Abstract
Heat shock factor 1 (HSF1) is one of the most important transcriptional molecules in the heat shock process; however, HSF1 can also regulate the expression of other proteins. Dystrophin Dp71 is one of the most widely expressed isoforms of the dystrophin gene family. In our experiments, we showed for the first time that HSF1 can function as a transcriptional factor for endogenous Dp71 expression in vivo and in vitro. We demonstrated that the messenger RNA (mRNA) and protein expression of Dp71 were significantly reduced in HSF1-knockout mice compared with wild-type mice in brain, lung, liver, spleen, and kidney. Overexpression of HSF1 significantly enhanced the mRNA and protein expression of Dp71 in HeLa cells. Inhibiting the expression of HSF1 in HeLa cells significantly reduced the expression of Dp71. By use of the EMSA technique, the chromatin immunoprecipitation assay, and the luciferase reporter system, we demonstrated that HSF1 can directly bind the HSE in the Dp71 promoter region. We concluded from our data that HSF1 functions as a transcriptional regulator of Dp71 expression.
Keywords: HSF1, Dp71, Transcription regulation
Introduction
As one of the most important transcriptional molecules in the heat shock process, heat shock factor 1 (HSF1) is the protein defined for decades by its ability to coordinate chaperone protein expression and enhance survival under thermal stress (Christians et al. 2002; Tanabe et al. 1997). Recent research on HSF1 revealed that it has versatile biological functions other than regulating thermal reaction. For example, HSF1 was found to be a regulator of longevity in several organisms (Chiang et al. 2012; Volovik et al. 2012). HSF1 contributes to the protein homeostasis that underlies many human diseases such as aging (Morimoto 2008). HSF1 has also been identified as one of only six potent metastasis-promoting genes in a genome-wide screen for enhancers of invasion by malignant melanoma cells (Scott et al. 2011). HSF1 is regarded as one of the transcription molecules in tumorigenesis, by taking part in a broad biological process that extends far beyond protein folding, and includes energy metabolism, cell cycle signaling, DNA repair, apoptosis, cell adhesion, extracellular matrix formation, and translation (Mendillo et al. 2012). The biological function of HSF1 extends far beyond its ability to coordinate chaperone protein expression.
Dystrophin Dp71 (Dp71) is one of the most widely expressed isoforms of the dystrophin gene family (Bar et al. 1990; Austin et al. 1995). It is expressed in pluripotent embryonic stem cells and is the first product of the gene detectable during development. Relatively high Dp71 promoter activity is associated with morphogenic events and terminal differentiation of several mouse tissues and organs, including the central and peripheral nervous systems, limb buds, lungs, blood vessels, eyes, inner ears, and nasal organs (Sarig et al. 1999; Lumeng et al. 1999). It also plays an important role in the adhesion and proliferation process of neuronal cells (García-Sierra et al. 2005).
In our project, one HSE was found via bio-informatics software analysis in the promoter region of Dp71. It was demonstrated that HSF1 can bind to this sequence in vitro and in vivo. All the experimental data indicated that HSF1 plays a critical role in regulating Dp71 expression.
Materials and methods
Animals
Hsf1 gene knockout mice (HSF1−/−) were obtained from Dr Ivor Benjamin (University of Utah, Salt Lake City, Utah, USA). Wild-type (WT, HSF1+/+) mice of the same age, strain, and sex were used as controls. All animal procedures were approved by the Animal Care and Use Committee of Central South University.
Antibodies
The following antibodies were used: mouse anti-dystrophin monoclonal antibody; rabbit anti-dystrophin polyclonal antibody (Abcam); rabbit anti-HSF1 polyclonal antibody (Cell Signaling); mouse antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (Sigma); and peroxidase-conjugated anti-mouse and anti-rabbit IgG (Boster Biological Technology).
Plasmids
The wild-type HSF1 expression plasmid, pcDNA3.1 (+)/HSF1-wt, and the constitutively activated HSF1 mutant plasmid pcDNA3.1 (+)/HSF1 (+) were kindly provided by Dr. Richard Voellmy at HSF Pharmaceuticals S.A., Switzerland. HSF1-specific siRNA construct, Psilencer-HSF1 plasmid and control plasmids were a kind gift from Dr Shunmei E. (Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences). Generation of the Dp71 promoter constructs: pGL3-Dp71 (−979 to +20), pGL3-Dp71-Mut (-979 to -832//-829 to +20), pGL3-Dp71-del (−712 to +20) were performed by RT-PCR and cloned into the pGL3 vector. The authenticity of the constructs was verified by sequencing (data not shown).
