Abstract
Background
Quality of life (QoL) is an increasingly important parameter in clinical practice as it predicts mortality and poor health outcomes. It is hypothesized that one may have a genetic predisposition for QoL. We therefore related 139 candidate genes, selected through a literature search, to QoL in healthy females.
Methods
In 5,142 healthy females, background characteristics (i.e. demographic, clinical, lifestyle, and psychological factors) were assessed. QoL was measured by the EORTC QLQ-C30, which consists of 15 domains. For all women genotype information was available. For each candidate gene, single nucleotide polymorphisms (SNPs) were identified based on their functional (n = 2,663) and physical annotation (n = 10,649). SNPs were related to each QoL-domain, while controlling for background characteristics and population stratification. Finally, gene-based analyses were performed relating the combined effect of 10,649 SNPs (selected based on physical annotation) for each gene, to QoL using the statistical software package VEGAS.
Results
Overall, we found no relation between genetic variations (SNPs and genes) and 14 out of 15 QoL-domains. The strongest association was found between cognitive functioning and the top SNP rs1468951 (p = 1.21E-05) in the GSTZ1 gene. Furthermore, results of the gene-based test showed that the combined effect of 11 SNPs within the GSTZ1 gene is significantly associated with cognitive functioning (p = 2.60E-05).
Conclusion
If validated, the involvement of GSTZ1 in cognitive functioning underscores its heritability which is likely the result of differences in the dopamine pathway, as GSTZ1 contributes to the equilibrium between dopamine and its neurotoxic metabolites via the glutathione redox cycle.
Introduction
During the last decades quality of life (QoL) is frequently measured as a subjective rating in research and clinical practice. It is a multifactorial concept that consists of a person’s perception of physical, psychological, and social functioning that is often subdivided into several domains (e.g. physical functioning, emotional functioning, cognitive functioning, social functioning, fatigue, and pain).[1] QoL is an increasingly important parameter in both research and clinical practice as it is predictive of mortality and poor health outcomes, such as morbidity, self-management, and health care.[2–4]
QoL is influenced by demographic characteristics (e.g. age, sex, and race), lifestyle factors (e.g. diet and physical activity) and psychological factors, such as mood states and stress.[5–8] A large proportion of variation between individuals remains unexplained. It is therefore suggested that individual genetic predisposition contributes to the perception of QoL.[9] There is increasing evidence for genetic determinants of depression, well-being, pain, and fatigue.[10–14] In addition, family and twin-studies indicate that the heritability for subjective well-being, depression, and anxiety ranges from thirty to as much as fifty percent.[15–19] Moreover, there is ample evidence that the hypothalamic-pituitary-adrenal axis, immune, neuroendocrine, and cardiovascular system are associated with various QoL-domains.[20]
In 2009 an international and interdisciplinary Consortium for Genetics and Quality of Life Research (GeneQol) was initiated.[9] Its main objective is to identify and investigate potential biological mechanisms, genes and genetic variants involved in QoL. The first studies relating genes to QoL have shown that various single nucleotide polymorphisms (SNPs) in cytokine genes and the glutathione metabolic pathway are related to QoL in different patient groups.[21–23] The previous studies are valuable with little generalizability, as they all include only patient samples. The current study is conducted in a sample of healthy women, which increases the generalizability, as the relation between genetics and QoL is examined without the confounding role of diseases. Recently, the GeneQol consortium has provided an overview of biological markers involved in overall QoL and related domains, such as fatigue, pain, negative and positive functioning.[24] They have identified several candidate genes based on an extensive literature search.[24] We aim to perform an empirical study relating these candidate genes to QoL in a healthy female sample.
The specific objectives are (1) to relate SNPs for each of the listed candidate genes to QoL; and (2) to relate the combined effect of SNPs within each gene to QoL.
Methods
Study population and procedure
This study utilized data from the Karolinska Mammography Project for Risk Prediction of Breast Cancer (KARMA) study, see www.karmastudy.org for a detailed description. Data used in this manuscript is available upon request through the KARMA Research Platform which can be found at www.karmastudy.org. In short, KARMA collects data and bio-samples each time a participating woman comes for mammography screening or a clinical mammography at one of four Swedish participating hospitals. In Sweden, the national screening program invites all women at 18 month intervals for those 40–55 years, and for those older than 56–74 years at 24 months. Every woman completes a comprehensive online survey. This survey entails more than 250 questions addressing breast cancer related issues such as reproductive history, cancer treatment, and family history of cancer; lifestyle factors (e.g. alcohol and tobacco use); previous medical conditions other than breast cancer; medication use; and QoL. Blood is donated at each visit and processed at the Karolinska biobank. Every six months data from several registries are linked to the KARMA data: the information network for cancer treatment which entails clinical information on breast cancer patients; the Swedish Cancer; Cause-of-Death; Prescription; and In- and Out-patient registers. The KARMA study was approved by the Swedish regional ethical board at the Karolinska Institutet and is conducted in accordance with the Declaration of Helskini.[25] All women gave written informed consent.
Measurements
Background characteristics. Demographic and clinical factors All women reported age, educational level, use of painkillers (e.g. paracetamol and ibuprofen) and being on hormone replacement therapy (yes/no) during the last year. Participants’ previous or ongoing medical conditions such as, high blood pressure, hyperlipidemia, myocardial infarction, angina, heart failure, stroke, polycystics ovary syndrome (PCOS), pre-eclampsia, depression, diabetes, bulimia, and anorexia were self-reported.
Life style factors Body Mass Index (BMI) was calculated based on women’s weight in kilogram divided by their squared height in meters. Current tobacco use (yes/no) was self-reported—if women either smoked cigarettes or used snuff (a typical Swedish tobacco in moist powder form).
Psychological factors The level of stress experienced during the last five years was assessed by one item “Please state how stressed you have been feeling in the past five years”. Answers could be given on a 4 point Likert-scale ranging from ‘never stressed’ to ‘always stressed’. All participants were asked whether they have experienced any of the following life stressors during the last five years: a close relative who died; own divorce or separation; a close friend who died; serious disease or injury; became unemployed; other very stressing event. Finally, the average number of hours of sleep per night was assessed.
