 2006;27:717. doi: 10.1002/humu.9432. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Plummer SJ, Merkulova A, Cheng I, Tucker TC, Casey G, Li L. No association between cyclooxygenase-2 and uridine diphosphate glucuronosyltransferase 1A6 genetic polymorphisms and colon cancer risk. World J Gastroenterol. 2009;15:2240–2244. doi: 10.3748/wjg.15.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougeron D, Sha D, Manthravadi S, Sinicrope FA. Aspirin and colorectal cancer: Back to the Future. Clin Cancer Res. 2013;20:1087–94. doi: 10.1158/1078-0432.CCR-13-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat Rev Cancer. 2006;6:130–140. doi: 10.1038/nrc1801. [DOI] [PubMed] [Google Scholar]
- Weber WW, Hein DW. Clinical pharmacokinetics of isoniazid. Clin Pharmacokinet. 1979;4:401–422. doi: 10.2165/00003088-197904060-00001. [DOI] [PubMed] [Google Scholar]
- Yong M, Schwartz SM, Atkinson C, Makar KW, Thomas SS, Stanczyk FZ, Westerlind KC, Newton KM, Holt VL, Leisenring WM, Lampe JW. Associations between polymorphisms in glucuronidation and sulfation enzymes and sex steroid concentrations in premenopausal women in the United States. J Steroid Biochem Mol Biol. 2011;124:10–18. doi: 10.1016/j.jsbmb.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


