Abstract
Background. Despite effective antiretroviral therapy (ART), patients with chronic human immunodeficiency virus (HIV) infection have increased microbial translocation and systemic inflammation. Alterations in the intestinal microbiota may play a role in microbial translocation and inflammation.
Methods. We profiled the fecal microbiota by pyrosequencing the gene encoding 16S ribosomal RNA (rRNA) and measured markers of microbial translocation and systemic inflammation in 21 patients who had chronic HIV infection and were receiving suppressive ART (cases) and 16 HIV-uninfected controls.
Results. The fecal microbial community composition was significantly different between cases and controls. The relative abundance of Proteobacteria, Gammaproteobacteria, Enterobacteriales, Enterobacteriaceae, Erysipelotrichi, Erysipelotrichales, Erysipelotrichaceae, and Barnesiella was significantly enriched in cases, whereas that of Rikenellaceae and Alistipes was depleted. The plasma soluble CD14 level (sCD14) was significantly higher and the endotoxin core immunoglobulin M (IgM) level lower in cases, compared with controls. There were significant positive correlations between the relative abundances of Enterobacteriales and Enterobacteriaceae and the sCD14 level; the relative abundances of Gammaproteobacteria, Enterobacteriales, and Enterobacteriaceae and the interleukin 1β (IL-1β) level; the relative abundances of Enterobacteriales and Enterobacteriaceae and the interferon γ level; and the relative abundances of Erysipelotrichi and Barnesiella and the TNF-α level. There were negative correlations between endotoxin core IgM and IL-1β levels.
Conclusions. Patients who have chronic HIV infection and are receiving suppressive ART display intestinal dysbiosis associated with increased microbial translocation and significant associations between specific taxa and markers of microbial translocation and systemic inflammation. This was an exploratory study, the findings of which need to be confirmed.
Keywords: HIV, microbiota, dysbiosis, inflammation, microbial translocation
In the current era of effective antiretroviral therapy (ART), human immunodeficiency virus (HIV) infection has become a chronic manageable disease. However, noninfectious complications such as cardiovascular disease, diabetes, metabolic syndrome, obesity, and accelerated aging, all associated with chronic inflammation, are being reported with increased frequency in these patients [1–3]. Although ART is effective at suppressing viral replication to below the limits of detection in blood, immune activation persists and is associated with HIV disease progression [4]. This ongoing immune activation and associated inflammation may also contribute to noninfectious complications in patients with chronic HIV infection who are receiving suppressive ART.
The mechanisms underlying immune activation in these patients have been attributed to persistent viral replication in the gut, residual immune dysregulation, or microbial translocation [5–9]. Increased microbial translocation, immune activation, and inflammation have been reported in HIV-infected individuals, compared with HIV-uninfected individuals [10–13]. Microbial translocation in these patients is thought to be associated with loss of mucosal barrier function and increased intestinal permeability secondary to immune dysregulation and/or alterations in the intestinal microbiome.
The gut microbiome is critical for maintaining intestinal homeostasis and plays vital roles in maintenance of mucosal barrier function and regulation of innate and adaptive immune responses [14]. Dysbiosis (an imbalance in the composition of the microbiota) has been implicated in chronic inflammation associated with conditions such as obesity, diabetes, and inflammatory bowel disease [7, 15–21]. Previous studies have reported alterations in the intestinal microbiota in HIV infection, with or without associated microbial translocation, immune activation, and inflammation [22–27]. However, few studies have addressed the role of the intestinal microbiota in microbial translocation and inflammation in patients with chronic HIV infection who are receiving suppressive ART.
To determine whether individuals with chronic HIV infection who are receiving suppressive ART display dysbiosis and whether there is an association between dysbiosis, microbial translocation, and systemic inflammation in these individuals, we profiled the intestinal microbiota and measured plasma markers of microbial translocation and systemic inflammation in an exploratory study of 21 patients who had chronic HIV infection, were receiving ART, and had an undetectable plasma HIV RNA level (cases) and 16 HIV-uninfected controls.