Quality of life. QoL was measured with the European Organization for Research and Treatment of Cancer Quality of Life questionnaire Core 30 (EORTC QLQ-C30), a cancer specific QoL-questionnaire.[26] It includes global health status, five functional scales (physical; role; emotional; cognitive; and social), three symptom scales (fatigue; nausea or vomiting; and pain), and six single items (dyspnea; insomnia; appetite loss; constipation; diarrhea; and financial difficulties). These scales and items are linearly transformed from 0 to 100. High scores on the global health status/QoL scale indicate a high level of QoL, and high scores on the functional scales indicate a high level of functioning. Conversely, high scores on the symptom scales/items indicate high levels of health problems. The EORTC QLQ-C30 has been validated and is considered to have good psychometric properties.[26]
Genotyping. Some of the KARMA participants were genotyped as part of the iCOGS project (www.nature.com/iCOGS) using the iCOGS array. This array was specifically designed to evaluate genetic variants associated with the risk of breast, ovarian and prostate cancer.[27,28] It comprises ~200,000 SNPs, which were selected in samples from large case-control studies in disease-based consortia. A genome-wide imputation of SNPs using the IMPUTE (V.2.0) software, based on the 1000 Genomes Project [(1KGP) Mar 2012 release (updated Apr 19, 2012)], was performed.[29] For the KARMA dataset, the genotypes of 4,310,392 SNPs were successfully called and passed quality control filter (INFO score from IMPUTE > = 0.8 and minor allele frequency > = 0.01). The imputed iCOGS chip has a 60% coverage of what the Illumnia HumanHap550 chip would cover.
Selection of single nucleotide polymorphisms The list of candidate genes derived by the GeneQol consortium is continuously updated based on current literature.[24] At the start of this study, the list entailed 139 candidate genes, which were all related to at least one QoL-domain (S1 Table: The list of 139 candidate genes, which are all related to at least one QoL-domain). SNPs for each candidate gene were selected based on both functional and physical annotation (build 37).[30] For the functional annotation, SNPs were selected according to their effects on expression levels, i.e. whether they are expressions of quantitative trait loci (eQTLs) for that gene. Based on the functional annotation 2,663 SNPs were selected for the 139 candidate genes. For the physical-based annotation, a 20 kb window was used where SNPs were categorized based on both their position and linkage disequilibrium (LD) pattern. For the 139 candidate genes 10,649 SNPs were selected based on their physical annotation.
Statistical analyses
Background characteristics and quality of life QoL scores of the selected KARMA women were compared to a Swedish reference population.[31] The QoL scores on the 15 domains were therefore transformed to standard scores based on the scores of an age-matched Swedish reference population. To compare, standard scores were calculated by dividing the difference between the mean scores of the KARMA women and the scores of the age-matched reference population, by the standard deviations of the reference population. The value of the standard scores can be interpreted according to Cohen’s effect size (d), where a score of <0.2 indicates a small, 0.5–0.8 a moderate and >0.8 a large difference. Analyses were performed in SPSS 16.0.
Relating single nucleotide polymorphisms to quality of life Initially, possible covariables for the relation between SNPs and QoL were identified. To do so, all background characteristics (listed in Table 1) were related to each of the QoL-domains separately by means of regression analyses. Background characteristics that were associated with QoL (p<0.10) were included as covariables in the subsequent analyses. To control for population stratification [32], principal components analysis (PCA) was performed by EIGENSTRAT V.4.2 (1,2). We visually inspected PCA plots for outliers in terms of ancestry from CEU (northern and western Europe) clusters. Five principal components were retained after inspection of a Scree plot, and included as covariables in subsequent analyses. For the main analyses, regression analyses were used to study the association between SNPs and QoL, while controlling for covariables and the five principal components. Analyses were performed for SNPs selected on functional and physical annotation separately and run in the statistical program PLINK.[33] Bonferonni corrected p-value was set at 3.76E-06 (0.05 divided by (2,663+10,649 SNPs)). For 10 of the 15 QoL-domains the distribution of scores was non-normal. Scores on the cognitive functioning scale were transformed using square root transformation [√(101-raw score)]. On the remaining nine domains a large percentage of women (range from 66.6% to 92.0%) reported the maximum score on the functional scales and the minimum score on the symptom scales/items. These domains were therefore dichotomized; minimum/maximum value versus the remaining answers.
Table 1. Background characteristics (demographic, clinical, lifestyle, and psychological factors) (n = 5,142).
| N (%) | |
|---|---|
| Demographic factors | |
| Age in mean years (range) a | 54.3 (22–88) |
| Educational level b | |
| Nine year school | 497 (9.7) |
| Gymnasium | 1688 (32.9) |
| University | 2525 (49.2) |
| Other | 419 (8.2) |
| Clinical factors | |
| Being on hormone replacement therapy (yes) | 1709 (33.2) |
| Using painkillers (yes) | 4931 (95.9) |
| Number of medical conditions c | |
| None | 2746 (53.4) |
| One | 1509 (29.3) |
| Two | 618 (12.0) |
| Three | 201 (3.9) |
| Four or more | 68 (1.3) |
| Lifestyle factors | |
| Body mass index (BMI) as mean score (range) d | 25.2 (17–52) |
| Using tobacco (yes) | 684 (13.3) |
| Psychological factors | |
| Stress in the last five years e | |
| Never stressed | 275 (5.4) |
| Seldom stressed | 1849 (36.4) |
| Often stressed | 2379 (46.9) |
| Always stressed | 571 (11.3) |
| Number of life stressors | |
| None | 1728 (33.6) |
| One | 2027 (39.4) |
| Two | 955 (18.6) |
| Three | 343 (6.7) |
| Four or five | 89 (1.7) |
| Hours of sleep f | |
| 5 hours or less | 207 (4.4) |
| 6 hours | 1103 (23.2) |
| 7 hours | 2170 (45.7) |
| 8 hours or more | 1269 (26.7) |
Note: Data is presented as frequencies (percentages) for 5,142 healthy women included in the KARMA study. Age and body mass index are provided in mean (range).
a = information is missing for 1 participant;
b = information is missing for 14 participants;
c = High blood pressure and depression are the most common conditions;
d = for 17 participants information was unavailable;
e = for 68 participants no information was available;
f = information is missing for 393 participants.
Gene-Based analyses Gene-based analyses were performed relating the combined effect of 10,649 SNPs (that were selected based on physical location) for each gene to QoL. Analyses were performed for each of the QoL-domains separately, using the Versatile Gene-based Association Study (VEGAS) software.[34] This software package applies a test by using simulations from the multivariate normal distribution by incorporating information on a set of SNPs within a gene while accounting for LD between SNPs. VEGAS uses HapMap populations to estimate patterns of LD for each gene.[34] Statistical significance is assessed adaptively. In the first step, 1000 simulations are run. If the empirical p-value is <0.01, another 10,000 simulations are performed. If, the empirical p-value is <0.001, another 1,000,000 simulations are performed. If an empirical p-value of 0 is reached, no more simulations will be performed.
