Abstract
Recently developed classification methods have enabled resolving multiple biological structures from cryo-EM data collected on heterogeneous biological samples. However, there remains the problem of how to base the decisions in the classification on the statistics of the cryo-EM data, to reduce the subjectivity in the process. Here, we propose a quantitative analysis to determine the iteration of convergence and the number of distinguishable classes, based on the statistics of the single particles in an iterative classification scheme. We start the classification with more number of classes than anticipated based on prior knowledge, and then combine the classes that yield similar reconstructions. The classes yielding similar reconstructions can be identified from the migrating particles (jumpers) during consecutive iterations after the iteration of convergence. We therefore termed the method “jumper analysis”, and applied it to the output of RELION 3D classification of a benchmark experimental dataset. This work is a step forward toward fully automated single-particle reconstruction and classification of cryo-EM data.
Keywords: cryogenic electron microscopy, single-particle reconstruction, iterative classification, class number, Bayesian agglomerative clustering, maximum a posteriori, ribosome
1. Introduction
Over the past three decades, cryogenic electron microscopy (cryo-EM) and single-particle three-dimensional (3D) reconstruction techniques have evolved into a powerful toolbox for determining biological macromolecular structures. There have been advances in computational tools, e.g. automated data collection (Suloway et al., 2005, Mastronarde et al., 2005) and the design of an image processing pipeline (Lander et al., 2009, Langlois et al., 2014, Scheres, 2012a), as well as significant improvements in hardware, particularly the direct electron detector (Milazzo et al., 2011, Bammes et al., 2012). Single-particle reconstruction for cryo-EM often deals with a large number of 2D images of biological macromolecules (“particles”). It employs automated particle selection, 2D alignment (transformation of the images, including in-plane rotation and translation, to bring them into optimal superimposition), and an iterative process of projection alignment and 3D reconstruction. In its original form (1990s - 2000s), single-particle reconstruction required a homogeneous sample, in which all particles represent identical copies of the macromolecule (see Frank, 2010). Because the signal-to-noise ratio (SNR) of individual particles is low, it needs to be increased by averaging multiple particles, but averaging is applicable only if the particles represent the same view of replicas of macromolecules. Therefore, the original single-particle reconstruction method was limited by sample heterogeneity, i.e., particles in a sample representing compositionally and conformationally different biological macromolecules (Frank, 2006).
Recently developed classification methods have enabled resolving multiple structures/conformations of the macromolecules from cryo-EM data obtained from heterogeneous biological samples (e.g. Scheres et al., 2007, Fischer et al., 2010, Agirrezabala et al., 2012). The classification methods can be divided into two categories, supervised and unsupervised methods (Frank, 2006). Supervised classification utilizes two or more 3D density maps as references, and separates the particles based on their similarities to these references. Therefore, the supervised classification methods pose the danger of reference bias: In the extreme case, images of mere noise can result in an averaged image resembling the reference (Shaikh et al., 2008). Unsupervised classification, in contrast, groups the particles based on their mutual similarities. Although a low-resolution 3D map may be needed for the initial 2D alignment, unsupervised classification methods are largely immune to the reference bias problem.
One important improvement in the classification methods is treating the class assignment of each particle (i.e., which structure/conformation of the macromolecule a particle represents) as a probability distribution among classes, instead of making an all-or-none class assignment. This idea forms the basis of the maximum-likelihood (ML)-based classification methods (Yin et al., 2003, Scheres et al., 2005, 2007, 2009). ML methods aim to find the values of a set of parameters which maximize the likelihood function of the parameters, given the observed data. In the case of cryo-EM and single-particle reconstruction, these parameters include each of the voxels of the 3D density map for each class, the class assignment, the projection angle relative to the 3D map, and the 2D rotation and translation of each particle image. The ML estimator is an intuitive and popular point estimator. However, because cryo-EM datasets are finite in size, noisy (SNR ~ 0.1 for low-dose exposure) (Baxter et al., 2009) and lack the projection angle information, the multi-parameter ML estimator for cryo-EM data is susceptible to the over-fitting problem, i.e., treating noise as signal erroneously (Scheres, 2012b).
