Abstract
Heat stress (HS) in hot climates is a major cause that strongly negatively affects milk yield in dairy cattle, leading to immeasurable economic loss. The heat stress response of bovine mammary epithelial cells (BMECs) is one component of the acute systemic response to HS. Gene networks of BMECs respond to environmental heat loads with both intra- and extracellular signals that coordinate cellular and whole-animal metabolism. Our experimental objective was to characterize the direct effects of heat stress on the cultured bovine mammary epithelial cells by microarray analyses. The data identified 2716 differentially expressed genes in 43,000 transcripts which were changed significantly between heat-stressed and normal bovine mammary epithelial cells (fold change ≥2, P ≤ 0.001). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that these differentially expressed genes are involved in different pathways that regulate cytoskeleton, cell cycle, and stress response processes. Our study provides an overview of gene expression profile and the interaction between gene expression and heat stress, which will lead to further understanding of the potential effects of heat stress on bovine mammary glands.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-014-0559-7) contains supplementary material, which is available to authorized users.
Keywords: Bovine mammary epithelial cells, Microarray analysis, Heat stress
Introduction
Heat stress (HS) in hot climates is a major cause that strongly negatively affects milk yield and composition in dairy cattle, leading to immeasurable economic loss (Rhoads et al. 2010, Smith et al. 2013, Hill and Wall 2014). As we all know, cellular heat stress response is one component of the acute systemic response to HS (Collier et al. 2008). Gene networks of bovine mammary epithelial cells (BMECs) respond to environmental heat loads above the thermoneutral zone with both intra- and extracellular signals that coordinate cellular and whole-animal metabolism. In addition, milk secretion and mammary function are regulated acutely by local autocrine feedback mechanisms that involve milk-borne factors which are sensitive to the frequency and efficiency of milking. So the response of BMECs to heat stress plays a vital role in regulating mammary function. To gain an overall view of the transcriptome profiling during BMEC heat stress, we employed the Illumina Genome Analyzer platform to perform transcriptome analysis, then systematically analyzed the gene expressional profiles in the heat-treated and control BMECs, including the up- and downregulated differentially expressed genes, functional clusters, Gene Ontology (GO) categories, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. These findings will facilitate further understanding of the potential effects of heat stress on bovine mammary glands.
Material and methods
BMEC isolation and cell culture
Mammary tissues were collected from a local abattoir in Nanjing (from Chinese Holstein cows). In vitro cultures of mammary epithelial cells were prepared in accordance with the established methods by Dairy Science Institute of Nanjing Agricultural University (Du et al. 2008, Zhao et al. 2010). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 mg/ml glucose and supplemented with 10 % fetal bovine serum (Life Technologies, Gaithersburg, MD, USA) and 1 % antibiotic-antimycotic solution (Sigma-Aldrich, St. Louis, MO, USA) in a humidified incubator, with 5 % CO2 at 37 °C. After the cells reached 80 % confluence, they were removed with 0.25 % trypsin and 0.15 % trypsin plus 0.02 % EDTA.
Heat treatment
Prior to heat treatment, the BMECs were cultured for 24 h under DMEM with serum-free conditions. Then, the dishes containing the cells were sealed with a paraffin membrane and put in a sterile, 40.5 °C water bath for 30 min. The dishes were immediately returned to the 37 °C CO2 incubator and recovered for 3–24 h.
Cell viability
The BMECs were treated as described above for the heat treatment. After 3–24 h of heat treatment, the medium was removed and 200 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) medium (0.5 mg/ml MTT reagent in fresh medium) was added to each well. After incubation for 4 h, the MTT reagent was removed and 150 μl of dimethyl sulfoxide (Jiancheng Bioengineering Institute, Nanjing, China) was added to each well, followed by 10 min of gentle shaking. Cell viability was assessed by a 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide trypan blue exclusion assay as the proportion of absorbance values to the control. The absorbance was read at a wavelength of 570 nm in a Multiskan MK3 (Thermo Electron Corporation, USA) as the value expressing the entity of proliferation.
Cell cycle analysis by FCM
After 6–24 h of heat treatment, the BMECs were collected, fixed in 75 % ice-cold ethanol overnight at 4 °C, and washed twice with ice-cold PBS. The cells were resuspended in 1 ml propidium iodide (PI) staining solution (PI 0.05 mg/ml, RNase 0.01 mg/ml, 0.025 % Triton X-100, and 1 mg/ml sodium citrate, pH 7.2–7.6) at 4 °C for 30 min. Ten thousand events were acquired in a FACSort (Becton Dickinson) and analyzed with Cell Quest software version 3.3 (Becton Dickinson).
RNA isolation
After 6 h of heat treatment, total RNA was extracted from BMECs using Trizol reagent (Invitrogen, Carlsbad, USA), according to the manufacturer’s description. The absorbance values at 260 and 280 nm were checked to assess the RNA concentration and purity for protein impurities in the samples. The RNA integrity was checked by electrophoresis on 2 % agarose gels (m/v).