Results
Background characteristics and quality of life
For 5,142 out of 68,334 KARMA women information on both QoL and genotype data was available, and they were therefore included in this study. Women diagnosed with breast cancer before entering KARMA were excluded. Characteristics of the participating women are presented in Table 1. Women’s scores on the QoL domains are presented in Table 2. Overall, the KARMA women reported a good QoL as they appear to function well and report few symptoms. Although the selected KARMA women scored significantly (p<0.01) different on many of the QoL-domains compared to the Swedish reference sample, differences had a small effect size (Cohen’s d<0.3) (S1 Fig. Comparing quality of life scores of the KARMA women to a Swedish reference sample).
Table 2. Mean quality of life scores.
| N = 5,142 | |
|---|---|
| EORTC QLQ-C30 DOMAINS | |
| Global health/ quality of life | 75.8 (22.2) |
| Functional scales | |
| Physical functioning (highest QoL) | 3427 (66.6) |
| Role functioning (highest QoL) | 3825 (74.5) |
| Emotional functioning | 76.1 (22.8) |
| Cognitive functioning a | 87.8 (19.2) |
| Social functioning (highest QoL) | 3826 (74.5) |
| Symptom scales/items | |
| Fatigue | 22.4 (20.8) |
| Nausea and vomiting (highest QoL) | 4486 (87.3) |
| Pain | 20.4 (26.5) |
| Dyspnoea | 19.3 (27.0) |
| Insomnia | 25.0 (30.2) |
| Appetite loss (highest QoL) | 4648 (90.4) |
| Constipation (highest QoL) | 4409 (85.8) |
| Diarrhea (highest QoL) | 4556 (88.7) |
| Financial difficulties (highest QoL) | 4725 (92.0) |
Note: For global health/quality of life and the functional scales a higher score indicates a better quality of life, whereas for the symptom scales/items a lower score indicates a better quality of life. For the continuous variables (i.e. global health/quality of life; emotional functioning; cognitive functioning; fatigue; pain; dyspnoea; and insomnia) mean scores (SD) are presented. For the dichotomized scales (i.e. physical functioning; role functioning; social functioning; nausea and vomiting; appetite loss; constipation; diarrhea; financial difficulties) frequencies and percentages for the category with the highest quality of life is provided. Please note that for 6, 3, 10, 1, 0, 6, 5, 3, 2, 13, 4, 3, 2, 4, 7 participants respectively information was missing.
a = cognitive functioning was transformed by using square root transformation [√(101-raw score)], ranging from 1–10 with low scores having a better cognitive functioning. The transformed mean score and standard deviation is 2.9 (2.4).
Results of the identification of possible covariables, relating background characteristics to each QoL-domain, are provided in S2 Table (S2 Table. The association between background characteristics and quality of life using Wald Chi Square test-statistic). As expected, age was positively related to mental QoL (e.g. emotional functioning) and negatively to physical QoL (e.g. physical functioning). Overall, the number of medical conditions, stress during the last five years, and the number of life stressors showed the strongest negative association, whereas the number of hours of sleep had the strongest positive relation with the QoL domains.
Single nucleotide polymorphisms and genes related to quality of life
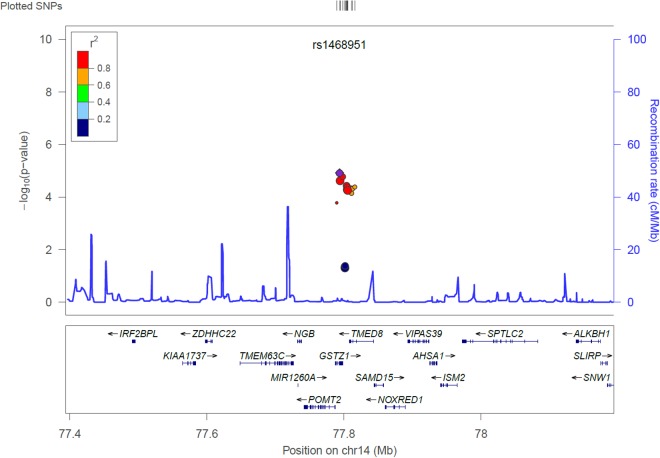
Results of the association study relating the SNPs selected by functional and physical annotation to QoL, while controlling for possible covariables (S2 Table. The association between background characteristics and quality of life using Wald Chi Square test-statistic) are provided in Table 3 and 4 respectively. None of the SNPs selected by functional annotation were significantly related to QoL (Table 3). For SNPs selected based on their physical annotation, there was no statistically significant relation between SNPs and QoL-domains (Table 4). The strongest association was found between cognitive functioning and the top SNP rs1468951 (p = 1.21E-05, Bonferonni-corrected p-value = 3.76E-06) in the GSTZ1 gene (Table 4), independent of background characteristics (i.e. age, using painkillers, number of medical conditions, using hormone replacement therapy, level of stress in the last five years, number of life stressors, and number of hours of sleep) and the five principal components (controlling for population stratification). This top SNP was an imputed marker, which is in high LD with the genotyped SNP rs1046428 (r2 = 0.99). A Manhattan plot of the relation between cognitive functioning and SNPs based on their physical annotation was prepared using Haploview and is displayed in (S2 Fig. Manhattan plot (p-values per chromosome) for the relation between cognitive functioning and the SNPs found based on physical location for the selected candidate genes).[35] The locus-specific association map centered at the top SNP rs1468951 showed low p-values for several SNPs on the GSTZ1 gene, indicating a relation with cognitive functioning (Fig. 1).[36] To examine the stability of the effect estimate, a sensitivity analysis was performed by sequential omission of individual covariables (leave-one-out analysis). Results revealed that the estimate of rs1468951 remained stable (data not shown).
