Abstract
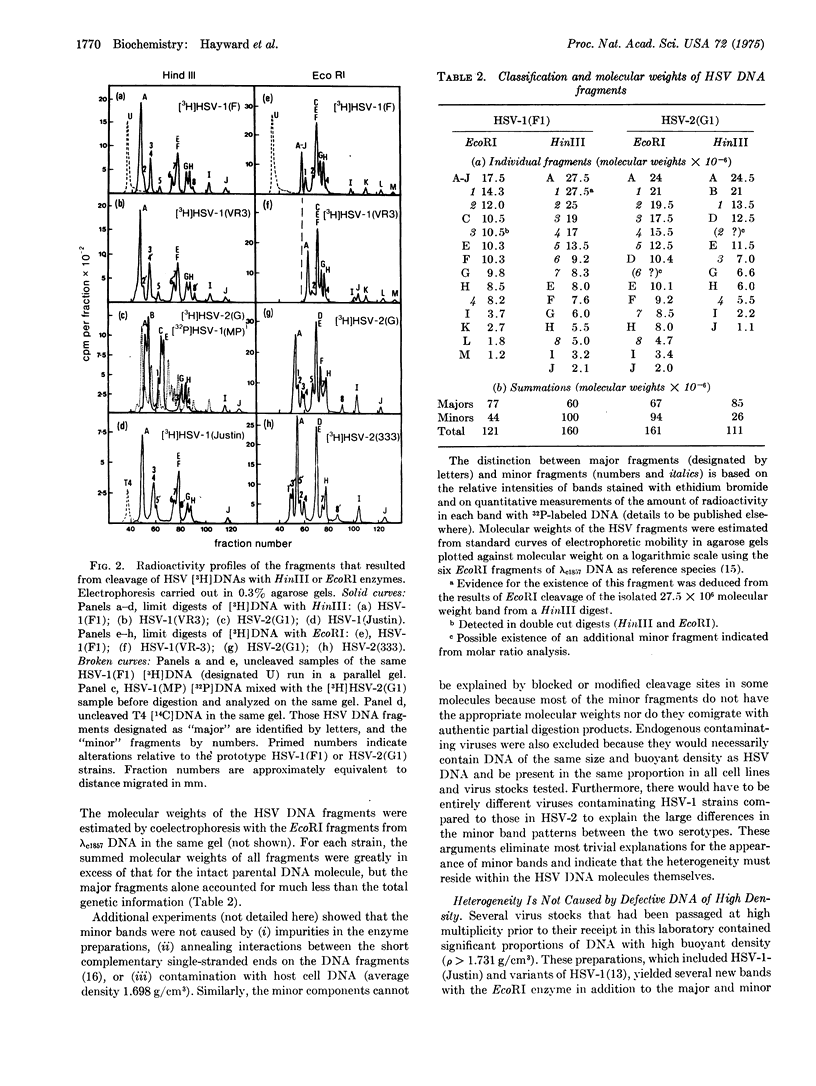
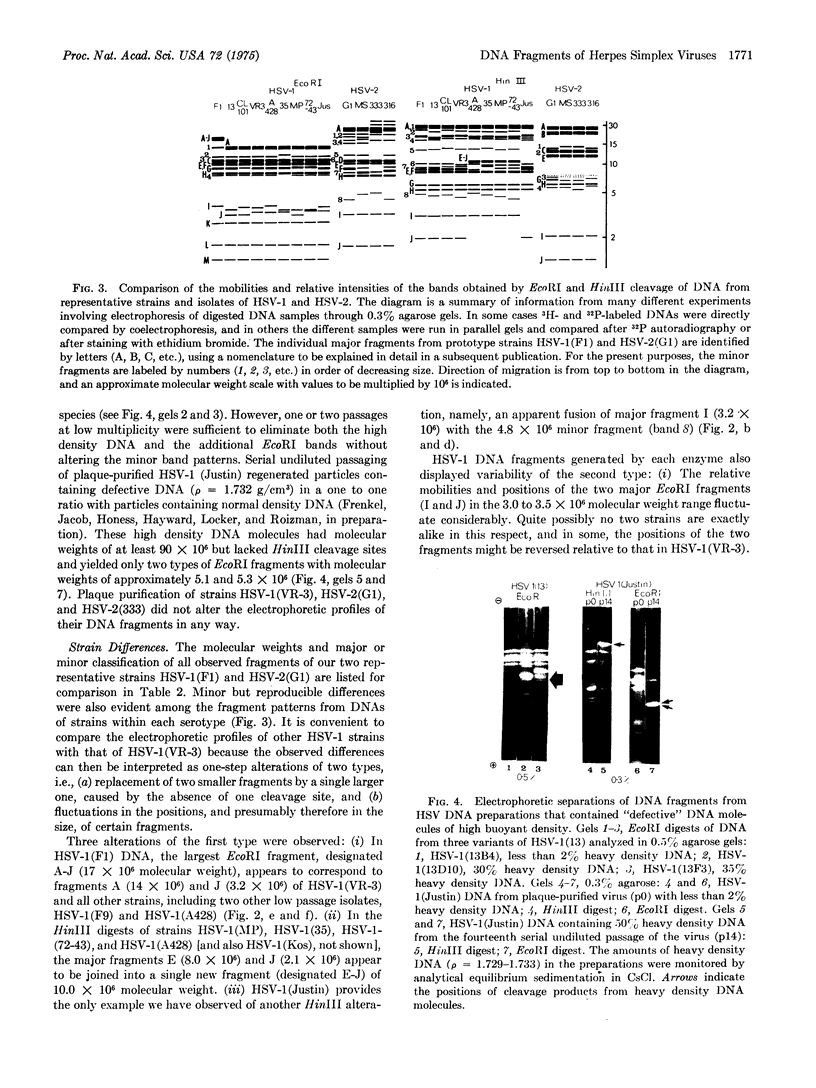
Digestion of herpes simplex virus DNA by the HinIII or Eco RI restriction endonucleases yielded 11 to 15 fragments with molecular weights between 1 x 10(6) and 28 x10(6). The electrophoretic profiles obtained in 0.3% agarose gels with DNA fragments from none different strains of herpes simplex virus type 1 could be readily differentiated from the patterns exhibited by the corresponding fragments from four separate strains of type 2 virus; however, with each serotype, the laboratory strains differed significantly among themselves and also from isolates passaged a minimum number of times outside the human host. Digestion of all DNAs of herpes simples virus with either enzyme reproducibly generated two classes of fragments (major and minor) which differed in molar ocncentration. Moreover, although the molecular weight of an intact herpes simplex 1(F1) DNA molecule is approximately 98 x 10(6), the summed molecular weights of all major and minor HinIII fragments totalled 160 x 10(6), and the seven major fragments alone accounted for only 60 x 10(6). These unusual features indicate the existence of limited heterogeneity in the positions of cleavage sitet along individual molecules. We have eliminated the possibility that minor fragments arose from contamination with the defective DNA of high byoyant density which appears on serial undiluted passage of the virus. In fact, this latter type of DNA was resistant to cleavage by HinIII and gave large amounts of only two species of EcoRI fragments; suggesting that the defective molecules consist of many tandem repeats of a small segment of viral DNA. The heterogeneity in the viral DNA of normal density appears to be related to the structural organization of the molecules and does not necessarily imply differences in genetic content.