Microarray assay
Gene chip analysis of the Bovine Genome Array was performed by an outside service provider (LC-Bio. CO., LTD). Total RNA from the BMECs was individually hybridized with gene chips. Briefly, in the first-strand complementary DNA (cDNA) synthesis reaction, 10 mg total RNA was used for reverse transcription using a T7-oligo (dT) promoter primer. Then, the double-stranded cDNA was synthesized from the first-strand cDNA using RNase H. After purification of the resulting DNA, an in vitro transcription reaction using the MEGAscript T7 Kit (Ambion, Inc., USA) produced biotin-labeled complementary RNA (cRNA). After the cRNA was cleaned and fragmented, it was hybridized to the probe array at 45 °C for 16 h. Thereafter, the probe array was washed and stained on the Fluidics Station, and the microarrays were scanned using the GeneChip Scanner 3000 (Affymetrix). The Affymetrix Micro Array Suite 5.0-Specific Terms GCOS version 1.4 was used for quantity analysis of the hybridization. The gene expression levels that had ≥2-fold difference between control and heat-treated BMECs were checked and further analyzed.
Analysis of the functional annotation cluster and KEGG pathway
The Molecule Annotation System (http://david.abcc.ncifcrf.gov/) was used to analyze the differentially expressed genes, using the Kyoto Encyclopedia of Genes and Genomes (KEGG) public pathway resource and the Gene Ontology (GO) consortium.
Real-time reverse transcription-polymerase chain reaction
Reverse transcription-polymerase chain reaction (RT-PCR) was performed to confirm the microarray results. Total RNA was extracted from control and heat-treated granulosa cells as described above, and total RNA was reverse transcribed using a revere transcription levels kit (TaKaRa, Dalian, China) according to the manufacturer’s protocols. The expression levels were checked for ten genes. The 18S ribosomal RNA (rRNA) gene was used as the invariant control. Primers were designed using Primer Premier 5.0 and are shown in Table 1. RT-PCR was performed with SYBR® Premix Ex TaqTM (TaKaRa Biotechnology Co., Ltd., Japan, DRR081A). The reaction solution was prepared on ice and comprised 10 μl of 2× SYBR® Premix Ex TaqTM, 0.8 μl of PCR Forward Primer (10 μM), 0.8 μl of PCR Reverse Primer (10 μM), 0.4 ml of 50× ROX Reference Dye, 2 μl of cDNA (100 ng μl−1), and dH2O to a final volume of 20 μl. The reaction mixtures were incubated in a 96-well plate at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s. All reactions were performed in triplicate. The gene expression levels in the normal and heat-treated BMECs were analyzed with the 2−ΔΔCT method.
Table 1.
Primer sequences for RT-PCR
| Gene | Primer sequence | GenBank access no. | Product size (bp) |
|---|---|---|---|
| FGF7 | F: AGGACAGTGGCTGTTGGAAT R: CTGGAACCCCCTTTTGATTT |
NM_001193131 | 220 |
| CHRM3 | F: CCAAGCTTCCCATCCAGTTA R: CCGCTTAGTGATCTGGCTTC |
NM_174270 | 161 |
| ROCK2 | F: TGAAGCCTGACAACATGCTC R: GTAACCATCACCCCCTTGTG |
NM_174452.2 | 167 |
| PIK3CA | F: GTCAATCGGTGACTGTGTGG R: GACGATCTCCAATTCCCAAA |
NM_174574 | 233 |
| BMPR1A | F: AGACACCAGAGCCCTGCTTA R: CCAGGTCAGCAATACAGCAA |
NM_001076800 | 167 |
| PPP1R12A | F: AAGCACCACATCAACACCAA R: ATGGTCACTGCCGTAGGAAC |
NM_001102217 | 192 |
| HSPB8 | F: GAGCCCTGGAAAGTGTGTGT R: TGCAGGAAGCTGGATTTTCT |
NM_001014955 | 162 |
| HSPA1A | F: CAAGATCACCATCACCAACG R: AAATCACCTCCTGGCACTTG |
NM_174550 | 239 |
| HSPA5 | F: TGCAGCAGGACATCAAGTTC R: AGCCTCAGCAGTTTCCTTCA |
NM_001075148 | 155 |
| HSP90AB1 | F: GATCACTTGGCAGTGAAGCA R: TCCGGAATCAACTCATCACA |
NM_001079637 | 176 |
| 18S rRNA | F: TCCAGCCTTCCTTCCTGGGCAT R: GGACAGCACCGTGTTGGCGTAGA |
NM_174841.2 | 116 |
Statistical analysis
All data were obtained from one independent experiment carried out in triplicate. Main and interactive effects were analyzed by the independent-samples t test using SPSS 16.0 software. P < 0.05 was considered statistically significant.