Table 3. Relation between quality of life and the single nucleotide polymorphisms selected by functional annotation (n = 2,663).
| FUNCTIONAL ANNOTATION | ||||||||
|---|---|---|---|---|---|---|---|---|
| top SNP | Chr | Position | Minor/major | MAF | Beta (SE) | p | GENE | |
| QUALITY OF LIFE | ||||||||
| Global health/ QoL | rs1603406 | 12 | 87887139 | G/A | 0.40 | -1.31 (0.41) | 1.52E-03 | GLDC |
| Functional scales | ||||||||
| Physical functioning | rs10750403 | 11 | 128477472 | C/T | 0.45 | 0.15 (0.05) | 1.36E-03 | PRKACA |
| Role functioning | rs12218712 | 10 | 24292743 | A/T | 0.31 | 0.18 (0.05) | 1.18E-03 | HLA-DRB1 |
| Emotional functioning | rs12415866 | 10 | 44686664 | G/A | 0.12 | 2.15 (0.60) | 3.34E-04 | RHBDF2 |
| Cognitive functioning a | rs17159612 | 7 | 84725413 | C/T | 0.24 | -0.16 (0.05) | 2.69E-03 | SLC6A4 |
| Social functioning | rs1380162 | 4 | 119970203 | A/G | 0.33 | 0.16 (0.06) | 4.71E-03 | HSN2 |
| Symptom scales/items | ||||||||
| Fatigue | rs1603406 | 12 | 87887139 | G/A | 0.40 | 1.46 (0.42) | 5.56E-04 | GLDC |
| Nausea and vomiting | rs1560580 | 2 | 137745374 | A/G | 0.45 | -0.26 (0.07) | 1.24E-04 | RHBDF2 |
| Pain | rs10150965 | 14 | 29018461 | G/C | 0.41 | 1.82 (0.52) | 5.02E-04 | WNK1 |
| Dyspnoea | rs1407818 | 1 | 192561712 | G/A | 0.19 | -2.21 (0.68) | 1.18E-03 | MYB |
| Insomnia | rs1185701 | 1 | 156419617 | C/G | 0.14 | -3.24 (0.79) | 4.46E-05 | LIPG |
| Apetite loss | rs10883690 | 10 | 83488792 | G/T | 0.29 | -0.28 (0.09) | 1.34E-03 | PRKACA |
| Constipation | rs1408808 | 9 | 12542187 | C/G | 0.36 | 0.20 (0.06) | 1.43E-03 | UMPS/DRD4 |
| Diarrhoea | rs11848780 | 14 | 34169150 | A/G | 0.13 | -0.34 (0.11) | 1.29E-03 | CD19/MIF/GSTP1 |
| Financial difficulties | rs13160478 | 5 | 118082740 | G/A | 0.11 | -0.51 (0.16) | 1.04E-03 | CASP8 |
Note: For the 139 candidate genes, 2,663 SNPs were selected based on functional annotation. Bonferonni p-value = 3.76E-06 (0.05/2,663+10,649 SNPs). For the continuous variables (i.e. global health/quality of life; emotional functioning; cognitive functioning; fatigue; pain; dyspnoea; and insomnia) linear regressions were performed. For the dichotomized variables (i.e. physical functioning; role functioning; social functioning; nausea and vomiting; appetite loss; constipation; diarrhea; financial difficulties) we used logistic regression analyses. Chr = chromosome; Position = position of the chromosome; Minor/major = minor and major alleles based on forward strand and minor allele frequencies in Europeans; MAF = minor allele frequency over all European controls in iCOGS; Beta = beta value for the minor allele relative to the major allele; SE = standard error; p = p-value.
a = cognitive functioning was transformed by using square root transformation [√(101-raw score)] ranging from 1–10, with low scores having a better cognitive functioning, therefore the direction of the relation is reversed.
Table 4. Relation between quality of life and the single nucleotide polymorphisms selected by physical annotation (n = 10,649).
| PHYSICAL ANNOTATION | ||||||||
|---|---|---|---|---|---|---|---|---|
| top SNP | Chr | Position | Minor/ major | MAF | Beta (SE) | p | GENE | |
| QUALITY OF LIFE | ||||||||
| Global health/ QoL | rs3783547 | 2 | 113533339 | G/A | 0.37 | -1.43 (0.42) | 6.66E-04 | IL1A |
| Functional scales | ||||||||
| Physical functioning | rs16080 | 7 | 24350966 | T/C | 0.08 | 0.35 (0.10) | 3.28E-04 | NPY |
| Role functioning | rs3889728 | 1 | 23084881 | C/T | 0.25 | -0.22 (0.06) | 1.14E-04 | AGT |
| Emotional functioning | rs2475376 | 10 | 96712400 | A/G | 0.15 | 2.04 (0.54) | 1.63E-04 | CYP2C9 |
| Cognitive functioning a | rs1468951 | 14 | 77793487 | A/C | 0.18 | 0.25 (0.06) | 1.21E-05 | GSTZ1 |
| Social functioning | rs57758950 | 8 | 105453992 | T/C | 0.13 | -0.32 (0.07) | 1.63E-05 | DPYS |
| Symptom scales/items | ||||||||
| Fatigue | rs2813555 | 6 | 152442582 | A/G | 0.20 | 1.76 (0.52) | 7.56E-04 | ESR1 |
| Nausea and vomiting | rs4950025 | 1 | 97717279 | C/A | 0.05 | -0.82 (0.19) | 1.95E-05 | DPYD |
| Pain | rs35258421 | 4 | 142545105 | A/G | 0.18 | 2.29 (0.66) | 4.86E-04 | IL15 |
| Dyspnoea | rs7648614 | 3 | 123328980 | T/C | 0.08 | -3.85 (1.08) | 3.59E-04 | MYLK |
| Insomnia | rs4298 | 17 | 61557200 | C/G | 0.05 | 4.09 (1.23) | 8.61E-04 | ACE |
| Apetite loss | rs6062900 | 20 | 61980125 | G/C | 0.12 | -0.43 (0.12) | 6.03E-04 | CHRNA4 |
| Constipation | rs324969 | 7 | 34791852 | G/A | 0.48 | -0.20 (0.06) | 9.11E-04 | NPSR1 |
| Diarrhoea | rs748190 | 10 | 131519274 | G/A | 0.46 | 0.26 (0.07) | 2.12E-04 | MGMT |
| Financial difficulties | rs496338 | 10 | 131412605 | A/T | 0.12 | -0.59 (0.15) | 8.86E-05 | MGMT |
Note: For the 139 candidate genes, 10,649 SNPs were selected based on physical annotation (build 37). Bonferonni corrected p-value = 3.76E-06 (0.05/2,663+10,649 SNPs). For the continuous variables (i.e. global health/quality of life; emotional functioning; cognitive functioning; fatigue; pain; dyspnoea; and insomnia) linear regressions were performed. For the dichotomized variables (i.e. physical functioning; role functioning; social functioning; nausea and vomiting; appetite loss; constipation; diarrhea; financial difficulties) we used logistic regression analyses. Chr = chromosome; Position = position of the chromosome; Minor/major = minor and major alleles based on forward strand and minor allele frequencies in Europeans; MAF = minor allele frequency over all European controls in iCOGS; Beta = beta value for the minor allele relative to the major allele; SE = standard error; p = p-value.
a = cognitive functioning was transformed by using square root transformation [√(101-raw score)] ranging from 1–10, with low scores having a better cognitive functioning, therefore the direction of the relation is reversed.