To address the over-fitting problem, one can instead use the maximum a posteriori (MAP) estimator, a Bayesian approach to statistics. MAP estimation considers the experimenter’s belief (prior knowledge) as well as the likelihood function of the parameters. In Bayesian statistics, the set of parameters is considered a quantity subject to variation that can be described by a probability distribution, called the prior distribution. The prior distribution is updated in light of the observed data to yield the posterior distribution, which is proportional to the product of the likelihood function and the prior distribution. As the size of the observed data increases, the MAP estimator gives more weight to the sample information, and less weight to the prior information (Casella and Berger, 2001).
The Expectation-Maximization algorithm is particularly well suited to find the ML/MAP estimator for a mathematically incomplete problem such as 3D reconstruction and classification of cryo-EM data, because cryo-EM data lack the information of class assignment and the projection angle for every particle. The Expectation-Maximization algorithm is based on the idea of alternately optimizing the set of parameters and the set of missing data (or hidden variables), while fixing the values of the other set. This optimization is performed iteratively, with its limit being the ML/MAP estimator for the original problem (Casella and Berger, 2001). The Expectation-Maximization algorithm for cryo-EM classification and 3D reconstruction has been implemented in ML3D (Scheres et al., 2007) and MLn3D (n stands for normalization) (Scheres et al., 2009) to find the ML estimator, and in RELION (REgularized Likelihood OptimizatioN) (Scheres, 2012b,a) to find the MAP estimator.
It has remained a question how to base the decisions made in the course of classification on the statistics of the data. The above-mentioned classification methods all can possibly give reliable solutions, if performed properly. The pit-fall, however, is that they all involve various amounts of subjective decisions made by researchers with various degrees of experience. Subjective decisions may be involved in many steps of 3D classification, such as particle selection, particle alignment, 3D reconstruction, and filtering. The employment of subjective decisions can limit the use of these methods by inexperienced researchers, and should therefore be minimized. RELION has set a good example for reducing user discretion (Scheres, 2012b), although the user is still responsible for choosing the number of classes, number of iterations of the Expectation-Maximization algorithm, and the initial reference volume.
In this work, to further curb the role of subjective decisions in classification, we propose the jumper analysis based on the statistics of cryo-EM particles, to determine the iteration of convergence and the number of distinguishable classes. Specifically, the iteration of convergence, i.e., from which point onwards the 3D reconstructions become trustworthy and stable for user examination, is indicated primarily based on the probability distribution of all the particles over the itera tions. The classes yielding similar 3D reconstructions are indicated by the migration behavior of the particles, i.e., change in class assignment, that occurs after the iteration of convergence. As we will show, this migration information can provide reliable criteria for determining which classes of particles represent the same conformation of the biological macromolecule, and can therefore be combined to obtain a better 3D reconstruction. We demonstrate the jumper analysis method by using the output of RELION classification on a well-characterized experimental cryo-EM dataset. Evidently, this analysis method can also be applied to other iterative classification schemes, e.g. the iterative classification algorithm implemented in FREALIGN (Lyumkis et al., 2013).
2. Methods
2.1. Image Formation Model for 3D Reconstruction and classification
Assume we collected N particles from a heterogeneous cryo-EM sample containing K structures. These K structures, v1, v2, …, vK, differ from one another in composition and/or in conformation. Each particle xi, i = 1, 2, …, N, is a 2D projection of one of the 3D structures vki, ki ∈ {1, 2, …, K}. According to the weak-phase-object approximation (WPOA), the image formation model in Fourier Space is:
| (1) |
where:
Xi is the 2D Fourier transform of xi, i = 1, 2, …, N. Xij is the j-th component of Xi, j = 1, 2, …, J and J = D2. D is the number of pixels in one dimension.