Results
Heat treatment affects cell viability
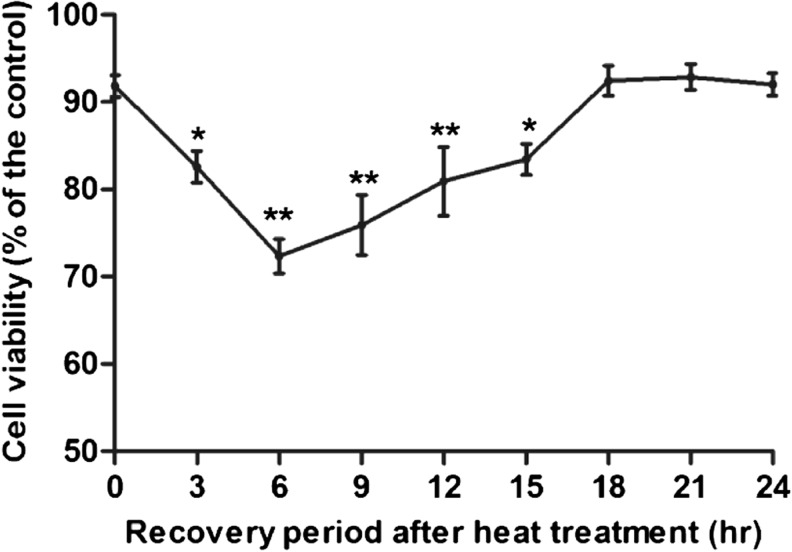
The cellular viability of heat-treated groups changed in response to the time of the recovery period; cell viability rate decreased significantly at the 3-h period after heat treatment (P < 0.05), arrived at the lowest at the 6-h period (P < 0.01), and then gradually increased. Moreover, the cell viability rates of the 18–24-h groups recovered to the normal level (Fig. 1).
Fig. 1.
Effect of heat treatment on cell viability. Cell viability was estimated by MTT, expressed as the rate of the value obtained with the control culture (37 °C). Asterisks and double asterisks separately represent values significantly different from the controls (P < 0.05 and P < 0.01)
Heat treatment affects the cell cycle phase
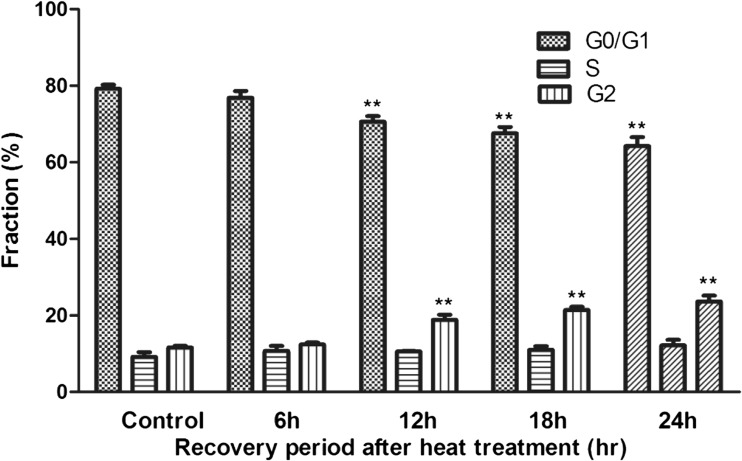
In the heat-treated groups, there was a significant decrease in the percentage of cells in the G0/G1 phase at the 12–24-h period after heat treatment (P < 0.01), and the G2/M subpopulation significantly increased (P < 0.01). The result showed that there was a slight G2/M arrest in cell cycle progression after heat treatment (Fig. 2).
Fig. 2.
Cell cycle progression by flow cytometry. Double asterisks represent values significantly different from the controls (P < 0.01)
Microarray identification of differentially expressed genes
The global gene expression profiles in bovine mammary epithelial cell samples representing heat-treated and control cells were identified with the microarray technique. Of a total of about 43,000 probe sets, we observed that the expression of 2716 genes was significantly different (P < 0.001) between control BMECs and heat-treated ones, 542 of which were altered ≥3-fold (Table 2). Both up- and downregulated genes are shown in Table S1.
Table 2.