Fig 1. Locus-specific association map generated from genotyped SNPs in the chromosome 14, centered at rs1468951 for cognitive functioning.
Note: Vertical axis is the—log10 of the p-value, the horizontal axis is the chromosomal position. Each dot represents a SNP tested for association with cognitive functioning. Linkage disequilibrium (LD) between the most significant SNP, listed at the top of the plot, and the other SNPs in the plot is shown by the r2 legend. Locus zoom software was used to prepare this figure.[36]
Furthermore, results of the gene-based test VEGAS are provided in Table 5. The GSTZ1 gene (11 SNPs) was significantly associated with cognitive functioning (p = 2.60E-05). For the other domains, none of the genes reached statistical significance after correction for multiple testing. The genotype specific sample and effect sizes for the 11 GSTZ1 SNPs are provided in S3 Table (S3 Table: The sample and effect sizes for the 11 SNPs in the GSTZ1 gene).
Table 5. Gene-based test for 139 candidate genes using the single nucleotide polymorphisms selected by physical location.
| QUALITY OF LIFE | Chr | Gene | nSNPs | Start pos | End pos | p |
|---|---|---|---|---|---|---|
| Global health/ QoL | 5 | IL12B | 57 | 158674368 | 158690059 | 1.20E-02 |
| Functional scales | ||||||
| Physical functioning | 7 | NPY | 5 | 24290333 | 24298002 | 8.38E-04 |
| Role functioning | 1 | AGT | 51 | 228904891 | 228916959 | 5.74E-04 |
| Emotional functioning | 5 | NR3C1 | 12 | 142637688 | 142795270 | 6.00E-03 |
| Cognitive functioning | 14 | GSTZ1 | 11 | 76857106 | 76867693 | 2.60E-05* |
| Social functioning | 10 | MGMT | 114 | 131155455 | 131455358 | 4.81E-03 |
| Symptom scales/items | ||||||
| Fatigue | 12 | NR3C1 | 12 | 142637688 | 142795270 | 1.31E-02 |
| Nausea and vomiting | 12 | GNB3 | 1 | 6819635 | 6826818 | 1.13E-03 |
| Pain | 1 | PER3 | 51 | 7767349 | 7827824 | 1.21E-02 |
| Dyspnoea | 20 | GNAS | 7 | 56848189 | 56919645 | 5.53E-03 |
| Insomnia | 12 | AVPR1A | 35 | 61826482 | 61832857 | 6.32E-03 |
| Apetite loss | 20 | CHRNA4 | 2 | 61445108 | 61463139 | 1.91E-03 |
| Constipation | 3 | UMPS | 56 | 125931902 | 125946730 | 9.73E-03 |
| Diarrhoea | 10 | BTRC | 58 | 103103814 | 103307060 | 1.75E-03 |
| Financial difficulties | 7 | NPY | 5 | 24290333 | 24298002 | 1.49E-03 |
Note: * p < Bonferonni corrected p-value of 3.60E-04 (0.05/139 candidate genes). Chr = Chromosome; nSNPs = number of SNPs; Test stat = test statistic; p = p-value.
Identification of causal variants
To identify the causal variant or variants we used two distinct approaches. First, forward selection regression analyses using the step function in R, adjusted for all covariates and principal components, was utilized. The full model included the genotypes of the 11 SNPs annotated to be +-20kb of GSTZ1 and retained in the VEGAS gene-based analysis. Due to high LD structure in the region a penalty term of five was used [37], with rs11845842 retained in the analysis. Second, we used the ICSNPathway software [38], to explore the coding variant likely responsible for the relation with cognitive functioning. Results showed one candidate causal SNP (rs1046428) and three candidate causal pathways (‘nitrogen compound metabolic process’, ‘anine metabolic process’, ‘amino acid metabolic process’, ‘carboxylic acid metabolic process’, and ‘oxidoreductase activity’). These results indicate the following hypothesis [rs1046428 (non-synonymous coding)-> GSTZ1 -> nitrogen compound metabolic process’/‘anine metabolic process’/‘amino acid metabolic process’/‘carboxylic acid metabolic process’/‘oxidoreductase activity’ pathways]. Both procedures identified different SNPs, which are in high LD with each other (r2 = 0.90), they are thus probably tagging the same causal variant. Furthermore, rerunning the gene-based analysis by excluding our top SNP rs1468951 and the eight GSTZ1 SNPs in high LD (r2>0.9) showed that GSTZ1 was no longer significantly related to cognitive functioning (p = 4.15E-02). This indicates the cumulative effects in the LD block surrounding rs1468951 within the GSTZ1 gene.
Discussion
Overall, we found no relation between genetic variations and 14 out of 15 QoL-domains investigated in this study. For cognitive functioning variations in the GSTZ1 gene were statistically significant, independent of background characteristics and population stratification.
There are various plausible reasons for the absence of associations between genetic variations and QoL in this study. It is likely that this is—at least in part—the result of limited variation in QoL, due to our healthy female sample. Second, adoption of a candidate gene approach may have resulted in a too limited selection of genes. Furthermore, genotyping was performed by using the iCOGS chip, which was originally built to identify the genetic risk for breast, ovarian and prostate cancer. Although, after imputation, the iCOGS chip covers 60% of what an Illumnia HumanHap550 chip covers, the dispersion over the entire genome may still be skewed. Third, for complex phenotypes a genetic predisposition may be the result of several genes working in concert or the effect of an entire pathway.