CTFij is the j-th component of the contrast transfer function (CTF) of particle Xi, which is assumed to be constant in the 3D classification and reconstruction process.
Vki is the 3D Fourier transform of the 3D structure vki, ki ∈ {1, 2, …, K}. Vkil is the l-th component of Vki, l = 1, 2, …, L and L = D3.
The operation for all j extracts a central slice of Vki at orientation φi. According to the projection-slice theorem, this operation is equivalent to the real-space projection operation. φi is the Fourier-space equivalent of the real-space position parameter, which comprises the 3D rotation and 2D translation of particle xi relative to vki in real space. For convenience, φi is referred to as the orientation of Xi.
Nij is the noise in Fourier space. Commonly used Wiener filters assume that the noise is independent and Gaussian distributed with mean 0 and variance .
The goal of 3D classification and reconstruction is to find a solution for the model of 3D structures with parameter set Θ = {V1, V2, …, VK }, given the observed data X = {X1, X2, …, XN }. In Bayesian statistics, we find the MAP estimate of the parameter set which maximizes the regularized likelihood:
| (2) |
where p(X|Θ) is the conditional probability, or likelihood, of observing the data X given the parameter set Θ. p(Θ) is the prior probability distribution of the parameter set.
However, 3D classification and reconstruction is a mathematically incomplete problem, because cryo-EM data lack the information on class assignment and the orientation of every particle. Such missing information can be treated as hidden variables, and integrated out in the solution of the model. Let φ = {φ1, φ2, …, φN } denote the orientation information of X. Let z = {z1, z2, …, zN } denote the class assignment of each particles, where zi = k if xi is a projection of vk. The true values of (z, φ ), denoted as (), are unknown. Furthermore, we treat the class assignment of each particle as a probability distribution among all K classes, instead of an all-or-none class assignment. Let cik = p(zi = k|Xi, Θ) denote the conditional probability of xi being a projection of vk given the parameter set Θ, where cik ≥ 0 and . We define the most-probable class assignment for a particle Xi as:
| (3) |
We can also estimate the probability of any one particle being a projection of vk using , where the indicator function I(ẑi = k)= 1, if ẑi = k; or 0, if ẑi ≠ k.
2.2. Iterative classification and Convergence
For a mathematically incomplete problem like 3D classification and reconstruction, the Expectation-Maximization algorithm is suited to find its MAP estimator. In each iteration, this algorithm alternately optimizes the parameter set Θ and the set of hidden variables {z, φ}, while fixing the values of the other set. Generally, this algorithm converges to a local optimum, which depends on the initial values of the parameters. RELION has successfully implemented the Expectation-Maximization algorithm to find the MAP estimator (Scheres, 2012b,a). The reader is referred to these papers and our summary of the method (Shen et al., 2014) for the detailed explanation of the algorithm. Thereafter we use the output of RELION 3D classification to demonstrate our jumper analysis of iterative classification.
We define the iteration of convergence as the smallest number of iterations from which onwards the 3D classification and reconstruction results become stable. In practice, we look for the iteration after which the likelihood function p(X|Θ) reaches a plateau, because the optimization criterion is p(X|Θ)p(Θ), and p(Θ) is negligible compared to p(X|Θ) with a large dataset. Notably, after the iteration of convergence, the orientation distribution for most particles should be close to the delta function, so only a few of orientations need to be considered when calculating the particle statistics (Scheres, 2012a).
2.3. Agglomerative classification after the Iteration of Convergence
In this section, we propose a way to identify classes that should be combined by analyzing the change in most-probable class assignment of the particles after the iteration of convergence. The rationale is the following: if there is a sizeable portion of the particles commuting between two classes after the iteration of convergence, then these two classes are likely to have similar 3D reconstructions and can be combined. This prediction is validated both by visually examining the 3D reconstructions and calculating the difference map between each pair of 3D reconstructions. The particles in the classes with similar reconstructions can then be combined to obtain a better-quality reconstruction. We use a simplified, ideal situation to explain the rationale of this approach.