The number of genes which were differentially regulated between control and heat-treated BMECs
| Fold | Upregulated | Downregulated | Total |
|---|---|---|---|
| ≥2 | 1376 | 1340 | 2716 |
| ≥3 | 123 | 419 | 542 |
Functional annotation cluster and Gene Ontology analysis
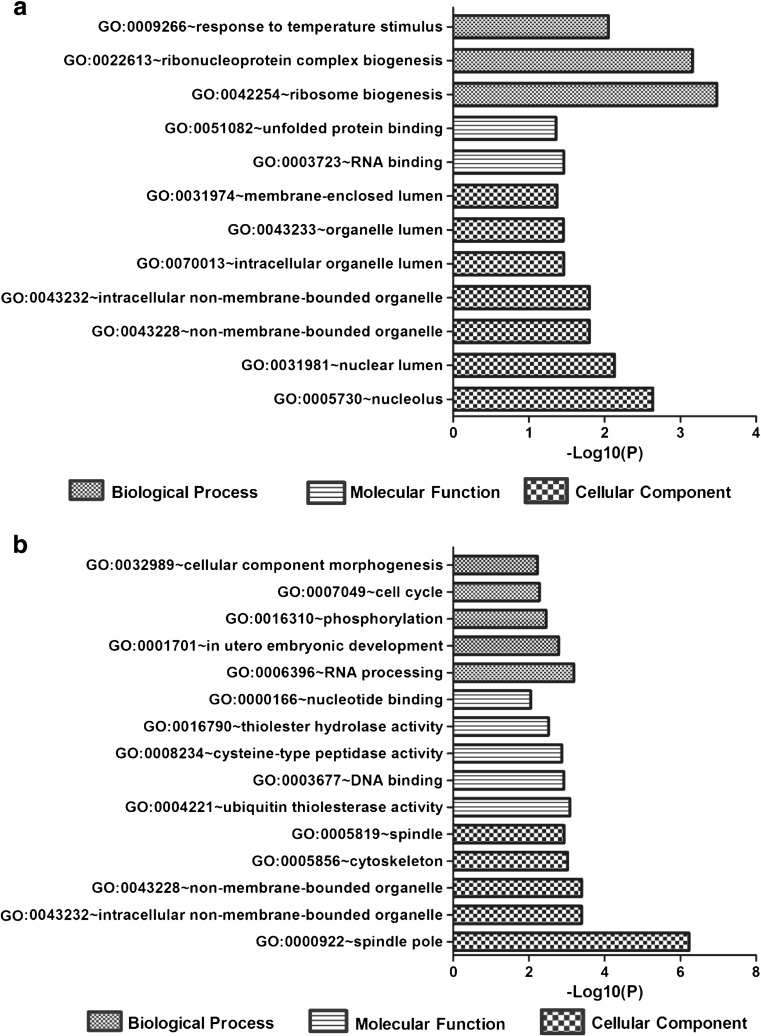
To categorize biological processes that are overrepresented in control and heat-treated BMECs, we classified all known differentially expressed genes using the Functional Annotation Cluster (FAC) tool available in the Database for Annotation, Visualization and Integrated Discovery (DAVID) [http://david.abcc.ncifcrf.gov/home.jsp]. DAVID FAC analysis of 123 upregulated genes (fold change ≥3) generated a total of five functional clusters using default parameters. All clusters were further filtered to one cluster by selecting only those clusters having an enrichment score ≥2, P values ≤0.05, and number of genes in each term ≥5 (Table 3). Similarly, the GO functional annotation chart was further filtered to top 12 records by selecting only the most specific subcategories of GO terms under biological process; cellular component and molecular function which were considered significant by the Benjamini-Yuketeli test (P < 0.05) were ranked (Fig. 3a). From these results, it can be seen that the GO terms are enriched in genes with functions necessary for actively proliferating cells such as cell structure and cell biogenesis (Table 2). The other major processes that exhibit higher levels of gene expression as a consequence of heat-treated BMECs include GO terms related to response to temperature stimulus and biogenesis processes (Fig. 3a). These data suggest that the process of response to temperature stimulus is the major processes upregulated by heat-treated BMECs and that their overrepresentation may be due to the response of BMECs under heat stress.
Table 3.
Functional annotation clustering of upregulated and downregulated genes (fold change ≥3)
| Annotation cluster | Term | Enrichment score | P value |
|---|---|---|---|
| Upregulated genes | |||
| The first class | Ribosome biogenesis | 3.094 | 3.29E−04 |
| Ribonucleoprotein complex biogenesis | 6.90E−04 | ||
| Nucleolus | 0.002304 | ||
| Downregulated genes | |||
| The first class | Embryonic development | 2.067 | 0.0012 |
| Regulation of cell cycle | 0.0487 | ||
| Organelle | 4.80E−04 | ||
| The second class | Cytoskeleton | 2.058 | 0.0332 |
| Cytoskeletal part | 0.0448 | ||
Fig. 3.
Highly enriched GO terms analyzed of upregulated (a) and downregulated (b) genes with fold change ≥3 by Gene Ontology analysis (P values <0.05). Only the most specific subcategories of GO terms under biological process, cellular component, and molecular function which were considered significant by the Benjamini-Yuketeli test (P < 0.05) were ranked
Functional analysis of 419 downregulated genes resulted in 28 clusters, and 2 clusters were filtered by selecting only those clusters having an enrichment score ≥2, P values ≤0.05, and number of genes in each term ≥5 (Table 2). From these results, it can be seen that the GO terms are enriched in genes with functions necessary for cell growth such as cell cycle and cytoskeleton (Table 3). The GO functional annotation chart reported 59 records (Table S2), of which 15 highly enriched GO terms were selected under biological process, cellular component, and molecular function. These processes that were significantly downregulated by heat-treated BMECs as shown by functional analysis include DNA/RNA binding, cytoskeleton, and regulation of cell cycle processes (Fig. 3b).
Identification of signaling pathways contributing to the normal bovine mammary epithelial cell growth
To determine the signaling pathways of upregulated and downregulated genes associated with mammary epithelial cell growth and function, all genes with more than 3-fold change were subjected to the KEGG pathway analysis. No directly related pathways were found in upregulated genes; there are four signaling pathways involved in downregulated genes, such as regulation of the actin cytoskeleton and TGF-β signaling pathway (Table 4).
Table 4.