The strongest association (p = 1.21E-05, Bonferonni-corrected p-value = 3.76E-06) was found between cognitive functioning and the top SNP rs1468951 in the GSTZ1 gene, while controlling for background characteristics and population stratification. The imputed marker is in almost perfect LD (r2 = 0.99) with the genotyped SNP rs1046428 the latter of which has been annotated in dbSNP as a non-synonymous missense mutation (M [Met] ⇒ T [Thr]), (S3 Fig. Predicted chromatin state, sequence conservation across mammals, and effect on regulatory motifs of rs1468951 and variants with r2 > = 0.8).[39] Mining the ENCODE [40] data via HaploReg[39], the intronic variant rs1468951 was predicted to be in DNase hypersensitivity regions in numerous cell lines; and altering predicted relative affinity of two transcription factors (EBF and FXR) (S4 Fig. Epigenetic road map for rs1468951 effect on regulatory motifs of rs1468951 and variants with r2 > = 0.8). Mining the RoadMap [41] data predicts rs1468951 to lie in regions in which modification of histone proteins is suggestive in several different cell types (LIV.A, PFM.3 and PFF.2) (S4 Fig. Epigenetic road map for rs1468951 effect on regulatory motifs of rs1468951 and variants with r2 > = 0.8). Results of the association between cognitive functioning and the top SNP (rs1468951) without adjusting for background characteristics showed that the relation was not significant [data not shown]. We opted for the inclusion of the covariables based on both literature and the significant findings in the preliminary analyses.[42–45]
In addition, we found that the combined effect of the 11 SNPs within the GSTZ1 gene were significantly related to cognitive functioning independent of background characteristics, indicating that the multiple smaller effects of the 11 individual GSTZ1 SNPs seem to be working in concert. This finding is in line with the general understanding that cognitive functioning (e.g. IQ, memory, and concentration) is heritable, and in concordance with the current knowledge of the GSTZ1 gene. GSTZ1 encodes multifunctional enzymes important in detoxification and several drugs by conjugation with glutathione. One of these enzymes is maleylacetoacetate isomerase (MAAI) which is involved in the catabolism of phenylalanine and tyrosine.[23,46] Defects in the tyrosine enzyme may lead to severe metabolic disorders including tyrosinaemia which leads to mental retardation and cognitive problems.[47] In experimental studies the administration of tyrosine to individuals under stress leads to improved cognitive functioning, including memory tasks.[48] The physiological basis of this beneficial effect of tyrosine is attributed to its role as precursor for the synthesis of dopamine, which is a major neurotransmitter widely distributed within the brain.[48,49] It is well-known that dopaminergic neurotransmission in the prefrontal cortex contributes to individual differences following a non-linear relation, a so-called reversed U-form.[50] Next to the catabolization of tyrosine into dopamine, GSTZ1 also contributes to the equilibrium between dopamine and its neurotoxic metabolites via the glutathione redox cycle.[51] Hypothesized is that dopamine and its metabolites have cytotoxic actions on neurons, thereby negatively impacting cognitive functioning, contributing to the U-shaped relation.[52] In a first study relating GSTZ1 to cognitive functioning among 64–68 aged 470 Scottish community volunteers, a significant association with SNP-1002 G>A was found. A-carriers showed a significantly lower mean score on cognitive functioning, supporting the hypothesis that dopamine disposal pathways may have a negative impact on cognitive functioning.[52]
Limitations and strengths
There are some noteworthy limitations to this study. As already mentioned, genotyping was performed by using the iCOGS chip, which was originally designed to identify risk factors for breast, ovarian, and prostate cancer. For the gene based analyses we used the VEGAS approach which utilizes HapMap populations to estimate patterns of LD for each gene. In this study, imputations of SNPs were performed by using the 1000 genome reference panel. HapMap has no complete coverage of the number of SNPs per candidate gene as compared to the 1000 genome reference panel.[53] Second, other factors that were not measured could also impact variation in QoL either directly or via genes. For a detailed description of possible relations between biological factors (including genetic markers), background characteristics, and QoL, see the adapted theoretical model of Wilson and Cleary.[20] In addition it is worth mentioning, that we measured subjective cognitive functioning which was assessed by two self-reported items concerning memory and concentration. This two-item scale can be considered limited, however, it has been found to yield high levels of reliability and validity.[26] Furthermore, there is a risk for reporting false positives given the number of tests; for each QoL-domain we related 13,312 SNPs and 139 genes. Finally, this candidate gene study is based on only one study, therefore further validation in independent datasets would be required to confirm the association between GSTZ1 and subjective cognitive functioning. Since this is a novel area of research the number of studies collecting both genetic and QoL-information are scarce. To the best of our knowledge, no external data are currently available that combine subjective cognitive functioning as assessed with the EORTC QLQ-C30 and genetic data. Since an increasing number of studies are now embarking on the assessment of QoL and genetic data, validation of these data will be possible in the future.
It is important to note that there is controversy in what constitutes QoL. Although QoL can be described as a uni-dimensional concept, we view it as multifactorial consisting of a person’s perception of several domains such as fatigue, physical, emotional, cognitive, and social functioning. One can hypothesize that the more ‘biological’ domains, such as physical and cognitive functioning may have a stronger genetic basis, than for example social functioning. Nevertheless, a recent review reported heritability for social functioning.[54] In our study, there was no significant relation between social functioning and genetic markers. Various possible reasons for this lack of association are provided at the beginning of the discussion. Contrary to our findings, another study examining the relation between genetics and cognitive functioning found a significant association with rs1046428.[55] The most likely explanation for this discrepancy is the difference in measuring cognitive functioning. Where we measured cognitive functioning by self-report examining perceived memory function and concentration, Harris et al. used tasks to examine general mental ability, non-verbal reasoning, verbal fluency and logical memory.[55] These two studies thus examine distinct, albeit related concepts, thereby impeding their comparison.
We would also like to stress the strengths. This is the first study relating QoL to genes in a large sample of healthy females, while statistically controlling for background characteristics including self-reported chronic diseases thereby minimizing the impact of medical conditions. Moreover, the used iCOGS chip has a fairly comprehensive genetic coverage. Furthermore, the included sample of healthy women is representative for the general Swedish population in terms of QoL, increasing generalizability of the results.
Conclusion and future directions
In conclusion, the involvement of GSTZ1 in cognitive functioning underscores its heritability which is likely the result of differences in the dopamine pathway. Findings support the hypothesis that dopamine can have negative effects via the neurotoxic by-products.[51] The obvious next step is to replicate the association between cognitive functioning and variations in the GSTZ1 gene to ensure it is not a chance finding. Although needed, validation is challenging as cognitive functioning is measured in varying ways. In this study measurement entailed two questions tapping into memory and concentration as specific aspects of cognitive functioning.
For future research relating various QoL-domains to genetic markers, a candidate gene approach may be too limited, possibly missing valuable associated genes. Therefore we also opt for a whole genome approach in future studies. Additional research is needed as findings will be valuable in clinical settings. Identified genes can be used as indicators for those who are susceptible to impairments in their QoL. This is particularly useful information for individuals who are experiencing high levels of stress, such as when being diagnosed with or treated for a life-threatening disease. Clinicians may be guided by this information, opting for treatments with the smallest negative impact on QoL and providing additional support to those patients who need it.