2.3.1. Ideal Situation
Assume at iteration t after the iteration of convergence t*, the 3D reconstructions from the K classes, V1,t, V2,t, …, VK,t are distinguishable from each other at the current resolution. Then each particle Xi has a distinct class assignment, ẑi,t = z̃i, and cik,t = 1, if k = z̃i; or 0, if k ≠ z̃i. Thus Xi only contributes to one reconstruction Vz̃i,t. Let Gk,t = {Xi| ẑi,t = k} denote all the particles with the most-probable class assignment being k at iteration t. Then the fraction population of class k is ĉk,t = # Gk,t /N, where # Gk,t denotes the number of elements of Gk,t. Furthermore, if each of the K classes is homogenous, then the selected number of classes K equals the maximum number of distinguishable classes K* in the dataset.
2.3.2. Situation with Jumper Particles
Since we do not know K* from the start, in practice we usually perform the 3D classification multiple times, each time starting with the number of classes that is supposed to be greater than or equal to K*. When the selected number of classes K is greater than K*, at iteration t after t*, there will be at least two classes with reconstructions indistinguishable at the current resolution, apart from translation and rotation offsets. For simplicity, we consider the situation where only two classes, r, s ∈ {1, 2, …, K}, r < s, have identical reconstructions, and the other (K − 2) classes have reconstructions distinguishable from the rest classes. Then for Xi ∈ {Xi| z̃i = r or z̃i = s}, cir,t + cis,t = 1, and cik,t = 0, if k ≠ r, s.
Looking at consecutive iterations after t*, if ẑi,t ≠ ẑi,t+1, we call Xi a jumper particle from class ẑi,t at iteration t to class ẑi,t+1 at iteration (t + 1). We can approximate the most-probable class assignment ẑi,t by using , which is an output of the RELION program. This is because after the iteration of convergence, the orientation distribution for most particles is close to the delta function, and therefore .
Furthermore, we can generalize the jumper particle analysis to multiple iterations in tandem after the iteration of convergence, and among any pair of classes. For iterations t1 through t2, t* ≤ t1 < t2, let , then ĉik,t1~t2 is another estimate of cik that is less dependent on the choice of iteration t.
Let Gr→s,t = {Xi|ẑi,t = r and ẑi,t+1 = s} denote all the particles with the most-probable class assignment being r at iteration t AND being s at iteration (t + 1), r, s ∈ {1, 2, …, K}, t ≥ t*. We use a transition matrix, defined as , to indicate the probability of having jumper particles in each class, where the element of the K × K matrix Mt, msr,t = # Gr→s,t /# Gr,t. In practice, the transition matrix is a sparse matrix, i.e., most of the elements off the diagonal have values close to zero, because most classes are distinguishable from the others.
We then rearrange TMt1~t2 into At1~t2, a K × K sparse matrix with approximate minimum degrees using an agglomerative method (Amestoy et al., 1996, 2004). The class numbers [1, 2, …, K] in TMt1~t2 are reordered into [q1, q2, …, qK ] in At1~t2, where qi ∈ {1, 2, …, K}, i = 1, 2, …, K. After rearrangement, the classes that share a sizeable portion of jumper particles can be distinguished (Algorithm 1) by choosing an empirical cutoff value of 1/3 (i.e., the number of jumper particles is half the number of particles staying in these two classes), and these classes may yield similar reconstructions and be combined. Moreover, we can reduce the dimension of transition matrix data into a 2D bar diagram, where , to visually examine the effectiveness of grouping the classes that have commuting jumper particles.
Algorithm 1.
Identify classes that share a sizeable portion of jumper particles.
