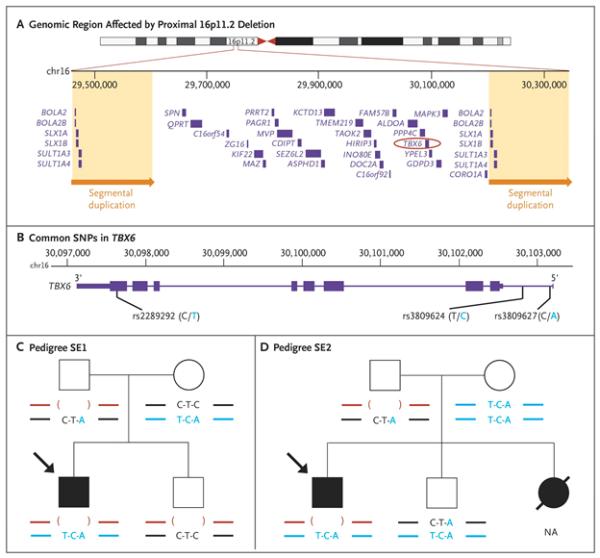
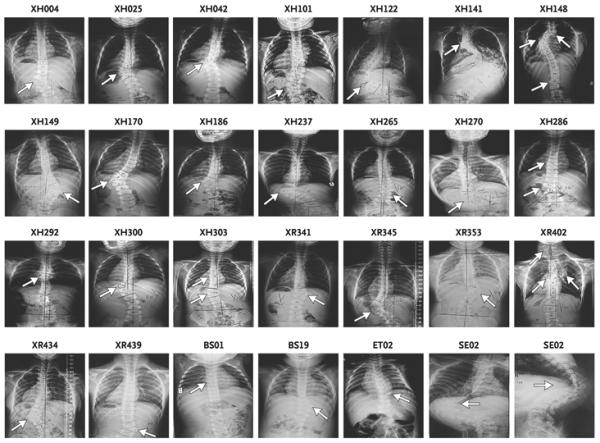
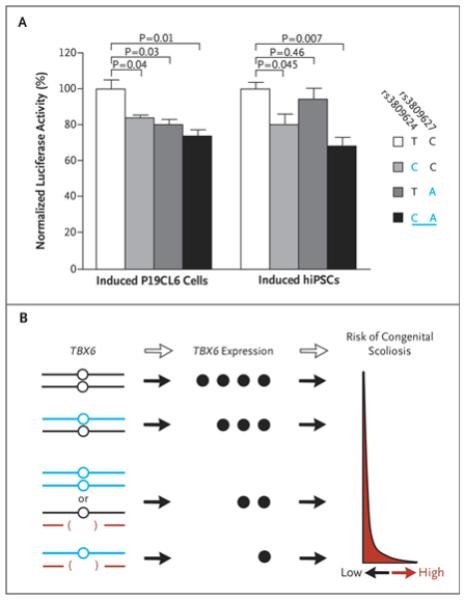
N Wu
N Wu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
X Ming
X Ming
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
J Xiao
J Xiao
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Z Wu
Z Wu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
X Chen
X Chen
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
M Shinawi
M Shinawi
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Shen
Y Shen
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
G Yu
G Yu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
J Liu
J Liu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
H Xie
H Xie
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
ZS Gucev
ZS Gucev
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
S Liu
S Liu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
N Yang
N Yang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
H Al-Kateb
H Al-Kateb
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
J Chen
J Chen
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Jian Zhang
Jian Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
N Hauser
N Hauser
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
T Zhang
T Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
V Tasic
V Tasic
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
P Liu
P Liu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
X Su
X Su
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
X Pan
X Pan
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
C Liu
C Liu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
L Wang
L Wang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Joseph Shen
Joseph Shen
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Jianxiong Shen
Jianxiong Shen
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Chen
Y Chen
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
T Zhang
T Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Jianguo Zhang
Jianguo Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
KW Choy
KW Choy
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Jun Wang
Jun Wang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Q Wang
Q Wang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
S Li
S Li
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
W Zhou
W Zhou
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
J Guo
J Guo
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Wang
Y Wang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
C Zhang
C Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
H Zhao
H Zhao
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y An
Y An
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Zhao
Y Zhao
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Jiucun Wang
Jiucun Wang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Z Liu
Z Liu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Zuo
Y Zuo
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Tian
Y Tian
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
X Weng
X Weng
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
VR Sutton
VR Sutton
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
H Wang
H Wang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
Y Ming
Y Ming
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