METHODS
Study Design, Subjects, and Data and Sample Collection
Patients with chronic HIV infection, were receiving suppressive ART, and had an undetectable plasma HIV RNA level (cases) and HIV-uninfected subjects (controls) were recruited from ongoing studies of cardiovascular risk during HIV/AIDS at Tufts University School of Medicine [28, 29] and from the Infectious Diseases Clinic at Tufts Medical Center by flyers at local community centers or by advertisement in the greater Boston area. All subjects were from eastern Massachusetts. Inclusion criteria for cases included documented HIV infection, current ART use, and a plasma HIV RNA level below the limit of detection. Inclusion criteria for controls included documented lack of HIV infection. Exclusion criteria for all subjects were based on factors known or likely to impact the intestinal microbiota or known to be associated with microbial translocation: age >60 years, body mass index (BMI; defined as the weight in kilograms divided by the height in meters squares) of >30, antibiotic or probiotic use within the previous 4 weeks, gastrointestinal morbidity (including irritable bowel syndrome, inflammatory bowel disease, history of gastrointestinal cancer or surgical resection, or acute, severe gastrointestinal symptoms requiring medical attention), opportunistic infection, and evidence of hepatitis B or C virus infection. A total of 21 cases and 16 controls were enrolled in the study. Sociodemographic data and past medical history were obtained at enrollment. Recreational drug, alcohol, and tobacco use and behavioral or lifestyle data were obtained via an audio computer assisted self-interview survey (http://acasi.tufts.edu) [30].
The study was approved by the Tufts Health Sciences Institutional Review Board, and informed consent was obtained from all participants.
Stool DNA Extraction, 16S rRNA Gene Amplicon Generation, and Pyrosequencing
Stool samples were collected and stored and DNA extracted as described elsewhere [27]. The V3-5 region of the gene encoding 16S ribosomal RNA (rRNA) was amplified by polymerase chain reaction (PCR). Amplicons were pooled in equimolar concentrations, purified, and sequenced on a Roche 454 Genome Sequencer GS FLX+ at the Tufts University Core facility Genomics Core as previously described [27].
Computational analyses were performed using QIIME, version 1.6 (http://qiime.org) [31]. Noise was removed from sequences, using Chimera Slayer (version 2010-12-12). Similar sequences were clustered into operational taxonomic units based on a minimum identity of 97%, using UClust [32]. The most frequent sequence within each operational taxonomic unit was used for alignment, using PyNAST [33] with Greengenes (http://greengenes.secondgenome.com/), and assigned to the lowest possible taxonomic level, using the Ribosomal Database Project Classifier (http://rdp.cme.msu.edu) [34]. The number of sequences was normalized, and alpha diversity measures, including equitability, number of observed species, Shannon diversity index, Chao-1, and PD (phylogenetic diversity), were determined. Sequences were submitted to the Sequence Read Archive in GenBank. The accession number for the project is SRP039076.
Markers of Microbial Translocation and Systemic Inflammation
Blood samples were collected, processed, and stored as described elsewhere [27]. Plasma levels of endotoxin core immunoglobulin M (IgM; EndoCAb IgM) and soluble CD14 (sCD14) were measured using enzyme-linked immunosorbent assay kits from Hycult (Uden, the Netherlands) and R&D (Minneapolis, MN), respectively. Plasma lipopolysaccharide (LPS) levels were measured as described elsewhere [35], using a Limulus amebocyte lysate assay (Associates of Cape Cod, East Falmouth, MA). 16S rRNA gene levels in plasma were measured by quantitative real-time PCR as described elsewhere [35]. Plasma levels of interferon γ (IFN-γ), interleukin 1β (IL-1β), interleukin 6 (IL-6), and tumor necrosis factor α (TNF-α) were measured using the Multi-Array system (Meso Scale Discovery, Rockville, MD).
Statistical Analyses
Statistical analyses were performed in GraphPad Prism, version 6.0 (La Jolla, CA) and in the R programming environment (http://www.r-project.org/), using the Mann–Whitney test for continuous variables, the Fisher exact test for dichotomous variables, and the Spearman test for correlations. Beta diversity was assessed by nonmetric dimensional scaling with the R package ecodist, using a Canberra dissimilarity matrix. The R package ellipse was used to generate 95% confidence intervals, and differences were determined using Adonis with 1000 permutations in the R package vegan. Median regression analysis was performed using the R package quantreg. Univariate analysis was performed first for all variables, using HIV status and age as sole predictors. The association of those variables at a P value of ≤.2 (Table 1) and/or those that may have an effect on the intestinal microbiota, microbial translocation, or inflammation was assessed in multivariate analysis. Given the number of variables and the small sample size, multivariate analyses with >3 predictors (HIV status, age, and 1 other parameter) were not performed, to avoid model instability. The standard errors (and P values) for each model were calculated using bootstrapping with R equal to 15 000. Linear discriminant analysis effect size (LEfSe) analysis was performed according to the methods of Segata et al, using default parameters (http://huttenhower.sph.harvard.edu/galaxy/) [36]. Since this was an exploratory study, corrections for multiple testing for other parameters were not performed [37].
Table 1.