3. Results and discussion
3.1. Benchmark experimental data of 70S ribosome
We used a standard benchmark cryo-EM dataset of 70S ribosome (Baxter et al., 2009) to illustrate the procedure of the jumper analysis. The benchmark dataset contains 10,000 ribosome particles, among which 5,000 were classified by supervised classification as 70S ribosome containing elongation factor G (EF-G), and the other 5,000 as 70S ribosome containing no EF-G in the original work. The 70S ribosome containing no EF-G was observed to be in a classical, non-rotated global conformation, and contains three tRNAs in the aminoacyl (A), peptidyl (P), and exit (E) sites. The 70S ribosome containing EF-G was observed to be in a rotated global conformation (i.e., the small subunit (30S) is rotated relative to the large subunit (50S), compared to the non-rotated global conformation), and also contains an E-site tRNA. Scheres performed RELION 3D classification on this benchmark dataset, and discovered a small class of particles representing the 50S subunit, demonstrating the intrinsic capability of non-supervised classification method, such as RELION, to detect unanticipated class(es) in a heterogeneous dataset (Scheres, 2012b).
We chose K = 6 as the number of classes to start the RELION 3D classification, because we knew that there are at least three classes: 50S subunit, 70S containing EF-G, and 70S containing three tRNAs but no EF-G, and wanted to be able to accommodate potential new classes. We ran the classification for 60 iterations to demonstrate how to determine the iteration of convergence.
3.2. Determining the iteration of convergence
The purpose of determining the iteration of convergence is two-fold: (1) to determine when to stop the iterative classification, and (2) to save the effort of manually examining the reconstructions of all classes from every iteration. We first note that the sum of likelihood functions of all classes reaches a plateau after a certain iteration, in this example iteration 24 (Fig. 1a). In practice, we wrote a MATLAB function to examine the change of the sum of likelihood function, to determine the iteration of convergence. If the change is less than 5% of the sum of likelihood function for 5 consecutive iterations, we deem that the classification has converged at the first of the five consecutive iterations. In addition, the likelihood function of each class is less ideal than the sum of likelihood functions of all classes for determining the iteration of convergence, because the likelihood function of different classes may reach a plateau after different iterations, and have more fluctuations than the sum of likelihood (Fig. 1b).
Figure 1.
Likelihood function and number of particles as a function of iteration number. (a) The sum of likelihood functions of all six classes in each iteration. The value of likelihood is in arbitrary unit (a.u.). Note that the sum of likelihood functions of all classes reaches a plateau after iteration 24. (b) The likelihood function of each class in each iteration. Class 1, yellow; Class 2, green; Class 3, blue; Class 4, purple; Class 5, gray; Class 6, magenta. (c) The number of particles of each class in each iteration. Color scheme is the same as in panel (b).
3.3. There are jumper particles after the iteration of convergence
The number of particles of each class may fluctuate even after the iteration of convergence, as exemplified in Fig. 1c. After iteration 24, Class 5 and Class 3 contain about 200 and 500 particles, respectively, much fewer than the other four classes, which each contains 2, 000 ~ 3, 000 particles. Furthermore, the numbers of particles of some pair of classes may be anti-correlated. From iteration 24 to 60, the correlation coefficient between the numbers of particles of each pair of classes is: ρ1,2 = −0.56, ρ1,3 = −0.45, ρ1,4 = −0.48, ρ1,5 = 0.48, ρ1,6 = −0.62, ρ2,3 = 0.72, ρ2,4 = −0.00, ρ2,5 = −0.74, ρ2,6 = 0.05, ρ3,4 = −0.14, ρ3,5 = −0.81, ρ3,6 = 0.31, ρ4,5 = −0.21, ρ4,6 = −0.01, ρ5,6 = −0.26. As shown in the next section, the correlation coefficient is not a robust indicator to determine which classes may be combined.