Cell lines and culture conditions
A human immortal cell line derived from a cervical carcinoma (HeLa) cell line and murine RAW264.7 macrophage cell lines were obtained from the Cell Culture Center, Chinese Academy of Medical Sciences (Shanghai, China). HeLa cells and RAW264.7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % (v/v) fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen). They were maintained at 37 °C in a humidified atmosphere containing 5 % CO2.
Lipofectamine-mediated plasmid transfection
Transfection of HeLa and RAW264.7 macrophages was carried out according to the manufacturer’s instructions (LIPOFECTAMINE 2000™, Invitrogen). Briefly, about 5 × 105 cells per bottle containing 5 ml appropriate complete growth medium were seeded and incubated at 37 °C with 5 % CO2 until the cells were 70–80 % confluent. After rinsing with serum- and antibiotics-free medium, the cells were transfected separately with 10 μg pcDNA3.1-HSF1 (experimental group) and 10 μg pcDNA3.1 (vector control), and the plasmids were mixed with 20 μl lipofectamine in serum- and antibiotics-free DMEM, and DNA/lipofectamine mixture was added into the cell culture medium and incubated at 37 °C in a CO2 incubator for 6 h. The medium was then replaced by DMEM culture medium containing 10 % FBS. Cells from each experimental group were collected for further experiments after 48 h.
RNA extraction and real-time PCR
Mice tissues (200–300 mg) and cultured cells were homogenized using the Trizol® Reagent (Invitrogen), and total RNA was isolated in accordance with the manufacturer’s protocol. One microgram of total RNA was then used as a template to synthesize the complimentary complementary DNA (cDNA) using a First Strand Synthesis Kit (TaKaRa Bio, Siga, Japan). The synthesized cDNA samples were subjected to quantitative PCR (qPCR) using a SYBR® Green Quantitative PCR kit (TaKaRa Bio, Siga, Japan). All experiments were performed in triplicate using the BioRad CFX96 C1000 System (BioRad, Hercules, CA, USA). RT-PCR data were normalized by measuring average cycle threshold (Ct) ratios between candidate genes and the control gene, GAPDH. The formula 2Ct (Candidate)/2Ct (Control) was used to calculate normalized ratios. The primers were designed using Primer 3 online software; the following primers were used in our study: forward: 5′-TTGGCAGTCAAACTTCGGACTC-3′, reverse: 5′-GTGTCCTCTCTCATTGGCTTTCCAG-3′ for human dystrophin Dp71 (157 bp); forward: 5′-GGTTGGCAGTCAAACTTC, reverse: 5′- TGGGGAG GACTCAGAAG ATCT-3′ for mouse dystrophin Dp71 (157 bp); and forward: 5′-ACGGATTTGGTCGTATTGGG-3′, reverse: 5′-CGCTCCTGGAAGATGGTGAT-3′ for GAPDH (214 bp).
Immunoblotting
Cells were centrifuged and resuspended in RIPA buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 % Triton X-100, 1 % sodium deoxycholate, 0.1 % sodium dodecyl sulfate (SDS), 0.25 % sodium ortho-vanadate, 50 mM sodium fluoride, 1 mM EDTA, and 1 μg/mL leupeptin). Homogenates were sonicated, and protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce, USA). An equal amount of whole-cell protein extracts (60 μg) was mixed with Tris–glycine SDS sample buffer, and proteins were denatured by boiling for 3 min. Lysates were then separated by 10 % SDS polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to nitrocellulose membranes. Membranes were incubated for 1 h in TBS-T (150 mM NaCl, 10 mM Tris–HCl, pH 8, 0.05 % Tween 20) containing 6 % low-fat dried milk and then incubated overnight with the corresponding primary antibody. After three washes with TBS-T, horseradish peroxidase-conjugated antirabbit or antimouse IgG was used as the secondary antibody. The immunoreactive bands were visualized using 3,3′-diaminobenzidine (Boster Biological Technology, Wuhang, China). An anti-GAPDH antibody was used to normalize for equal amounts of proteins and calculate the relative induction ratio.
Bioinformatics analysis of the mouse promoter region
Dp71 promoter region of mouse (GenBank accession number: NT-039706.8) was analyzed by the TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess) program to predict transcription factor binding sites.