Supporting Information
(XLS)
(DOCX)
(DOCX)
[31]. Note: QL = global health/quality of life; PF = physical functioning; RF = role functioning; CF = cognitive functioning; EF = emotional functioning; SF = social functioning; PA = pain; FA = fatigue; NV = nausea and vomiting; SL = insomnia; DY = dyspnoea; AP = appetite loss; CO = constipation; DI = diarrhea; FI = financial difficulties *p<0.01. For example, KARMA women reported better physical functioning, yet more sleeping problems than the Swedish reference sample.[31]
(TIF)
Note: The Bonferonni corrected value is—log10(3.76E-06) = 5.42.This Manhattan plot was prepared using Haploview.[35]
(TIF)
Note: This figure is a print shot of the haploreg database, see http://www.broadinstitute.org/mammals/haploreg/haploreg.php.[39]
(TIF)
Note: This figure is a print shot of the haploreg database, see http://www.broadinstitute.org/mammals/haploreg/haploreg.php.[39]
(TIF)
Funding Statement
The authors have no funding or support to report.
References
- 1. Sprangers MA (2002) Quality-of-life assessment in oncology. Achievements and challenges. Acta Oncol 41: 229–237. [DOI] [PubMed] [Google Scholar]
- 2. Benyamini Y, Idler EL (1999) Community studies reporting association between self-rated health and mortality—Additional studies, 1995 to 1998. Research on Aging 21: 392–401. [Google Scholar]
- 3. Farkas J, Nabb S, Zaletel-Kragelj L, Cleland JG, Lainscak M (2009) Self-rated health and mortality in patients with chronic heart failure. Eur J Heart Fail 11: 518–524. 10.1093/eurjhf/hfp038 [DOI] [PubMed] [Google Scholar]
- 4. Mathews WC, May S (2007) EuroQol (EQ-5D) measure of quality of life predicts mortality, emergency department utilization, and hospital discharge rates in HIV-infected adults under care. Health Qual Life Outcomes 5: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kullowatz A, Kanniess F, Dahme B, Magnussen H, Ritz T (2007) Association of depression and anxiety with health care use and quality of life in asthma patients. Respir Med 101: 638–644. [DOI] [PubMed] [Google Scholar]
- 6. Pelle AJ, Pedersen SS, Szabo BM, Denollet J (2009) Beyond Type D personality: reduced positive affect (anhedonia) predicts impaired health status in chronic heart failure. Qual Life Res 18: 689–698. 10.1007/s11136-009-9485-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wald A, Sigurdsson L (2011) Quality of life in children and adults with constipation. Best Pract Res Clin Gastroenterol 25: 19–27. 10.1016/j.bpg.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 8. Schoormans D, Mulder BJ, van Melle JP, Pieper PG, van Dijk AP et al. (2014) Illness perceptions of adults with congenital heart disease and their predictive value for quality of life two years later. Eur J Cardiovasc Nurs 13: 86–94. 10.1177/1474515113481908 [DOI] [PubMed] [Google Scholar]
- 9. Sprangers MA, Sloan JA, Veenhoven R, Cleeland CS, Halyard MY et al. (2009) The establishment of the GENEQOL consortium to investigate the genetic disposition of patient-reported quality-of-life outcomes. Twin Res Hum Genet 12: 301–311. 10.1375/twin.12.3.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barsevick A, Frost M, Zwinderman A, Hall P, Halyard M (2010) I’m so tired: biological and genetic mechanisms of cancer-related fatigue. Qual Life Res 19: 1419–1427. 10.1007/s11136-010-9757-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bower JE, Ganz PA, Irwin MR, Castellon S, Arevalo J et al. (2013) Cytokine genetic variations and fatigue among patients with breast cancer. J Clin Oncol 31: 1656–1661. 10.1200/JCO.2012.46.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Illi J, Miaskowski C, Cooper B, Levine JD, Dunn L et al. (2012) Association between pro- and anti-inflammatory cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression. Cytokine 58: 437–447. 10.1016/j.cyto.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kao CF, Jia P, Zhao Z, Kuo PH (2012) Enriched pathways for major depressive disorder identified from a genome-wide association study. Int J Neuropsychopharmacol 15: 1401–1411. 10.1017/S1461145711001891 [DOI] [PubMed] [Google Scholar]
- 14. Shi Q, Cleeland CS, Klepstad P, Miaskowski C, Pedersen NL (2010) Biological pathways and genetic variables involved in pain. Qual Life Res 19: 1407–1417. 10.1007/s11136-010-9738-x [DOI] [PubMed] [Google Scholar]
- 15. Nes RB, Roysamb E, Tambs K, Harris JR, Reichborn-Kjennerud T (2006) Subjective well-being: genetic and environmental contributions to stability and change. Psychol Med 36: 1033–1042. [DOI] [PubMed] [Google Scholar]
- 16. Hettema JM, Neale MC, Kendler KS (2001) A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry 158: 1568–1578. [DOI] [PubMed] [Google Scholar]
- 17. Sullivan PF, Neale MC, Kendler KS (2000) Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 157: 1552–1562. [DOI] [PubMed] [Google Scholar]
- 18. Ask H, Torgersen S, Seglem KB, Waaktaar T (2014) Genetic and environmental causes of variation in adolescent anxiety symptoms: A multiple-rater twin study. J Anxiety Disord 28: 363–371. 10.1016/j.janxdis.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 19. Jang KL, Livesley WJ, Taylor S, Stein MB, Moon EC (2004) Heritability of individual depressive symptoms. J Affect Disord 80: 125–133. [DOI] [PubMed] [Google Scholar]
- 20. Sprangers MA, Sloan JA, Barsevick A, Chauhan C, Dueck AC et al. (2010) Scientific imperatives, clinical implications, and theoretical underpinnings for the investigation of the relationship between genetic variables and patient-reported quality-of-life outcomes. Qual Life Res 19: 1395–1403. 10.1007/s11136-010-9759-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rausch SM, Clark MM, Patten C, Liu H, Felten S et al. (2010) Relationship between cytokine gene single nucleotide polymorphisms and symptom burden and quality of life in lung cancer survivors. Cancer 116: 4103–4113. 10.1002/cncr.25255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schoormans D, Radonic T, de Witte P, Groenink M, Azim D et al. (2012) Mental quality of life is related to a cytokine genetic pathway. PLoS One 7: e45126 10.1371/journal.pone.0045126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sloan JA, de Andrade M, Decker P, Wampfler J, Oswold C et al. (2012) Genetic variations and patient-reported quality of life among patients with lung cancer. J Clin Oncol 30: 1699–1704. 10.1200/JCO.2010.34.5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sprangers MA, Thong MS, Bartels M, Barsevick A, Ordonana J et al. (2014) Biological pathways, candidate genes and molecular markers associated with quality-of-life domains: an update. Qual Life Res 23: 1997–2013. 10.1007/s11136-014-0656-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rickham PP (1964) Human experimentation. Code of ethics of the World Medical Association. Declaration of Helsinki. Br Med J 2: 177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aaronson NK, Ahmedzai S, Bergman B, Bullinger M, Cull A et al. (1993) The European Organization for Research and Treatment of Cancer QLQ-C30: a quality-of-life instrument for use in international clinical trials in oncology. J Natl Cancer Inst 85: 365–376. [DOI] [PubMed] [Google Scholar]