Sociodemographic Characteristics of Human Immunodeficiency Virus (HIV)-Infected Subjects Who Were Receiving Suppressive Antiretroviral Therapy and Had Undetectable Plasma HIV RNA (Cases) and HIV-Uninfected Subjects (Controls)
| Characteristic | Cases (n = 21) | Controls (n = 16) | P Valuea |
|---|---|---|---|
| Age, y | 50.6 (45.5–54.5) | 45.6 (34.4–50.3) | .04b |
| Male sex | 17 (80.9) | 12 (75) | .70 |
| High school education | 8 (38.1) | 4 (25) | .49 |
| Post–high school education | 11 (52.4) | 11 (68.8) | .50 |
| Employed | 6 (28.6) | 9 (56.3) | .11 |
| Homeless | 2 (9.5) | 5 (31.3) | .20 |
| BMIc | 25 (22.6–27.7) | 27.1 (24.2–28.9) | .20b |
| Alcohol consumptiond | 4 (19) | 1 (6.3) | .36 |
| Recreational drug usee | 10 (47.6) | 3 (18.8) | .09 |
| Current smoker | 14 (66.7) | 7 (43.8) | .20 |
Data are median (interquartile range) or no. (%) of subjects. P values of <.05 are considered statistically significant.
a By the Fisher exact test, unless otherwise indicated.
b By the Mann–Whitney test.
c Body mass index (BMI) is calculated as the weight in kilograms divided by the height in meters squared.
d In the past month.
e Defined as use of ≥1 of the following: heroin, cocaine, marijuana, sedatives, and/or poppers.
RESULTS
Sociodemographic Characteristics
Cases were significantly older than controls (P = .04; Table 1). There were no significant differences in other parameters between cases and controls. The HIV infection status of the cases is shown in Table 2. The median duration of HIV infection was 15.9 years (interquartile range [IQR], 10.2–20.4 years) from first diagnosis, indicating the chronic nature of the disease. The median duration of ART was 13.3 years (IQR, 5.8–15.1 years) with protease inhibitors and/or nonnucleoside reverse transcriptase inhibitors. The median CD4+ T-cell count was 668 cells/mm3 (IQR, 424–870 cells/mm3), and HIV RNA was below the level of detection (<400 copies/mL), confirming effective suppression of HIV infection by ART in all cases.
Table 2.
Characteristics of Human Immunodeficiency Virus (HIV) Infection Among Subjects Who Were Receiving Suppressive Antiretroviral Therapy (ART) and Had Undetectable Plasma HIV RNA (Cases)
| Parameter | Value | Cases, no. |
|---|---|---|
| HIV infection duration, y | 15.9 (9.48–19.8) | 21 |
| CD4+ T-cell count, cells/mm3 | 668 (424–870) | 21 |
| HIV RNA load, copies/mL | <400a | 21 |
| ART duration, y | 12.6 (5.14–15.1) | 17b |
| ART component(s) | ||
| PI | … | 8 |
| NNRTI | … | 11 |
| PI plus NNRTI | … | 2 |
Data are median (interquartile range), unless otherwise indicated.
Abbreviations: NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
a Considered undetectable.
b Unknown for 4 subjects.
Intestinal Microbiota
Pyrosequencing of 16S rRNA gene amplicons from fecal DNA yielded 122 470 sequences with an average of 3310 sequences per sample and 29 782 operational taxonomic units in all samples.
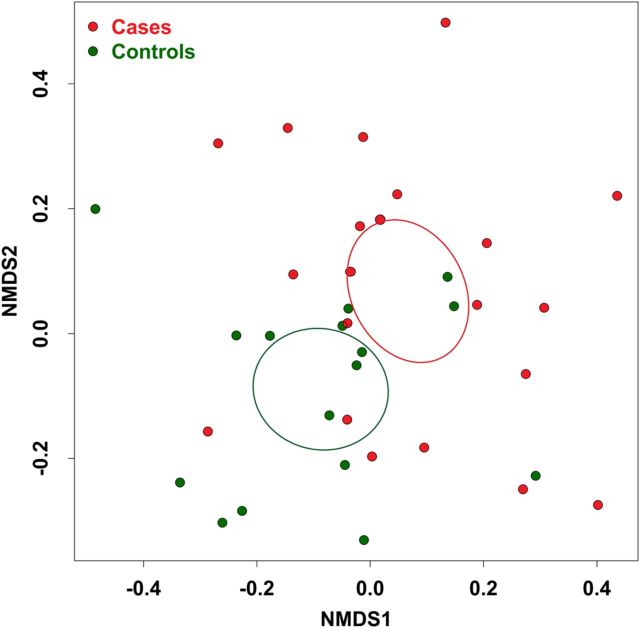
There were no significant differences in alpha diversity between cases and controls (Supplementary Table 1). Beta diversity analysis, performed using nonmetric dimensional scaling with a Canberra community dissimilarity matrix, revealed significant clustering of cases and controls (Adonis, P < .05; Figure 1). In both groups, the 2 dominant phyla were Bacteroidetes and Firmicutes, with the exception of 1 case, who had an unusually high relative abundance of fusobacteria (Figure 2). The relative abundance of Proteobacteria was significantly greater in cases than controls in univariate analysis (Supplementary Table 2). This difference remained significant in multivariate analysis when controlling for age, BMI, smoking status, alcohol use, or recreational drug use. There were no significant differences in the relative abundance of other phyla between cases and controls.