Electrophoretic mobility shift assay and super shift analysis
Electrophoretic mobility shift assay (EMSA) was performed on nuclear extracts from HeLa cells according to the manufacturer’s instructions (LightShift Chemiluminescent EMSA Kit, PIERCE). The extract preparation and binding reaction were performed as previously described (Zhang et al. 2011). DNA probes were generated according to the HSE sites at positions −840 to −817 bp of the mouse Dp71 promoter as double-stranded, HRP-labeled oligonucleotides corresponding to the wild-type sequences (5′- actgttagTTCTAGAAggacattt -3′) and mutant sequence (5′- actgttagCAGTATGGggacattt -3′). Specific binding was confirmed by competition experiments with 200-fold excess of unlabeled mutant oligonucleotides (mutant cold probe); 200-fold excess of unlabeled, identical oligonucleotides (cold probe) were also added to prove the binding specificity. The nuclear extracts were incubated with oligonucleotides before being separated on a non-denaturating PAGE gel. Target bands were detected by enhanced chemiluminescent (ECL) assay kit (Pierce).
Luciferase reporter assay
For the luciferase reporter assay, exponentially growing HeLa cells were seeded in 24-well culture dishes. Transfection of HeLa cells was carried out according to the manufacturer’s instructions (LIPOFECTAMINE 2000™, Invitrogen). Each transfection experiment contained 0.5 μg of reporter plasmid with 0.5 μg of transcription factor expression vector and with 20 ng of PRL-TK-Renilla vector (Promega) as an internal transfection control. Luciferase activities were measured with the dual luciferase system according to the manufacturer’s instructions (Promega). Transfections were performed in triplicate. A LUMIstar Luminometer (BMG, Germany) was used to quantify light signals. The full-length pGL3-Dp71, pGL3-Dp71-Mut, and pGL3-Dp71-del reporter construct (the construction process was described in “Plasmids”) activities were normalized by comparison with activity from the PRL-TK-Renilla vector.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) analysis was made following a protocol provided by Beyotime Biotech (Shanghai, China). Precipitated DNA samples were analyzed by PCR to detect the HSF1 binding site at position −1.0 kb within the Dp71 promoter (GenBank accession number: NT-039706.8), the sequences of the PCR primers used in the PCRs were as follows: 5′-ATGACAGCTTCTGATATGAGTC- 3′ (sense) and 5′-GAGACAGGGTCTCTCACATAGTTC-3′ (antisense).
Statistical analysis
Data in the figures and text were expressed as the means ± standard errors of the mean. Each experiment was performed at least three times, and statistical analysis was performed using a two-tailed Student’s t test or Fisher’s least significant difference test; otherwise, representative data were shown. P < 0.05 was considered significant.
Results
Dp71 mRNA and protein expression were reduced in HSF1-KO mice
In order to determine whether Dp71 is under the transcriptional control of HSF1, protein and messenger RNA (mRNA) expression were analyzed in HSF1-KO and WT mice (HSF1+/+ mice of the same age, strain, and sex were used as controls) in various tissues. Quantitative real-time polymerase chain reaction (QRT-PCR) revealed that Dp71 mRNA level expressed in brain, lung, kidney, liver, and spleen was statistically reduced in HSF KO mice (Fig. 1a). Western blot also revealed that the Dp71 protein expression of brain, lung, liver, spleen, and kidney was reduced compared with wild-type mice (Fig. 1b, c). All these data suggest that the knockdown of HSF1 expression can affect the expression of Dp71 in vivo.
Fig. 1.
Dp71 mRNA (a) and protein expression (b) in brain, lung, kidney, liver, and spleen in HSF1 knock out (KO) and wild-type (WT) mice. c Statistical analysis of protein expression in b, *vs HSF1+/+, P < 0.01, n = 5
Overexpression of the constitutively activated HSF1 mutant plasmid enhances Dp71 expression in HeLa cells
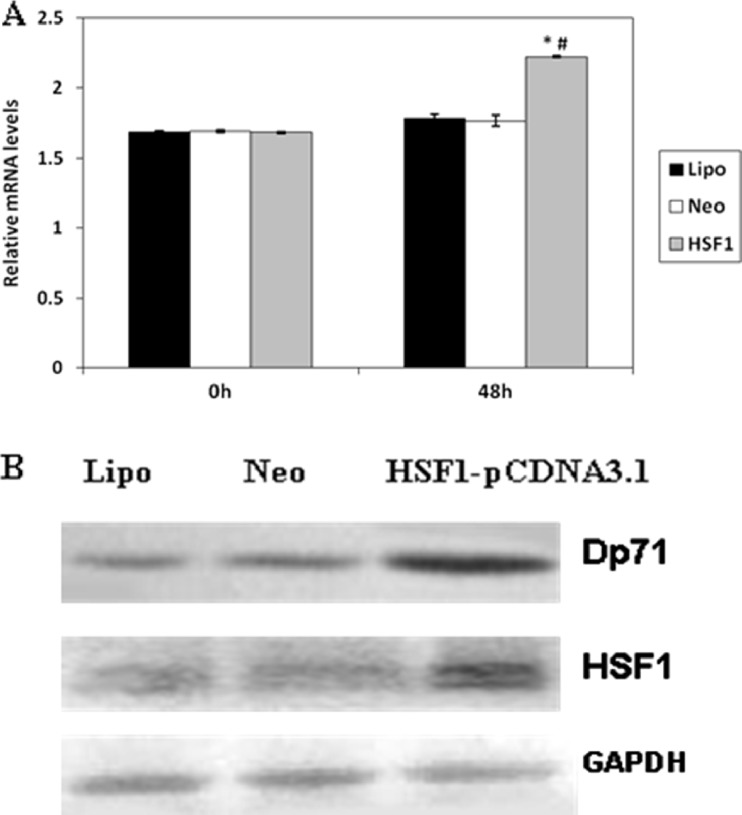
HeLa cells were used in our experiment to further test whether HSF1 regulated the expression of the endogenous Dp71 gene. Then, pcDNA3.1 (+)/HSF1 (+), which deletes amino acids 203–315 that are located within the trimetric domain while maintaining the DNA binding activity and transcriptional activity in the absence of thermal stress, was overexpressed in HeLa cells via transfection. Forty-eight hours after transfection, overexpression of exogenous HSF1 was detected by Western blot (2B). QRT-PCR and Western blot were used to analyze the expression of Dp71, as displayed in Fig. 2a and b; overexpression of HSF1 alone increased both Dp71 mRNA and protein levels.