S Kulkarni
S Kulkarni
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
TP Zhong
TP Zhong
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
PF Giampietro
PF Giampietro
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
SL Dunwoodie
SL Dunwoodie
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
SW Cheung
SW Cheung
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
X Zhang
X Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
L Jin
L Jin
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
JR Lupski
JR Lupski
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
G Qiu
G Qiu
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1,
F Zhang
F Zhang
1Department of Orthopedic Surgery, Peking Union Medical College Hospital (N.W., Z.W., J.L., S. Liu, J.C., X.S., Jianxiong Shen, Jianguo Zhang, S. Li, Y.W., H.Z., Y. Zhao, Z.L., Y. Zuo, Y.T., X.W., G.Q.), the PET-CT Center, Cancer Hospital (Y M.), and McKusick–Zhang Center for Genetic Medicine and State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences (X.Z.), Peking Union Medical College and Chinese Academy of Medical Sciences, and the Beijing Municipal Key Laboratory of Child Development and Nutriomics (X.C., H.X., T.Z. [M.D.], J.G.), and Department of Neurology, Affiliated Children’s Hospital (L.W., Jun Wang), Capital Institute of Pediatrics, Beijing, the State Key Laboratory of Genetic Engineering and Ministry of Education Key Laboratory of Contemporary Anthropology (X M., J.X., G.Y., N.Y., Jian Zhang, X.P., Y.C., Q.W., W.Z., C.Z., Y.A., Jiucun Wang, H.W., T.P.Z., L.J., F.Z.), and Collaborative Innovation Center for Genetics and Development (J.X., H.W., T.P.Z., L.J., F.Z.), School of Life Sciences, Fudan University, and the School of Medicine (Y.S.) and the Department of Gastroenterology, Hepatology, and Nutrition, Shanghai Children’s Hospital (T.Z. [M.D., Ph.D.]), Shanghai Jiao Tong University, Shanghai, the Department of Biotechnology, School of Basic Medical Science, Nanjing Medical University, Nanjing (C.L.), the Department of Obstetrics and Gynaecology, Chinese University of Hong Kong, Hong Kong (K.W.C.), and Fudan–Taizhou Institute of Health Sciences, Taizhou (Jiucun Wang, L.J., F.Z.) — all in China; the Department of Pediatrics, Division of Genetics and Genomic Medicine (M.S.), and the Department of Pathology and Immunology (H.A.-K., S.K.), Washington University School of Medicine, St. Louis; the Department of Pathology, Harvard Medical School, and the Department of Laboratory Medicine, Boston Children’s Hospital — both in Boston (Y.S.); the Department of Endocrinology and Genetics, University Children’s Hospital, Medical Faculty Skopje, Skopje, Macedonia (Z.S.G., V.T.); the Department of Medical Genetics and Metabolism, Children’s Hospital Central California, Madera (N.H., Joseph Shen); the Departments of Molecular and Human Genetics (P.L., V.R.S., S.W.C., J.R.L.) and Pediatrics (J.R.L.), Baylor College of Medicine, and Texas Children’s Hospital (V.R.S., J.R.L.) — both in Houston; the Department of Pediatrics, University of Wisconsin–Madison, Madison (P.F.G.); and the Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, and St. Vincent’s Clinical School Faculty of Medicine and the School of Biotechnology and Biomolecular Sciences Faculty of Science, University of New South Wales — all in Sydney (S.L.D.)
1