Figure 1.
Stool microbial community composition in cases, compared with that in controls. Community composition dissimilarity was analyzed using nonmetric dimensional scaling (NMDS) based on a Canberra community dissimilarity matrix. Each dot represents the microbiota of a single subject. Ellipses represent 95% confidence intervals for the standard error of weighted NDMS score means of cases and controls. Community differences were verified using Adonis (P = <.05).
Figure 2.
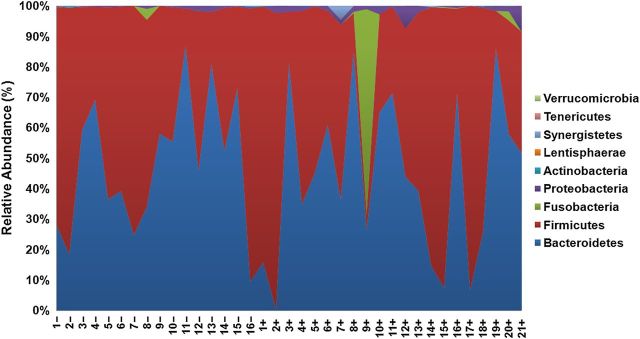
Relative abundance of major phyla in cases (+) and controls (−). In both groups, the 2 dominant phyla were Bacteroidetes and Firmicutes, which made up >95% of each individual's microbiota, with the exception of case 9, who had an unusually high relative abundance of fusobacteria in lieu of Firmicutes.
To identify differentially abundant taxa in cases and controls, we used the LEfSe algorithm, which identifies genomic features characterizing the differences between ≥2 biological conditions [36]. It emphasizes statistical significance, biological consistency, and effect relevance. Since there was a significant difference in age between cases and controls, we included age (above and below the median) as a subclass. LEfSe analysis confirmed enrichment in cases of Proteobacteria and revealed enrichment of Gammaproteobacteria, Enterobacteriales, and Enterobacteriaceae, enrichment of Erysipelotrichi, Erysipelotrichales, and Erysipelotrichaceae of the phylum Firmicutes, and enrichment of the genus Barnesiella in the phylum Bacteroidetes in cases, whereas Rikenellaceae and Alistipes in the phylum Bacteroidetes were enriched in controls (Figure 3A and 3B). Standard statistical analysis (using the Mann–Whitney test) performed on the relative abundance of each taxon that was identified by LEfSe as being differentially abundant confirmed that the relative abundance of all these taxa were significantly different between cases and controls (Figure 3C).
Figure 3.
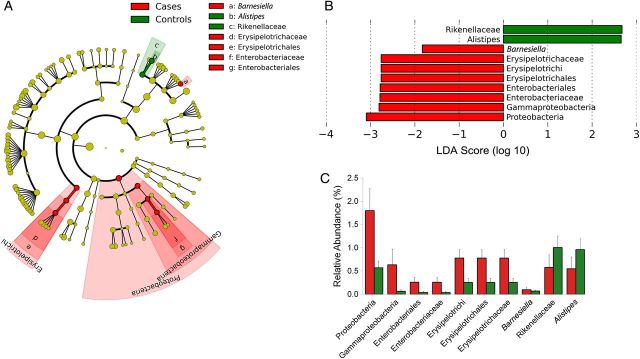
Linear discriminant analysis (LDA) effect size (LEfSe) detects specific differentially abundant taxa in cases and controls. A, Cladogram showing differentially abundant taxonomic clades with an LDA score >2.0 among cases and controls. B, LDA scores of differentially abundant taxa among cases and controls. The LDA score indicates the effect size and ranking of each differentially abundant taxon. C, The relative abundance of taxa identified by LEfSe as being differentially abundant in cases and controls was compared using the Mann–Whitney test: Proteobacteria, P = <.05; Gammaproteobacteria, P = .04; Enterobacteriales, P = <.05; Enterobacteriaceae, P = <.05; Erysipelotrichi, P = .01; Erysipelotrichales, P = .01; Erysipelotrichaceae, P = .01; Barnesiella, P = .01; Alistipes, P = .01; and Rikenellaceae, P = .01.