Fig. 2.
Over-expression of HSF1 can increase both the Dp71 mRNA and protein expression in HeLa cells. Relative Dp71 mRNA level was analyzed via QRT-PCR after transfection of HSF1-pCDNA3.1(HSF1, vector pcDNA3.1 (Neo), and lipofectamine (Lipo) in HeLa cells (a). The Dp71 mRNA between HSF1 and Neo, HSF1 and Lipo 48 h after transfection were statistically different; *P < 0.05, vs Neo group; # P < 0.05, vs Lipo group. There was no statistical difference between the Neo and Lipo group. HSF1 and Dp71 protein expression in HeLa cell (b) 48 h after transfection were displayed
HSF1 knockdown decreases Dp71 expression in HeLa cells
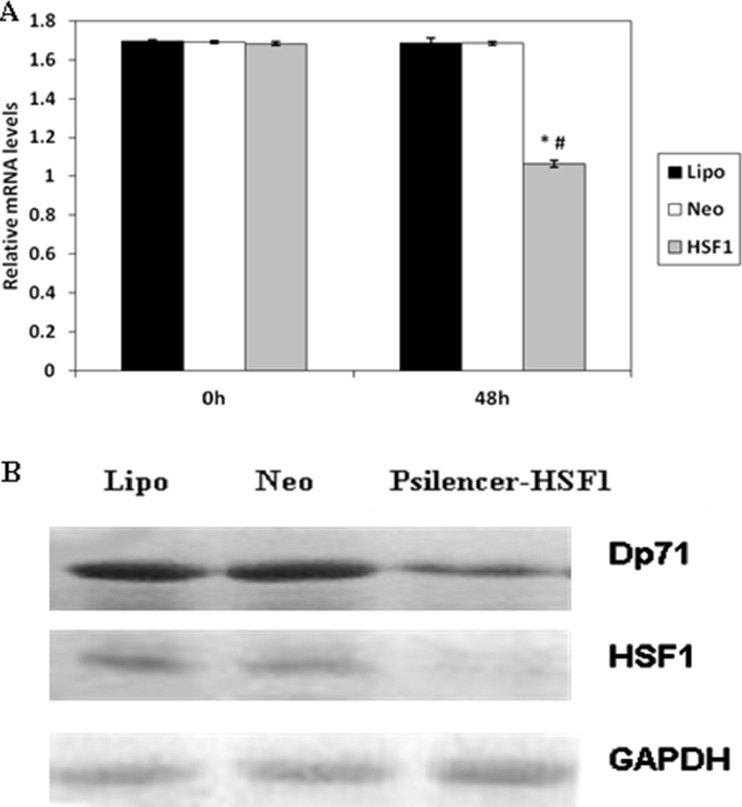
A psilencer1.0-u6 construct expressing specific siRNA targeting HSF1 transcripts (psilencer-HSF1) (Shunmei et al. 2010) was used to further test whether HSF1 is an endogenous regulator of Dp71 expression. Sub-confluent HeLa cells were transfected with psilencer-HSF1 or empty vectors. Western blot was carried out for identification of endogenous HSF1 inhibition 48 h after the transfection. After the expression of HSF1 was inhibited, the expression of Dp71 was determined by quantitative real-time polymerase chain reaction and Western blot. As shown in Fig. 3b, the level of Dp71 protein in the HSF1-KO cells decreased significantly compared with the group transfected with control vectors (Ctrl) and compared with the group treated only with lipofectamine (Lipo) under the normal conditions. As expected, the Dp71 mRNA in HSF1-KO cells was dramatically reduced by half in control groups under normal conditions (Fig. 3a). These results further suggested that HSF1 can regulate Dp71 expression.