Significantly enriched KEGG pathways of downregulated genes (fold change ≥3-fold)
| Term | No. of genes | Genes | P value | Fold enrichment |
|---|---|---|---|---|
| bta04810, regulation of actin cytoskeleton | 8 | FGF7, CHRM3, ROCK1, ROCK2, PPP1R12A, PIK3CA, ITGB1, PIK3R1 | 0.00914 | 3.30 |
| bta04350, TGF-β signaling pathway | 5 | SP1, ROCK1, ROCK2, ZFYVE16, BMPR1A | 0.01676 | 4.96 |
| bta04510, focal adhesion | 7 | ROCK1, ROCK2, PPP1R12A, PIK3CA, BIRC3, ITGB1, PIK3R1 | 0.02562 | 3.02 |
| bta05200, pathways in cancer | 9 | FGF7, HIF1A, PIK3CA, PTCH2, TPR, BIRC3, ITGB1, MMP1, PIK3R1 | 0.03423 | 2.33 |
Quantitation of gene expression by RT-PCR
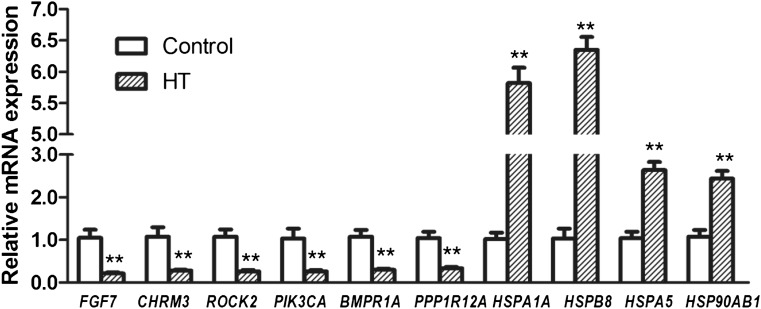
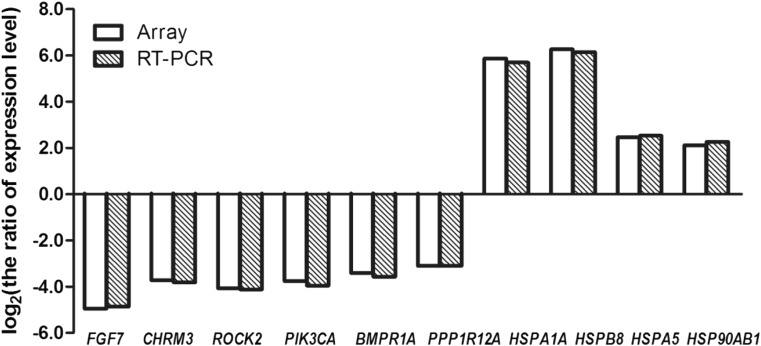
Figure 4 shows the quantitative RT-PCR results for some of the genes which were significantly affected by heat treatment of bovine mammary epithelial cells: six genes associated with the cytoskeleton and TGF-β signaling pathway, including FGF7, CHRM3, ROCK2, PIK3CA, PPP1R12A, and BMPR1A, and other genes associated with heat stress, including HSPB8, HSPA1A, HSPA5, and HSP90AB1 (Table 5). Six genes associated with the cytoskeleton and TGF-β signaling pathway were downregulated by heat treatment. The HSP family-related genes are known to be active during cell response to various stressors (Setroikromo et al. 2007, Noonan et al. 2007). HSPB8, HSPA1A, HSPA5, and HSP90AB1 were upregulated by heat treatment. The RT-PCR results were in accordance with the gene chip findings (Fig. 5).
Fig. 4.
Validation of differentially expressed genes by real-time qPCR. Relative quantification of ten representative genes was performed. qRT-PCR values were determined from the ΔΔCt for the target genes relative to 18S. Note: double asterisks indicate a statistical difference (P < 0.01)
Table 5.
Effects of heat stress on the expression of selected genes involved in the heat shock protein response (fold change ≥2-fold)
| Gene symbol | Full gene name | Fold change |
|---|---|---|
| HSPB8 | Heat shock 22 kDa protein 8 | 6.3 |
| HSPA1A | Heat shock 70 kDa protein 1A | 5.9 |
| DNAJA4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 3.4 |
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 3.4 |
| DNAJC5 | DnaJ (Hsp40) homolog, subfamily C, member 5 | 2.9 |
| DNAJC30 | DnaJ (Hsp40) homolog, subfamily C, member 30 | 2.7 |
| DNAJC11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | 2.7 |
| HSPA5 | Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 2.5 |
| DNAJC17 | DnaJ (Hsp40) homolog, subfamily C, member 17 | 2.1 |
| DNAJA3 | DnaJ (Hsp40) homolog, subfamily A, member 3 | 2.1 |
| HSPB2 | Heat shock 27 kDa protein 2 | 2.1 |
| HSP90AB1 | Heat shock protein 90 kDa alpha (cytosolic), class B member 1 | 2.1 |
Fig. 5.