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Jeppesen P. G., Katagiri K. J., Delius H. Mapping the DNA fragments produced by cleavage by lambda DNA with endonuclease RI. Nature. 1973 Jan 12;241(5385):120–123. doi: 10.1038/241120a0. [DOI] [PubMed] [Google Scholar]
- Becker Y., Dym H., Sarov I. Herpes simplex virus DNA. Virology. 1968 Oct;36(2):184–192. doi: 10.1016/0042-6822(68)90135-9. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Herpes vimplex virus: genome size and redundancy studied by renaturation kinetics. J Virol. 1971 Oct;8(4):591–593. doi: 10.1128/jvi.8.4.591-593.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOGGAN M. D., ROIZMAN B. The isolation and properties of a variant of Herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959 Sep;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- Hayward G. S. Unique double-stranded fragments of bacteriophage T5 DNA resulting from preferential shear-induced breakage at nicks. Proc Natl Acad Sci U S A. 1974 May;71(5):2108–2112. doi: 10.1073/pnas.71.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine J. W., Honess R. W., Cassai E., Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974 Sep;14(3):640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Laithier M., Lando D., Ryhiner M. L. Infectious DNA from herpes simplex virus: infectivity of double-stranded and single-stranded molecules. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3621–3625. doi: 10.1073/pnas.70.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Spear P. G., Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972 Jan;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]