Fig. 3.
HSF1 knockdown suppressed the expression of Dp71 in HeLa cells. Relative Dp71 mRNA level in HeLa cell (a) was analyzed via QRT-PCR after transfection of psilencer-HSF1 plasmids (HSF1), psilencer plasmid (Neo) and lipofectamine (Lipo). The Dp71 mRNA expressions between HSF1 and Neo, HSF1 and Lipo were statistically different 48 h after transfection; *P < 0.05, vs Neo group; # P < 0.05, vs Lipo group. There was no statistical mRNA expression difference between the Neo and Lipo group. HSF1 and Dp71 protein expression were also analyzed in HeLa cells (b)
HSF1 activates the Dp71 promoter
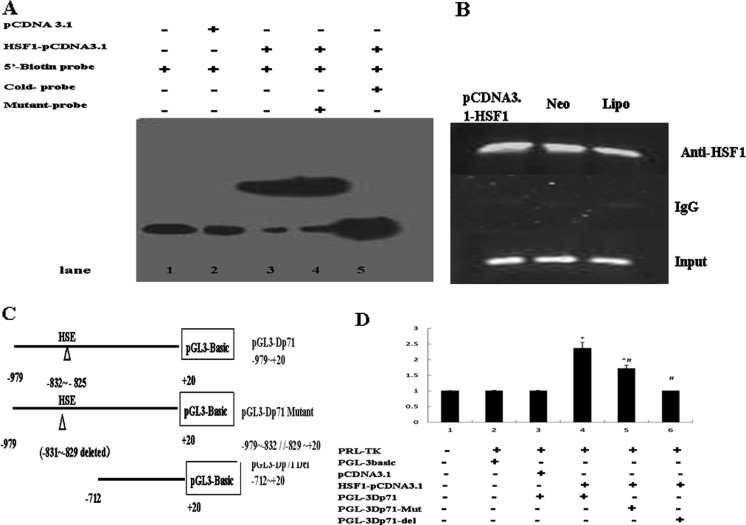
EMSA was used to determine whether the potential HSF1 binding element found at −840 to −817 bp of the promoter region of Dp71 was capable of binding to HSF1. As shown in Fig. 4a, the biotin-labeled probe designed according to the Dp71 promoter could bind to the HSF1 protein in the nuclear extract of HeLa cells. Specificity of binding was verified using mutant cold oligonucleotides, which failed to compete for binding with HSF1. These data provide direct evidence that HSF1 is able to bind to the promoter region of Dp71.
Fig. 4.
Transcription activity of HSF1 on Dp71 promoter. The potential HSF1 binding sites of Dp71 promoter were assessed by EMSA (a), CHIP (b), and dual luciferase reporter system (d). a PcDNA3.1: the vector control group; HSF1-pcDNA3.1: HSF1 over-expression group; cold probe: competition with cold probe (200-fold excess concentration); mutant probe: competition with mutant cold probe (200-fold excess concentration). b ChIP analysis of HSE/Dp71 from Raw 264.1 cells transfected with HSF1-pcDNA3.1 or Neo: the vector control group, and lipofectamine (Lipo). Prior to immunoprecipitation using antibody specific for HSF1, input was used as an internal control; mouse IgG was used as negative control. These figures were the representative of three independent experiments. c Schematic composition of Dp71 promoter constructs. d The constructs depicted in c were used, the ability of HSF1 to regulate the promoter activity of each Dp71 construct was assessed by transient transfection studies. *P < 0.05, statistically significant vs the vector control group. # P < 0.05, statistically significant vs the Dp71 construct group (−979 to +20)
To further understand how HSF1 induces Dp71 transcription, we assessed its effect on Dp71 promoter activity via luciferase reporter assay. For this purpose, we used three different fragments of the mouse Dp71 promoter (Fig. 4c) linked to the promoterless coding domain of the luciferase gene. The reporter construct was then co-transfected with either the control pcDNA3.1 or the pcDNA3.1 (+)/HSF1-wt expression vector into HeLa cells. Figure 4d showed that HSF1 was able to activate the promoter activity of Dp71 at levels (above twofold) compared with the control vector. This trans-activation, however, was almost completely lost upon further deletion up to the −712 to +20 bp construct. These data suggest that the critical regulatory element necessary for mediating HSF1 effects on the Dp71 promoter lies within the −979 to −712 bp region. A mutant construct with three nucleotides (−831 to −829) of the HSE core sequences deleted was also constructed to further test the transcriptional effect of HSF1. As is revealed in Fig. 4d, deletion of partial HSE can reduce the transcription to a significantly lower level. Together with the results of bioinformatics analysis and EMSA, it is suggested that the HSF1 DNA binding sites at −712 to −979 bp play a major role in the regulation of Dp71 promoter activity by HSF1.