Markers of Microbial Translocation and Systematic Inflammation
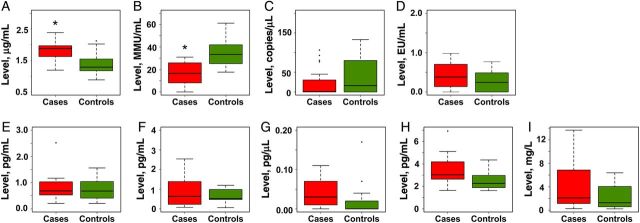
We used 4 biomarkers to assess microbial translocation. Three are markers of translocation of gram-negative bacteria: LPS (or endotoxin); the soluble form of the LPS coreceptor CD14 (sCD14), which is upregulated in response to LPS stimulation and is a marker of monocyte activation [38]; and IgM levels to the LPS core antigen (EndoCAb), which decrease following LPS binding [38–42]. The fourth, bacterial 16S rRNA gene copy number, is a marker of translocation of both gram-negative and gram-positive bacteria [43, 44]. In univariate analysis, plasma levels of sCD14 were significantly higher (P < .01), and, correspondingly, levels of EndoCAb were significantly lower (P < .01) in cases, compared with controls (Supplementary Table 3). This difference remained significant in multivariate median regression analysis after adjustment for possible confounding factors (Figure 4). There was a trend toward significantly higher levels of plasma LPS (P = .08) in cases, compared with controls (data not shown). However, there was no significant difference in plasma 16S rRNA gene copy numbers between the groups (Figure 4).
Figure 4.
Markers of microbial translocation and systemic inflammation in cases and controls. Levels of markers of microbial translocation (soluble CD14 [sCD14], endotoxin core antibody [EndoCAb], 16S ribosomal RNA [rRNA] gene, and lipopolysaccharide [LPS]) and systemic inflammation (interleukin 6 [IL-6], interferon γ [IFN-γ], interleukin 1β [IL-1β], tumor necrosis factor α [TNF-α], and high-sensitivity C-reactive protein [hsCRP]) were measured in cases and controls, as described in “Methods” section, and compared using the Mann–Whitney test. *P = <.01.
We assayed 5 markers of systemic inflammation: proinflammatory cytokines IL-6, IFN-γ, IL-1β, and TNF-α, as well as a nonspecific marker of inflammation, high-sensitivity C-reactive protein (hsCRP). Plasma IL-1β and TNF-α levels were significantly increased in cases (P = .02 and .03, respectively), compared with controls, in univariate analysis (data not shown). However, these differences were not significant in multivariate analysis. There were no significant differences in IL-6, IFN-γ, and hsCRP levels between the 2 groups (Figure 4).
Correlations Between Dysbiosis, Microbial Translocation, and Systemic Inflammation
We found statistically significant positive correlations between LPS and IL-6 levels, LPS and TNF-α levels, and LPS and hsCRP levels (P < .01, P < .01, P = .01, respectively; data not shown), as well as between sCD14 and IL-1β levels (P = .03; data not shown).
Since the microbiota of cases was significantly enriched or depleted in specific taxa, we examined possible correlations between the relative abundance of these taxa and markers of microbial translocation and systemic inflammation. There were significant positive correlations between the relative abundance of both Enterobacteriales and Enterobacteriaceae and sCD14 levels (P < .01 for both; Table 3). There were also significant positive correlations between the relative abundance of Gammaproteobacteria, Enterobacteriales, and Enterobacteriaceae and IL-1β levels (P < .01 for all). The relative abundance of Enterobacteriales and Enterobacteriaceae were also significantly positively correlated with IFN-γ levels (P = <.05 for both). The relative abundance of Erysipelotrichi correlated negatively with EndoCAb levels (P = .04) and positively with IL-1β levels (P = .03). Finally, there was a significant positive correlation between the relative abundance of Barnesiella and TNF-α levels (P = .01). The reasons why the correlations were identical for taxa in the same clade but at different levels is because there is only 1 family in that order or because taxa in that order are driving the correlation.
Table 3.
Correlation Between Relative Abundance of Specific Taxa and Markers of Microbial Translocation and Systemic Inflammation Among Human Immunodeficiency Virus (HIV)–Infected Subjects Who Were Receiving Suppressive Antiretroviral Therapy and Had Undetectable Plasma HIV RNA
| Marker | Taxon | Category | Spearman R | P Value |
|---|---|---|---|---|
| Microbial translocation | ||||
| EndoCAb | Erysipelotrichaceae | Family | −0.35 | .04 |
| EndoCAb | Erysipelotrichi | Class | −0.35 | .04 |
| sCD14 | Enterobacteriaceae | Family | 0.49 | <.01 |
| sCD14 | Enterobacteriales | Order | 0.49 | <.01 |
| Systemic inflammation | ||||
| IL-1β | Enterobacteriaceae | Family | 0.54 | <.01 |
| IL-1β | Enterobacteriales | Order | 0.54 | <.01 |
| IL-1β | Gammaproteobacteria | Class | 0.47 | <.01 |
| IL-1β | Erysipelotrichaceae | Family | 0.36 | .03 |
| IL-1β | Erysipelotrichi | Class | 0.36 | .03 |
| IFN-γ | Enterobacteriaceae | Family | 0.34 | <.05 |
| IFN-γ | Enterobacteriales | Order | 0.34 | <.05 |
| TNF-α | Barnesiella | Genus | −0.42 | .01 |
P values of <.05 are considered statistically significant.