Comparison of RT-PCR findings to microarray analysis results by fold change of ten selected genes
Discussion
Heat stress can be defined as a condition that occurs when an animal cannot adequately dissipate body heat in order to maintain thermal balance (Grover 2002, Bernabucci et al. 2014). Some reports showed that Holstein milk yield decreased during HS; feed intake and management factors partly account for the decreased milk yield during heat stress (Rhoads et al. 2010, Smith et al. 2013, Hill and Wall 2014). Several studies also suggested that transcription and translation of RNA in mammary epithelial cells are inhibited, as is cell cycling growth; membrane permeability is also changed during heat stress (Sonn et al. 2002, Collier et al. 2006&2008, Han et al. 2014). But the global effect of heat on gene expression in cultured bovine mammary epithelial cells is unclear. Our experimental results showed that heat stress triggers a dramatic and complex program of altered gene expression in BMECs. Transcriptional activity indicated a downregulation of a number of genes associated with the cell cycle and cytoskeleton activity. Meanwhile, the results of the cell cycle also show that there was a slight G2/M arrest after heat treatment and were in accordance with the gene chip findings (Fig. 2), thereby suggesting a repression of the genomic signals responsible for promoting ductal growth and networking. Overall, the transcriptome profile indicated downregulation of genes involved in cellular component morphogenesis, cell cycle, and focal adhesion. Of the upregulated genes, the majority were involved in response to temperature stimulus, protein repair, and biogenesis, thereby suggesting an activation of the genomic signals responsible for promoting heat tolerance of BMECs. These data indicate that the heat tolerance of mammary epithelium might depend upon the expression profile of a core set of genes and that the response to the temperature stimulus is under a positive mode of regulation.
The actin cytoskeleton and TGF-β signaling pathway are involved in downregulated genes. The cytoskeleton plays a major role in the process which governs the cellular mechanism by which cells dissociate with their microenvironment (Martin et al. 2014). Meanwhile, TGF-β signaling usually participates in regulating various biological processes including cell growth, differentiation, apoptosis, extracellular matrix modeling, and immune response (Su et al. 2010; Ramamoorthi 2014). In breast cancer, some studies show TGF-β-induced SMAD-dependent signaling as the major pathway involved in breast cancer metastasis and attenuated TGF-β signaling promotes metastasis (Flanders et al. 2014; Novitskiy et al. 2014). The TGF-β/miR-424/503 axis is part of the mechanism that regulates the proliferation of hormone receptor-positive mammary epithelial cells in vivo (Llobet-Navas et al. 2014). These research reports show that the TGF-β signaling pathway has the vital role on growth, progression, and function of mammary epithelial cells. Our gene array also indicated that heat treatment affected the TGF-β signaling through downregulating the expression of the bone morphogenetic protein receptor (BMPR1A), which is involved in the TGF-β/SMAD signaling pathway (Table S1, Figure S1).
In this study, some genes of the HSP family involved in upregulated genes included HSP70, HSP90, and some small HSPs, such as HSP22/27/40 (Table 5). In these genes, highly differential expression was observed for HSPA1A (heat shock 70 kDa protein 1A, 5.9-fold induction) and HSPB8 (heat shock 22 kDa protein 8, 6.3-fold induction). In all HSPs, the most pronounced stress-related changes and involvement in different cell functions have been demonstrated for HSP70 (Grover 2002; Jego et al. 2010; Sirotkin and Bauer 2011). The bovine two-cell embryo is capable of increasing transcription of HSPA1A and synthesis of HSP70 in response to heat shock (Sakatani et al. 2013). The mRNA and protein expression of HSP70 increased after 42 °C heat treatment in cultured bovine mammary epithelial cells (Han et al. 2014). Heat stress also induced HSP90 expression in bovine BMECs and the bovine oviduct (Kobayashi et al. 2013; Deb et al. 2014). Furthermore, we also found that heat stress increased the expression of HSP40 (also known as DNAJ proteins) in BMECs. The function and role of hsp40 are poorly understood. Several studies have revealed that the availability of DNAJ proteins, the co-chaperones to the Hsp70 machine, could be a rate-limiting factor in handling diseased proteins within the cell (Kakkar et al. 2012; McConnell and McAlpine 2013). The role of HSP40 in BMECs needs further investigation.
Heat stress compromised mammary growth and induced the change of gene expression. As reported, these changes were related to mammary function and include 1) inhibited milk secretion by upregulating an endogenous milk enzymatic system (Rucker et al. 2006, Silanikove et al. 2009), 2) disrupted tight junction of mammary gland epithelial cells, increased casein degradation and decreased the protein synthesis (Stelwagen et al. 1998, Silanikove et al. 2000, Shamay et al. 2003), and 3) disrupted cytoskeletal components and inhibited cell cycle of BMECs (Collier et al. 2006, Du et al. 2008). In this study, we also found that these genes, SERPINE, PLAU, and PLAUR, coding an endogenous milk enzymatic system were upregulated; some genes, such as RICTOR, PIK3 and ELF2, involving in mTOR signaling pathway of mediated protein synthesis were down-regulated (Table S1). Meanwhile, the genes coding the cytoskeletal components were downregulated (Table 4) and G2/M phase arrest was found (Fig. 2). The present results regarding the effect of HS on BMECs are consistent with the above-noted response to acute stress.
In summary, we have reported a gene expression analysis which could offer an improved understanding of the molecular mechanism responsible for the reduced milk yield in cattle in hot climate. Further research is needed to assess the function of individual differentially expressed genes that may be involved in heat stress and the milk secretion process and to identify these pathways and the components of the inward signal transduction.