To substantiate the activity of this HSE in vivo, we performed a ChIP assay using a specific antibody against HSF1. Normal goat IgG was used as a negative control. DNA associated with the chromatin immunoprecipitated by these antibodies was then amplified by PCR with primers specific for the putative HSE region of the Dp71 promoter. As expected, almost no DNA fragments were detected when normal IgG was used. By contrast, anti-HSF1 antibody specifically precipitated the Dp71 promoter fragment. In addition, we found that transfection of constitutively active HSF1 mutant significantly increased the amount of Dp71 promoter PCR fragments (Fig. 4b). These findings together with EMSA results strongly support that the −979 to −712 sequence of Dp71 gene contains a high affinity HSE for HSF1.
Discussion
For the first time, in vivo and in vitro evidence is provided supporting HSF1’s function as a transcriptional factor for Dp71. Dp71 mRNA and protein expression in various tissues were significantly reduced in HSF1 KO mice. In HeLa cells, overexpression and knockdown HSF1 expression was performed via transfection of constitutively active HSF1 mutant plasmid pcDNA3.1 (+)/HSF1 and HSF1-specific siRNA construct. Dp71 mRNA and protein expression were increased or reduced significantly in HeLa cells after the expression of HSF1 was changed. To further clarify the relationship between HSF1 and Dp71, EMSA studies indicated that HSF1 could bind the HSE at −840 to −817 bp of Dp71 with high affinity, and the specific bands in EMSA only showed up in the presence of both HSF1 and the binding site at −840 to −817 bp of Dp71 gene. A luciferase reporter assay showed that HSF1 activated significantly the transcription of the Dp71 gene. Deletion or mutation of the HSE at −840 ∼ −817 bp could reduce or abolish the Dp71 promoter activity induced by HSF1. A ChIP assay provided the in vivo evidence for the transcriptional regulation of Dp71 by HSF1. To sum up, our experiment demonstrated that HSF1 was capable of binding the promoter region of the Dp71 gene and promoting the transcription of the Dp71.
For many years, HSF1 has been regarded as the master regulator of the heat shock process via regulation of heat shock protein expression. The classic responses of HSF1 under environmental stressors are conserved from yeast to humans. Current research of HSF1 revealed that its functions are far more versatile than serving as stress regulators. Previous research found that yHSF (HSF1 in yeast) binds many genes involved in a wide-range of core cellular functions even at basal temperatures (Hahn and Thiele 2004). These transcriptional targets allow yeast not only to adapt to environmental contingencies but also to modulate metabolism and maintain proliferation under normal growth conditions. HSF1 functions as a critical regulator of longevity in some organisms (Chiang et al. 2012; Volovik et al. 2012). Consistent with their findings, recent work indicates that HSF1 helps cells accommodate the protein homeostasis that underlie many human diseases, especially those associated with aging (Morimoto 2008). Cancer programs have revealed that HSF1 can regulate energy metabolism, cell cycle signaling, DNA repair, apoptosis, cell adhesion, extracellular matrix formation, and translation by regulating a series of multifunctional proteins. The broad biological functions of HSF1 suggest that it has more downstream targets other than that of heat shock proteins.
Evidence from HSF1 knockout mice has proved that the aberrant transcriptional regulation of HSF1 plays an important role in the pathogenesis and pathophysiology of neuropsychiatric disorders (Uchida et al. 2011; Zhu et al. 2008; Xiao et al. 1999). The hsf1-null mice exhibit a reduction in basal anxiety levels and exploratory behavior and working memory deficits. It has been shown that the transcription factor HSF1 is essential to neurogenesis and spinogenesis in the dentate gyrus of the hippocampus and to the development of normal emotional and social behavior (Uchida et al. 2011). As one of the most widely expressed isoforms of dystrophin (Jin et al. 2007), Dp71 forms the dystrophin-associated protein complex (DAPC) in non-muscle tissues via association with dystroglycans, sarcoglycans, dystrobrevins, syntrophins, and accessory proteins. Mental retardation and retinal dysfunction are the two main phenotypes closely associated with Dp71 involved in DMD. Dp71-null mice display selective behavioral disturbances characterized by reduced exploratory and novelty-seeking behavior (Daoud et al. 2008), and exhibit high similarity with the HSF1−/− mice. As the most richly expressed isoforms in the central nervous system, Dp71mRNA and protein have been identified in different brain structures and neuron cell types. In the tissue differentiation process, relatively high Dp71 promoter activity was detected in the central and peripheral nervous systems, especially in the forebrain area, which gives rise to the hippocampus (Gorecki and Barnard 1995). Loss of Dp71 affects synaptic maturation and function, resulting in deficits in selective cognitive functions. Via association with the multi-protein scaffolds that cluster glutamate receptors and organize signaling proteins required for synaptic transmission, the reduced expression of Dp71 in HSF1 knockout mice may contribute to neuropsychiatric disorders in these animals.