Abbreviations: EndoCAb, endotoxin core antibody; IFN-γ, interferon γ; IL-1β, interleukin 1β; sCD14, soluble CD14; TNF-α, tumor necrosis factor α.
Since there were no significant differences in LPS levels or 16S rRNA copy numbers between cases and controls, and because sCD14 could also be considered a marker of inflammation, the relationship of these observations to microbial translocation may be questioned. However, the finding of a significant difference in EndoCAb levels (which are directly related to increased LPS levels) in addition to differences in sCD14 levels supports the association with microbial translocation.
DISCUSSION
In this study, we found dysbiosis characterized by enrichment or depletion of specific taxa and increased biomarkers of microbial translocation in subjects with chronic HIV infection who were receiving suppressive ART and had undetectable HIV loads (cases), compared with HIV-uninfected controls. In addition, we found significant correlations between biomarkers of microbial translocation and systemic inflammation and the relative abundance of specific bacterial taxa in cases.
Within the past year, 4 studies comparing the gut microbiota of HIV-infected subjects to that of HIV-uninfected subjects (using microarray or next-generation sequencing technology) were published. Vujkovic-Cvijin et al compared the rectal mucosa-associated microbiota of HIV-infected and uninfected men [25]. Lozupone et al compared the stool microbiota of untreated, recently HIV-infected subjects, or chronically HIV-infected subjects receiving short-term or long-term ART to that of non–HIV-infected controls [45]. Dillon et al compared the fecal and colonic mucosal microbiota of subjects with chronic untreated HIV infection with that of HIV-uninfected subjects [46]. Mutlu et al compared the ileal and colonic mucosal microbiota of HIV-infected subjects who were receiving highly active ART (HAART) to that of HIV-uninfected subjects [47]. We compared the fecal microbiota of subjects with chronic HIV infection who were receiving suppressive ART to that of uninfected controls. These studies differed in the sex, duration, and treatment status of the HIV-infected subjects, matching status of the HIV-uninfected controls, the nature of the samples studied, the technology used to characterize the microbiota, and the computational and statistical methods used to identify differences in microbial community composition and differentially abundant taxa. Although these may account for some of the differences found among these studies, all the studies, including ours, found significant differences in the community composition of the microbiota assessed by beta diversity measures in HIV-infected subjects, compared with uninfected subjects.
The differences among the studies appear to be in the microbiota profiles of treated or untreated subjects with chronic HIV infection, compared with uninfected controls. Thus, Vujkovic-Cvijin et al found that the community composition of subjects with virological suppression who were receiving HAART varied considerably, with the composition for some patients more similar to that of the viremic untreated group, whereas the composition for others was more similar to that of the HIV-uninfected group [25]. Similarly, Lozupone et al reported that the microbiota of some of the individuals with chronic HIV infection who were receiving ART clustered with that of HIV-uninfected subjects, whereas the microbiota for others clustered with that of untreated individuals with chronic HIV infection [45]. Dillon et al showed a difference in community composition between untreated HIV-infected subjects, compared with HIV-uninfected subjects [46]. Mutlu et al found differences between HIV-infected subjects who were receiving HAART and uninfected controls [47]. However, the CD4+ T-cell counts and viral loads of the HIV-infected subjects on ART varied. [47]. Our study (in which all HIV-infected subjects had virological suppression) found significant differences in the microbial community composition of subjects with chronic HIV infection who were receiving ART, compared with HIV-uninfected subjects.
In our study, as in that by Vujkovic-Cvijin et al [25] and Dillon et al [46] there were no significant differences in alpha diversity measures between subjects with chronic HIV infection who were receiving ART, compared with HIV-uninfected subjects. Surprisingly, however, Lozupone et al [45] found increased alpha diversity in subjects with untreated chronic HIV infection, compared with HIV-uninfected subjects or treated subjects with chronic HIV infection, whereas Mutlu et al reported decreased alpha diversity in the HIV-infected subjects [47].
Intestinal dysbiosis in our study was characterized by significant differences in taxa in 3 of the major phyla in cases, compared with controls. The first was a significant enrichment in the Proteobacteria phylum, specifically of taxa in the Gammaproteobacteria class, including Enterobacteriales and Enterobacteriaceae, in cases. Of interest, the Enterobacteriaceae family includes inflammogenic enteric pathogens, which may be relevant to the increased levels of the proinflammatory cytokine IFN-γ, which were seen in cases.