There are several reasons for the existence of jumper particles: (1) Projection angle step size is too large. The projection angle in real space is parameterized by three Euler angles (Scheres, 2012a), and the first two Euler angles are discretized using the HEALPix framework (Gorski et al., 2005) to achieve an approximately uniform sampling. As the projection angle step size decreases, fewer particles will fall between the sampled projection angles. Therefore, more particles will find their most-probable class assignment and orientation with high confidence, i.e., having high . (2) Limited number of particles in some class results in low-quality reconstruction of the class, likely because of missing projection angles and/or noisy averaged images in some projection angles. (3) A portion of the particles contain local conformational/compositional differences compared to the averaged 3D reconstruction. Such differences may not be distinguishable at the current resolution, partly due to the small number of particles containing these features.
3.4. Using jumper analysis to find the classes that may be combined
Particles commuting between classes after the iteration of convergence indicate that these classes may have similar reconstructions, and in that case their particles can be combined to yield a better-quality reconstruction. We use the jumper analysis to find the classes that may be combined. To reduce the dependence of the analysis on the choice of iteration, we monitor the change of particle assignment along several iterations after the iteration of convergence, rather than just between two consecutive iterations.
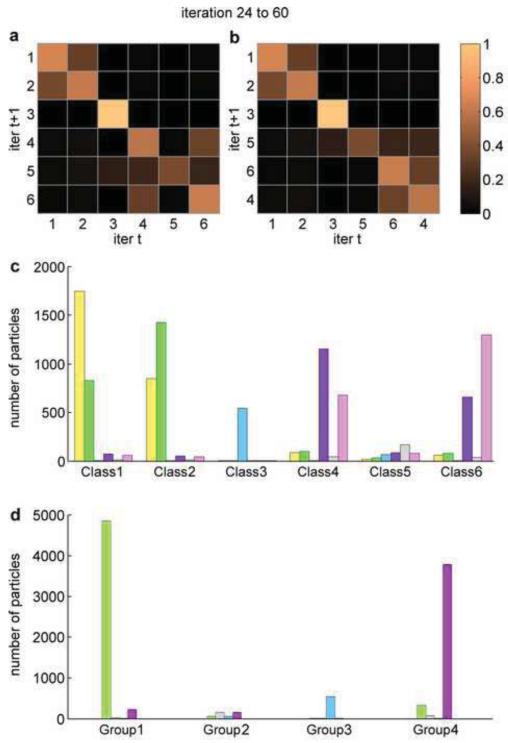
We now introduce the transition matrix to show the fraction of jumper particles in each class along several iterations (Fig. 2a). The elements on the diagonal represent the fraction of the particles that remain in each class in two consecutive iterations, averaged from iteration 24 to 60. The elements off the diagonal represent the fraction of jumper particles between each pair of classes. From this transition matrix, we are able to recognize readily that Class 3 and Class 5 are distinct classes, whereas Class 1 and Class 2 share ~ 40% of particles, and Class 4 and Class 6 share ~ 35% of particles. However, the transition matrix is more difficult to analyze by eye as the number of classes increases. We therefore applied an agglomerative algorithm to reorganize the transition matrix (Amestoy et al., 1996, 2004), so that the high values off the diagonal in the transition matrix are close to the diagonal (Fig. 2b). We then identify the two classes that share the highest portion of particles and combine them, and do this iteratively (Algorithm 1). By choosing a cutoff value of 35%, we combined Class 1 and Class 2 into new Group 1, Class 5 as new Group 2, Class 3 as new Group 3, and combined Class 4 and Class 6 into new Group 4.
Figure 2.
Transition matrix and bar diagram after the iteration of convergence. (a,b) Transition matrix before (T M24~60, panel (a)) and after (A24~60, panel (b)) agglomerative rearrangement, respectively. The order of classes in panel (b) is represented by [q] in the text. Heatmap color scheme from dark to light corresponds to values 0% to 100% in the transition matrix. There are notable size of commuting jumper particles between Class 1 and Class 2, and between Class 6 and Class 4. (c,d) Bar diagram before (c) and after (d) combining classes into new groups. Class 1 and Class 2 are combined into Group 1. Class 5 is assigned as Group 2. Class 3 is assigned as Group 3. Class 4 and Class 6 are combined into Group 4.