- 27. Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK et al. (2013) Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet 45: 392–398. 10.1038/ng.2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J et al. (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45: 353–361. 10.1038/ng.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO et al. (2010) SCAN: SNP and copy number annotation. Bioinformatics 26: 259–262. 10.1093/bioinformatics/btp644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michelson H, Bolund C, Nilsson B, Brandberg Y (2000) Health-related quality of life measured by the EORTC QLQ-C30—reference values from a large sample of Swedish population. Acta Oncol 39: 477–484. [DOI] [PubMed] [Google Scholar]
- 32. Humphreys K, Grankvist A, Leu M, Hall P, Liu J et al. (2011) The genetic structure of the Swedish population. PLoS One 6: e22547 10.1371/journal.pone.0022547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR et al. (2010) A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87: 139–145. 10.1016/j.ajhg.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 36. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K et al. (2013) Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet 92: 489–503. 10.1016/j.ajhg.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang K, Chang S, Cui S, Guo L, Zhang L et al. (2011) ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework. Nucleic Acids Res 39: W437–W443. 10.1093/nar/gkr391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–D934. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. ENCODE project consotrium (2011) A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chadwick LH (2012) The NIH Roadmap Epigenomics Program data resource. Epigenomics 4: 317–324. 10.2217/epi.12.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Canderelli R, Leccesse LA, Miller NL, Unruh DJ (2007) Benefits of hormone replacement therapy in postmenopausal women. J Am Acad Nurse Pract 19: 635–641. [DOI] [PubMed] [Google Scholar]
- 43. Coker TR, Elliott MN, Wallander JL, Cuccaro P, Grunbaum JA et al. (2011) Association of family stressful life-change events and health-related quality of life in fifth-grade children. Arch Pediatr Adolesc Med 165: 354–359. 10.1001/archpediatrics.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michelson H, Bolund C, Brandberg Y (2000) Multiple chronic health problems are negatively associated with health related quality of life (HRQoL) irrespective of age. Qual Life Res 9: 1093–1104. [DOI] [PubMed] [Google Scholar]
- 45. Schlarb AA, Reis D, Schroder A (2012) Sleep Characteristics, Sleep Problems, and Associations to Quality of Life among Psychotherapists. Sleep Disord 2012: 806913 10.1155/2012/806913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blackburn AC, Woollatt E, Sutherland GR, Board PG (1998) Characterization and chromosome location of the gene GSTZ1 encoding the human Zeta class glutathione transferase and maleylacetoacetate isomerase. Cytogenet Cell Genet 83: 109–114. [DOI] [PubMed] [Google Scholar]
- 47. Tanguay RM, Lambert M, Grompe M, Mitchell GA (2013) Chapter 79: Hypertyrosinemia In: Scriver S, Beaudet AL, Valle D, Sly WS, Vogelstein B et al. , editors. The Online Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill. [Google Scholar]
- 48. Lieberman HR (2003) Nutrition, brain function and cognitive performance. Appetite 40: 245–254. [DOI] [PubMed] [Google Scholar]
- 49. Bjorklund A, Dunnett SB (2007) Dopamine neuron systems in the brain: an update. Trends Neurosci 30: 194–202. [DOI] [PubMed] [Google Scholar]
- 50. Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM et al. (2001) Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A 98: 6917–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Starr JM, Quinn C (2008) GSTZ1 genotype and cognitive ability. European Neurological Review 3: 15–17. [Google Scholar]
- 52. Starr JM, Fox H, Harris SE, Deary IJ, Whalley LJ (2008) GSTz1 genotype and cognitive ability. Psychiatr Genet 18: 211–212. 10.1097/YPG.0b013e328304dea8 [DOI] [PubMed] [Google Scholar]
- 53. Buchanan CC, Torstenson ES, Bush WS, Ritchie MD (2012) A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data. J Am Med Inform Assoc 19: 289–294. 10.1136/amiajnl-2011-000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ordonana JR, Bartels M, Boomsma DI, Cella D, Mosing M et al. (2013) Biological pathways and genetic mechanisms involved in social functioning. Qual Life Res 22: 1189–1200. 10.1007/s11136-012-0277-5 [DOI] [PubMed] [Google Scholar]
- 55. Harris SE, Fox H, Wright AF, Hayward C, Starr JM et al. (2007) A genetic association analysis of cognitive ability and cognitive ageing using 325 markers for 109 genes associated with oxidative stress or cognition. BMC Genet 8: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(DOCX)
(DOCX)
[31]. Note: QL = global health/quality of life; PF = physical functioning; RF = role functioning; CF = cognitive functioning; EF = emotional functioning; SF = social functioning; PA = pain; FA = fatigue; NV = nausea and vomiting; SL = insomnia; DY = dyspnoea; AP = appetite loss; CO = constipation; DI = diarrhea; FI = financial difficulties *p<0.01. For example, KARMA women reported better physical functioning, yet more sleeping problems than the Swedish reference sample.[31]
(TIF)
Note: The Bonferonni corrected value is—log10(3.76E-06) = 5.42.This Manhattan plot was prepared using Haploview.[35]
(TIF)
Note: This figure is a print shot of the haploreg database, see http://www.broadinstitute.org/mammals/haploreg/haploreg.php.[39]
(TIF)
Note: This figure is a print shot of the haploreg database, see http://www.broadinstitute.org/mammals/haploreg/haploreg.php.[39]
(TIF)