In a recent study of the stool microbiota in HIV-infected cocaine users, we also found a significant increase in the relative abundance of Proteobacteria in HIV-infected subjects, compared with uninfected subjects [27]. Vujkovic-Cvijin et al found that taxa in the Proteobacteria phylum, particularly Enterobacteriaceae, were significantly enriched in the viremic untreated group, compared with HIV-uninfected controls. However, no differences in the relative abundance of Proteobacteria or Enterobacteriaceae between HIV-uninfected controls and subjects receiving HAART were reported [25]. Dillon et al found that the relative abundance of Proteobacteria was significantly higher in untreated subjects with chronic HIV infection, compared with uninfected subjects [46]. Mutlu et al reported enrichment of Enterobacteriaceae in HIV-infected subjects [47].
We also found a significant enrichment of taxa in the Erysipelotrichaceae family in cases. Taxa in this family were the most enriched in the untreated HIV-infected subjects in the study by Vujkovic-Cvijin et al [25]. Lozupone et al also reported enrichment in the Erysipelotrichaceae family in untreated subjects with chronic HIV infection, compared with HIV-uninfected individuals [45].
A third differentially abundant taxon in cases was Barnesiella in the phylum Bacteroidetes. Interestingly, Dillon et al found that the relative abundance of Barnesiella was lower in HIV-infected subjects, compared with uninfected subjects [46], and Mutlu et al reported enrichment of Barnesiellaceae in HIV-negative controls [47].
We found enrichment of the Alistipes genus and Rikenellaceae family in the controls. Vujkovic-Cvijin et al also found that these taxa were depleted in viremic untreated men, compared with uninfected men [25]. These taxa were also enriched in HIV-negative subjects in the study by Lozupone et al [45]. In addition, Dillon et al reported that the relative abundance of the Alistipes genus was lower in untreated subjects with chronic HIV infection, compared with uninfected subjects [46], and Mutlu et al found enrichment of Rikenellaceae in HI- negative subjects [47]. All studies, including ours, found an association between dysbiosis and markers of microbial translocation, immune activation, and/or inflammation, although the biomarkers and the methods used to measure them varied.
There are a few limitations to our study. The numbers of cases and controls were small, limiting our ability to control for potential confounding factors (although the numbers were similar to those in the studies discussed above); we did not include untreated HIV-infected individuals or those in whom ART was not suppressive, and we did not measure markers of immune activation. Since this was an exploratory study, we did not correct for multiple comparisons, and the results need to be confirmed by other studies. Nevertheless, our data showed that individuals with chronic HIV infection who were receiving suppressive ART displayed intestinal dysbiosis characterized by a significant enrichment in inflammogenic taxa and a significant association with increased microbial translocation and systemic inflammation, compared with HIV-uninfected controls. Additional, larger and longitudinal studies including functional metagenomics and metabolomics approaches to identify specific metabolic pathways of the microbiome that are impacted by HIV infection are needed to develop targeted interventions that reduce immune activation and systemic inflammation in these patients.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the subjects, study coordinators, and technical staff who participated in this study; Nicola Segata and Curtis Huttenhower (Harvard School of Public Health, Boston, MA), for help with LEfSe; and Mkaya Mwamburi (Tufts University School of Medicine), for statistical advice.