Electronic supplementary material
TGF-beta signaling pathway (List genes are noted by red starts). (GIF 232 kb)
Up- and down-regulated genes lists (fold change≥2) (XLS 407 kb).
GO functional annotation chart of down-regulated genes(fold change≥3).Top ten lists of GO functional annotation chart records under biological process, cellular component and molecular function, respectively. Only those terms that have p-values≤0.05 and number of genes in each term > 5 are shown (XLS 23.0 kb).
Acknowledgments
This work received funding from the National Supporting Projects for Science and Techniques of China (2012BAD12B10) and the Natural Science Foundation of China (31372290).
References
- Bernabucci U, Biffani S, Buggiotti L, Vitali A, Lacetera N, Nardone A. The effects of heat stress in Italian Holstein dairy cattle. J Dairy Sci. 2014;97(1):471–486. doi: 10.3168/jds.2013-6611. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Stiening CM, Pollard BC, VanBaale MJ, Baumgard LH, Gentry PC, Coussens PM (2006) Use of gene expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J Anim Sci Suppl: E1–13 [DOI] [PubMed]
- Collier R, Collier J, Rhoads RP, Baumgard L. Genes involved in the bovine heat stress response. J Dairy Sci. 2008;91:445–454. doi: 10.3168/jds.2007-0540. [DOI] [PubMed] [Google Scholar]
- Daly SEJ, Owens RA, Hartmann PE. The short-term synthesis and infant-regulated removal of milk in lactating women. Exp Physiol. 1993;78:209–220. doi: 10.1113/expphysiol.1993.sp003681. [DOI] [PubMed] [Google Scholar]
- Deb R, Sajjanar B, Singh U, Kumar S, Singh R, Sengar G, Sharma A. Effect of heat stress on the expression profile of Hsp90 among Sahiwal (Bos indicus) and Frieswal (Bos indicus × Bos taurus) breed of cattle: a comparative study. Gene. 2014;536(2):435–440. doi: 10.1016/j.gene.2013.11.086. [DOI] [PubMed] [Google Scholar]
- Du J, Di HS, Guo L, Li ZH, Wang GL. Hyperthermia causes bovine mammary epithelial cell death by a mitochondrial-induced pathway. J Therm Biol. 2008;33(1):37–47. doi: 10.1016/j.jtherbio.2007.06.002. [DOI] [Google Scholar]
- Flanders KC, Heger CD, Conway C, Tang B, Sato M, Dengler SL, Goldsmith PK, Hewitt SM, Wakefield LM. Brightfield proximity ligation assay reveals both canonical and mixed transforming growth factor-β/bone morphogenetic protein Smad signaling complexes in tissue sections (2014) J Histochem Cytochem. Aug 20. doi:10.1369/0022155414550163. [DOI] [PMC free article] [PubMed]
- Grover A. Molecular biology of stress responses. Cell Stress Chaperones. 2002;7(1):1–5. doi: 10.1379/1466-1268(2002)007<0001:MBOSR>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZY, Mu T, Yang Z. Methionine protects against hyperthermia-induced cell injury in cultured bovine mammary epithelial cells. Cell Stress Chaperones [Epub ahead of print] DOI. 2014 doi: 10.1007/s12192-014-0530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DL, Wall E. Dairy cattle in a temperate climate: the effects of weather on milk yield and composition depend on management. Animal. 2014;15:1–12. doi: 10.1017/S1751731114002456. [DOI] [PubMed] [Google Scholar]
- Jego G, Hazoumé A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer Lett. 2010;2332(2):275–85. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Kakkar V, Prins LC, Kampinga HH. DNAJ proteins and protein aggregation diseases. Curr Top Med Chem. 2012;12(22):2479–2490. doi: 10.2174/1568026611212220004. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Wakamiya K, Kohka M, Yamamoto Y, Okuda K. Summer heat stress affects prostaglandin synthesis in the bovine oviduct. Reproduction. 2013;146(2):103–110. doi: 10.1530/REP-12-0479. [DOI] [PubMed] [Google Scholar]
- Llobet-Navas D, Rodriguez-Barrueco R, de la Iglesia-Vicente J, Olivan M, Castro V, Saucedo-Cuevas L, Marshall N, Putcha P, Castillo-Martin M, Bardot E, Ezhkova E, Iavarone A, Cordon-Cardo C, Silva JM. The microRNA 424/503 cluster reduces CDC25A expression during cell cycle arrest imposed by transforming growth factor β in mammary epithelial cells. Mol Cell Biol. 2014;34(23):4216–4231. doi: 10.1128/MCB.00611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SK, Kamelgarn M, Kyprianou N. Cytoskeleton targeting value in prostate cancer treatment. Am J Clin Exp Urol. 2014;2(1):15–26. [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, McAlpine SR. Heat shock proteins 27, 40, and 70 as combinational and dual therapeutic cancer targets. Bioorg Med Chem Lett. 2013;23(7):1923–1928. doi: 10.1016/j.bmcl.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Giardina C, Hightower LE. Hsp70B' regulation and function. Cell Stress Chaperones. 2007;12(3):219–229. doi: 10.1379/CSC-278.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitskiy SV, Forrester E, Pickup MW, Gorska AE, Chytil A, Aakre M, Polosukhina D, Owens P, Yusupova DR, Zhao Z, Ye F, Shyr Y, Moses HL. Attenuated transforming growth factor beta signaling promotes metastasis in a model of HER2 mammary carcinogenesis. Breast Cancer Res. 2014;16(5):425. doi: 10.1186/s13058-014-0425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthi G, Sivalingam N. Molecular mechanism of TGF-β signaling pathway in colon carcinogenesis and status of curcumin as chemopreventive strategy. Tumour Biol. 2014;35(8):7295–305. doi: 10.1007/s13277-014-1840-1. [DOI] [PubMed] [Google Scholar]