HSF1 is found to be activated in a wide variety of malignant cells (Vydra et al. 2013; Santagata et al. 2011 ). The commonly held view is that HSF1 exerts a broad influence on cancer simply by allowing cells to manage the imbalances in protein homeostasis that cause malignancy. The pattern of DNA occupancy, however, differs fundamentally from the HSF1 program induced by thermal stress. The cellular processes HSF1 regulates in cancer including energy metabolism, cell cycle signaling, DNA repair, apoptosis, cell adhesion, and extracellular matrix (Marc et al. 2012). As the most ubiquitously expressed isoforms of dystrophin, Dp71 participates in various cellular processes, including cell adhesion, water homeostasis, and cell division in PC12 cells (Fort et al. 2008; Acosta et al. 2004). Compelling evidence supporting Dp71 and DAPC components in the nucleus suggests its possible biological function via the continuous network linking the plasma membrane to the nuclear envelope (Villarreal-Silva et al. 2011). Current research in our group found that the up-regulation of Dp71 can increase the proliferation, invasion, and mobility capability of the normal HBE cells (data not shown). Concerning the rewired transcriptome of HSF1 in cancer, the enhanced expression of Dp71 regulated by HSF1 probably plays an important role in different phenotypes of tumors.
In conclusion, our findings concerning the transcriptional regulation of HSF1 of Dp71 may have important biological significance in exploring classical and newly expanded biological functions of HSF1. Our project dealing with the relation between HSF1 and Dp71 sheds light on the investigation, clinical observation, and treatment of HSF1 related biological functions.
Acknowledgments
This study is supported by the National Natural Science Fund of China (Grant No. 30800550) and Hunan Natural Science Fund of China (Grant No.10JJ4016). pcDNA3.1/HSF1 (+) expressing construct was kindly provided by Dr. Richard Voellmy at HSF Pharmaceuticals S.A., Switzerland. Hsf1 gene knockout mice were from Dr. Ivor Benjamin (Medical College of Wisconsin, Milwaukee. WI, USA).
Footnotes
J. Tan and S. Tan contributed equally to the paper.
References
- Acosta R, Montañez C, Fuentes-Mera L, Gonzalez E, Gómez P, Quintero-Mora L, Mornet D, Alvarez-Salas LM, Cisneros B. Dystrophin Dp71 is required for neurite outgrowth in PC12 cells. Exp Cell Res. 2004;296(2):265–75. doi: 10.1016/j.yexcr.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Austin RC, Howard PL, D’Souza VN, Klamut HJ, Ray PN. Cloning and characterization of alternatively spliced isoforms of Dp71. Hum Mol Genet. 1995;4:1475–1483. doi: 10.1093/hmg/4.9.1475. [DOI] [PubMed] [Google Scholar]
- Bar S, Barnea E, Levy Z, Neuman S, Yaffe D, Nudel U. A novel product of the Duchenne muscular dystrophy gene which greatly differs from the known isoforms in its structure and tissue distribution. Biochem J. 1990;272:557–560. doi: 10.1042/bj2720557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. 2012;148:322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians ES, Yan LJ, Benjamin IJ. Heat shock factor 1 and heat shock proteins: critical partners in protection against acute cell injury. Crit Care Med. 2002;30(1 Supp):S43–S50. doi: 10.1097/00003246-200201001-00006. [DOI] [PubMed] [Google Scholar]
- Daoud F, Candelario-Martínez A, Billard JM, Avital A, Khelfaoui M, Rozenvald Y, Guegan M, Mornet D, Jaillard D, Nudel U, Chelly J, Martínez-Rojas D, Laroche S, Yaffe D, Vaillend C. Role of mental retardation-associated dystrophin-gene product Dp71 in excitatory synapse organization, synaptic plasticity and behavioral functions. PLoS One. 2008;4(8):e6574. doi: 10.1371/journal.pone.0006574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort PE, Sene A, Pannicke T, Roux MJ, Forster V, Mornet D, Nudel U, Yaffe D, Reichenbach A, Sahel JA, Rendon A. Kir4.1 and AQP4 associate with Dp71- and utrophin-DAPs complexes in specific and defined microdomains of Müller retinal glial cell membrane. Glia. 2008;56(6):597–610. doi: 10.1002/glia.20633. [DOI] [PubMed] [Google Scholar]