Financial support. This work was supported by the National Institutes of Health (NIH; grants RO1 HL096585 and RO1 HL6594, grant T32 AI07389 [to D. D., C. D., and S. B.], and grant T32AI007438 [to G. V.]), the Lifespan/Tufts/Brown Center for AIDS Research (NIH grant P30AI042853), the Tufts Nutrition Collaborative-Center for Drug Abuse and AIDS Research (NIH grant P30DA013868), and the Tufts Clinical and Translational Science Institute (NIH grant UL1 TR000073).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Triant VA, Grinspoon SK. Vascular dysfunction and cardiovascular complications. Curr Opin HIV AIDS. 2007;2:299–304. doi: 10.1097/COH.0b013e3281e7e831. [DOI] [PubMed] [Google Scholar]
- 2.Currier JS, Lundgren JD. Guidelines for managing cardiovascular risk: an evolving area. Curr Opin HIV AIDS. 2008;3:205–6. doi: 10.1097/COH.0b013e3282fc75fe. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG. HIV: How to escape treatment. Nature. 2011;477:36–7. doi: 10.1038/477036a. [DOI] [PubMed] [Google Scholar]
- 4.Hatano H. Immune activation and HIV persistence: considerations for novel therapeutic interventions. Curr Opin HIV AIDS. 2013;8:211–6. doi: 10.1097/COH.0b013e32835f9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Marle G, Gill MJ, Kolodka D, McManus L, Grant T, Church DL. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology. 2007;4:87. doi: 10.1186/1742-4690-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lederman MM, Funderburg NT, Sekaly RP, Klatt NR, Hunt PW. Residual immune dysregulation syndrome in treated HIV infection. Adv Immunol. 2013;119:51–83. doi: 10.1016/B978-0-12-407707-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30:149–73. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012;10:655–66. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 9.Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 11.Hunt PW, Brenchley J, Sinclair E, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–33. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauce D, Larsen M, Fastenackels S, et al. HIV disease progression despite suppression of viral replication is associated with exhaustion of lymphopoiesis. Blood. 2011;117:5142–51. doi: 10.1182/blood-2011-01-331306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalayjian RC, Machekano RN, Rizk N, et al. Pretreatment levels of soluble cellular receptors and interleukin-6 are associated with HIV disease progression in subjects treated with highly active antiretroviral therapy. J Infect Dis. 2010;201:1796–805. doi: 10.1086/652750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–95. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 16.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammer HF. Gut microbiota and inflammatory bowel disease. Dig Dis. 2011;29:550–3. doi: 10.1159/000332981. [DOI] [PubMed] [Google Scholar]
- 19.Kim MS, Hwang SS, Park EJ, Bae JW. Strict vegetarian diet improves the risk factors associated with metabolic diseases by modulating gut microbiota and reducing intestinal inflammation. Environ Microbiol Rep. 2013;5:765–75. doi: 10.1111/1758-2229.12079. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Shen D, Fang Z, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim JO. Gut microbiota in inflammatory bowel disease. Pediatr Gastroenterol Hepatol Nutr. 2013;16:17–21. doi: 10.5223/pghn.2013.16.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gori A, Rizzardini G, Van't Land B, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol. 2011;4:554–63. doi: 10.1038/mi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gori A, Tincati C, Rizzardini G, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46:757–8. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis CL, Ma ZM, Mann SK, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–70. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. Dysbiosis of the gut microbiota is associated with hiv disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Santiago J, Gianella S, Massanella M, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. Aids. 2013;27:1921–31. doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volpe GE, Ward H, Mwamburi M, et al. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J Stud Alcohol Drugs. 2014;75:347–57. doi: 10.15288/jsad.2014.75.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangili A, Polak JF, Skinner SC, et al. HIV infection and progression of carotid and coronary atherosclerosis: the CARE study. J Acquir Immune Defic Syndr. 2011;58:148–53. doi: 10.1097/QAI.0b013e31822d4993. [DOI] [PubMed] [Google Scholar]
- 29.Volpe GE, Tang AM, Polak JF, Mangili A, Skinner SC, Wanke CA. Progression of carotid intima-media thickness and coronary artery calcium over 6 years in an HIV-infected cohort. J Acquir Immune Defic Syndr. 2013;64:51–7. doi: 10.1097/QAI.0b013e31829ed726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooley PC, Miller HG, Gribble JN, Turner CF. Automating telephone surveys: using T-ACASI to obtain data on sensitive topics. Comput Human Behav. 1998;14:195–207. [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, Lederman MM, Hunt P, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199:1177–85. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–6. [PubMed] [Google Scholar]
- 38.Landmann R, Knopf HP, Link S, Sansano S, Schumann R, Zimmerli W. Human monocyte CD14 is upregulated by lipopolysaccharide. Infect Immun. 1996;64:1762–9. doi: 10.1128/iai.64.5.1762-1769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda K, Kumagai N, Yamamoto K, Fujitsu Y, Chikamoto N, Nishida T. Potentiation of lipopolysaccharide-induced chemokine and adhesion molecule expression in corneal fibroblasts by soluble CD14 or LPS-binding protein. Invest Ophthalmol Vis Sci. 2005;46:3095–101. doi: 10.1167/iovs.04-1365. [DOI] [PubMed] [Google Scholar]
- 40.Kitchens RL, Thompson PA. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J Endotoxin Res. 2005;11:225–9. doi: 10.1179/096805105X46565. [DOI] [PubMed] [Google Scholar]
- 41.Balagopal A, Philp FH, Astemborski J, et al. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135:226–33. doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasternak BA, D'Mello S, Jurickova II, et al. Lipopolysaccharide exposure is linked to activation of the acute phase response and growth failure in pediatric Crohn's disease and murine colitis. Inflamm Bowel Dis. 2010;16:856–69. doi: 10.1002/ibd.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14:908–34. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 44.Salipante SJ, Sengupta DJ, Rosenthal C, et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One. 2013;8:e65226. doi: 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–39. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon SM, Lee EJ, Kotter CV, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–94. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–86. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.