- Rhoads ML, Kim JW, Collier RJ, Crooker BA, Boisclair YR, Baumgard LH, Rhoads RP. Effects of heat stress and nutrition on lactating Holstein cows: II. Aspects of hepatic growth hormone responsiveness. J Dairy Sci. 2010;93(1):170–179. doi: 10.3168/jds.2009-2469. [DOI] [PubMed] [Google Scholar]
- Rucker M, Schafer T, Scheuer C, Harder Y, Vollmar B, Menger MD. Local heat shock priming promotes recanalization of thromboembolized microvasculature by upregulation of plasminogen activators. Arterios Thrombos Vasc Biol. 2006;26:1632–1639. doi: 10.1161/01.ATV.0000223144.65958.c3. [DOI] [PubMed] [Google Scholar]
- Sakatani M, Bonilla L, Dobbs KB, Block J, Ozawa M, Shanker S, Yao J, Hansen PJ. Changes in the transcriptome of morula-stage bovine embryos caused by heat shock: relationship to developmental acquisition of thermotolerance. Reprod Biol Endocrinol. 2013;11:3. doi: 10.1186/1477-7827-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setroikromo R, Wierenga PK, van Waarde MA, Brunsting JF, Vellenga E, Kampinga HH. Heat shock proteins and Bcl-2 expression and function in relation to the differential hyperthermic sensitivity between leukemic and normal hematopoietic cells. Cell Stress Chaperones. 2007;12(4):320–330. doi: 10.1379/CSC-279.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamay A, Shapiro F, Leitner G, Silanikove N. Infusions of casein hydrolyzates into the mammary gland disrupt tight junction integrity and induce involution in cows. J Dairy Sci. 2003;86:1250–1258. doi: 10.3168/jds.S0022-0302(03)73709-6. [DOI] [PubMed] [Google Scholar]
- Silanikove N, Shamay A, Shinder D, Moran A. Stress down-regulates milk yield in cows by plasmin induced beta-casein product that blocks K+ channels on the apical membranes. Life Sci. 2000;67:2201–2212. doi: 10.1016/S0024-3205(00)00808-0. [DOI] [PubMed] [Google Scholar]
- Silanikove N, Shapiro F, Shinder D. Acute heat stress brings down milk secretion in dairy cows by up-regulating the activity of the milk-borne negative feedback regulatory system. BMC Physiol. 2009;9:13. doi: 10.1186/1472-6793-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin AV, Bauer M. Heat shock proteins in porcine ovary: synthesis, accumulation and regulation by stress and hormones. Cell Stress and Chaperones. 2011;16:379–387. doi: 10.1007/s12192-010-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Smith T, Rude BJ, Ward SH. Short communication: comparison of the effects of heat stress on milk and component yields and somatic cell score in Holstein and Jersey cows. J Dairy Sci. 2013;96(5):3028–3033. doi: 10.3168/jds.2012-5737. [DOI] [PubMed] [Google Scholar]
- Sonn L, Fujita J, Gaffin SL. Invited review: Effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- Stelwagen K, van Espen DC, Verkerk GA, McFadden HA, Farr VC. Elevated plasma cortisol reduces permeability of mammary tight junctions in the lactating bovine mammary epithelium. J Endocrinol. 1998;159:173–178. doi: 10.1677/joe.0.1590173. [DOI] [PubMed] [Google Scholar]
- Su E, Han X, Jiang G. The transforming growth factor beta 1/SMAD signaling pathway involved in human chronic myeloid leukemia. Tumori. 2010;96(5):659–666. doi: 10.1177/030089161009600503. [DOI] [PubMed] [Google Scholar]
- Zhao K, Liu HY, Zhou MM, Liu JX. Establishment and characterization of a lactating bovine mammary epithelial cell model for the study of milk synthesis. Cell Biol Int. 2010;34(7):717–721. doi: 10.1042/CBI20100023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TGF-beta signaling pathway (List genes are noted by red starts). (GIF 232 kb)
Up- and down-regulated genes lists (fold change≥2) (XLS 407 kb).
GO functional annotation chart of down-regulated genes(fold change≥3).Top ten lists of GO functional annotation chart records under biological process, cellular component and molecular function, respectively. Only those terms that have p-values≤0.05 and number of genes in each term > 5 are shown (XLS 23.0 kb).