To examine the effectiveness of grouping classes, we further reduced the dimension of the data in the transition matrix by using the bar diagram. The bar diagram illustrates the average number of particles that stay in a class, or jump to another class, during the given iterations. The bar diagram in this example (Fig. 2c) clearly shows that Class 1 and Class 2 share a sizeable portion of the particles in these two classes, and so do Class 4 and Class 6, whereas Class 3 shares almost no particles with the other classes, and Class 5 have too few particles to yield a reliable reconstruction. After combining the classes, the new bar diagram (Fig. 2d) shows that the new groups do not share a sizeable portion of particles, indicating successful grouping. Furthermore, the new bar diagram indicates that the new Group 1 and Group 4 may have similar reconstructions, because they still share a detectable number of particles.
3.5. Examining the maps to confirm the jumper analysis results
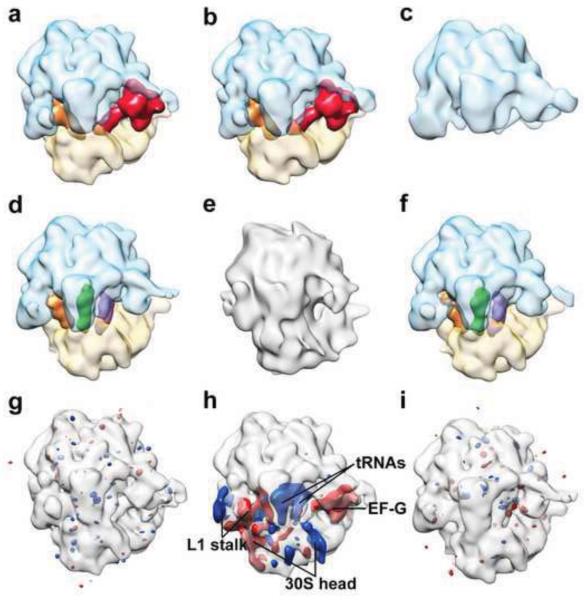
To verify the conclusion of the jumper analysis, we examined the reconstructions of the classes at the iteration of convergence (Fig. 3a-f). The Class 3 map is a ribosome large subunit, distinct from the other classes. The Class 1 and Class 2 maps are both 70S ribosome complexes containing EF-G, different from the Class 4 and Class 6 maps, which are 70S ribosome complexes containing three tRNAs but no EF-G. Class 5 yields a low-quality reconstruction that is not comparable with the other classes. This classification result is in good agreement with the study by Scheres (Scheres, 2012b). However, another study by Lyumkis et al. using FREALIGN identified a class of 70S ribosome complex with strong density for A- and P-site tRNAs and weak density for E-site tRNA, indicating that there may be 70S ribosome lacking E-site tRNA in this dataset (Lyumkis et al., 2013). The fact that such class of 70S containing only A- and P-site tRNAs was not identified in our experiment is likely due to differences in the performance of the classification algorithms with the given choices of classification parameters.
Figure 3.
Cryo-EM map of each class and comparison between maps using difference map. (a-f) Cryo-EM maps of Class 1-6, respectively. Ribosome large and small subunit, transparent blue and transparent yellow, respectively; elongation factor G (EF-G), red; A-, P-, and E-site tRNAs, purple, green, and orange, respectively. The resolutions of Class 1-6 maps are: 18.3 Å, 19.3 Å, 30.5 Å, 19.3 Å, 30.5 Å, 19.3 Å, respectively. (g-i) The difference maps of (Class 1 – Class 2), (Class 1 – Class 4), and (Class 4 – Class 6), respectively, shown at the threshold of ±5× standard deviation of each difference map. The maps being subtracted from are shown in transparent gray for viewing aid. Red mass represents positive difference; blue mass represents negative difference.