- García-Sierra F, González E, Mornet D, Cisneros B. Dystrophin Dp71 in PC12 cell adhesion. Neuroreport. 2005;16(3):235–238. doi: 10.1097/00001756-200502280-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecki DC, Barnard EA. Specific expression of G-dystrophin (Dp71) in the brain. Neuroreport. 1995;6(6):893–896. doi: 10.1097/00001756-199504190-00017. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem. 2004;279:5169–5176. doi: 10.1074/jbc.M311005200. [DOI] [PubMed] [Google Scholar]
- Jin H, Tan S, Hermanowski J, Böhm S, Pacheco S, McCauley JM, Greener MJ, Hinits Y, Hughes SM, Sharpe PT, Roberts RG. The dystrotelin, dystrophin and dystrobrevin superfamily: new paralogues and old isoforms. BMC Genomics. 2007;8:19. doi: 10.1186/1471-2164-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Hauser M, Brown V. Chamberlain JS expression of the 71 kDa dystrophin isoform (Dp71) evaluated by gene targeting. Brain Res. 1999;830(1):174–178. doi: 10.1016/S0006-8993(99)01201-9. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–62. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, Ince TA. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci U S A. 2011;108(45):18378–83. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarig R, Mezger-Lallemand V, Gitelman I, Davis C, Fuchs O, Yaffe D, Nudel U. Targeted inactivation of Dp71, the major non-muscle product of the DMD gene: differential activity of the Dp71 promoter during development. Hum Mol Genet. 1999;8(1):1–10. doi: 10.1093/hmg/8.1.1. [DOI] [PubMed] [Google Scholar]
- Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu CJ, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shunmei E, Zhao Y, Huang Y, Lai K, Chen C, Zeng J, Zou J. Heat shock factor 1 is a transcription factor of Fas gene. Mol Cells. 2010;29(5):527–31. doi: 10.1007/s10059-010-0065-4. [DOI] [PubMed] [Google Scholar]
- Tanabe M, Nakai A, Kawaoe Y, Nagata K. Different thresholds in the responses of two heat shock transcription factors, HSF1 and HSF3. J Biol Chem. 1997;272:15389–15395. doi: 10.1074/jbc.272.24.15389. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Fujimoto M, Otsuki K, Yamagata H, Hobara T, Abe N, Higuchi F, Shibata T, Hasegawa S, Kida S, Nakai A, Watanabe Y. Impaired hippocampal spinogenesis and neurogenesis and altered affective behavior in mice lacking heat shock factor 1. Proc Natl Acad Sci. 2011;108(4):1681–6. doi: 10.1073/pnas.1016424108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal-Silva M, Centeno-Cruz F, Suárez-Sánchez R, Garrido E, Cisneros B. Knockdown of dystrophin Dp71 impairs PC12 cells cycle: localization in the spindle and cytokinesis structures implies a role for Dp71 in cell division. PLoS One. 2011;6(8):e23504. doi: 10.1371/journal.pone.0023504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volovik Y, Maman M, Dubnikov T, Bejerano-Sagie M, Joyce D, Kapernick EA, Cohen E, Dillin A. Temporal requirements of heat shock factor-1 for longevity assurance. Aging Cell. 2012;11:491–499. doi: 10.1111/j.1474-9726.2012.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vydra N, Toma A, Glowala-Kosinska M, Gogler-Piglowska A, Widlak W. Overexpression of Heat Shock Transcription Factor 1 enhances the resistance of melanoma cells to doxorubicin and paclitaxel. BMC Cancer. 2013;13:504. doi: 10.1186/1471-2407-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang M, Wang Q, Liu M, Liang Q, Zhang H, Xiao X. HSF1 regulates expression of G-CSF through the binding element for NF-IL6/CCAAT enhancer binding protein beta. Mol Cell Biochem. 2011;352(1–2):11–7. doi: 10.1007/s11010-010-0624-1. [DOI] [PubMed] [Google Scholar]
- Zhu X, Cheng M, Peng M, Xiao X, Yao S, Zhang X. Basal behavioral characterization of hsf1 deficient mice and its cellular and behavioral abnormalities underlying chronic unpredictable stressors. Behav Brain Res. 2008;193(2):225–229. doi: 10.1016/j.bbr.2008.05.024. [DOI] [PubMed] [Google Scholar]