Furthermore, we calculated the difference maps between Class 1 and Class 2, and between Class 4 and Class 6, showing that each pair of maps are identical at the current resolution (Fig. 3g,i). In contrast, the Class 4 map is different from Class 1 map (Fig. 3h). The Class 4 map lacks EF-G, but contains additional A- and P-site tRNAs, and its ribosomal L1 stalk and small subunit head are in different conformations compared to the Class 1 map. Therefore, we conclude that the jumper analysis results are consistent with examining the maps visually, and that it can facilitate the identification of classes that have similar reconstructions in the iterative classification of cryo-EM data.
3.6. Computational cost of the jumper analysis
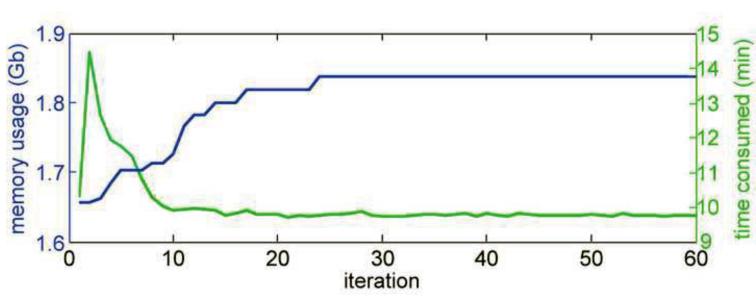
The jumper analysis requires moderate amount of extra computational cost of RELION 3D classification. Fig. 4 shows the memory usage and computational time for the benchmark dataset containing 10,000 particles with 130-pixel window size, on a 104 CPU × 2GB/CPU cluster. The first 24 iterations consumed ~ 40% of the total computational time from iteration 1 to 60, indicating that the jumper analysis, which requires extra iterations of classification after the iteration of convergence, adds ~ 1× of computational cost. The reason for shorter computational time to calculate expectation in the later iterations of RELION 3D classification is mainly due to the “spiky” angular search: in the early iterations, the probability distribution among projection angles for each particle is flat, thus many projection angles need to be considered when calculating the particle statistics; in later iterations, the probability distribution among projection angles becomes sharper, approaching the delta function, therefore only a few projection angles need to be considered.
Figure 4.
Memory usage and computation time to calculate expectation as a function of iteration number. The memory usage (blue) reaches a plateau after iteration 24, the iteration of convergence. The computation time to calculate expectation (green) peaks before iteration 5, and remains low after iteration 10.
3.7. Toward fully automated classification
In this work, we have demonstrated the usefulness of the jumper analysis in an iterative classification of cryo-EM data for determining the iteration of convergence and the number of distinguishable classes. Our method has great potential for further development into a fully automated classification process, which is an essential step toward a fully automated pipeline for cryo-EM and single-particle reconstruction–from data collection, particle picking, to 3D reconstruction and classification. Such a fully automated pipeline will greatly facilitate processing high-quality large datasets collected from direct detection devices, and will make cryo-EM more friendly to less-experienced users.
Acknowledgments
This work is supported by the Howard Hughes Medical Institute and the National Institute of Health Grant R01GM55440 to J.F..
Appendix A. MATLAB Functions
The MATLAB functions for running the jumper analysis are accessible at (http://franklab.cpmc.columbia.edu/franklab/wp-content/uploads/2014/08/JumperAnalysis.zip).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirrezabala X, Liao H, Schreiner E, Fu J, Ortiz-Meoz R, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proceedings of the National Academy of Sciences. 2012;109(16):6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amestoy PR, Davis TA, Duff IS. An approximate minimum degree ordering algorithm. SIAM Journal on Matrix Analysis and Applications. 1996;17(4):886–905. [Google Scholar]
- Amestoy PR, Davis TA, Duff IS. Algorithm 837: AMD, an approximate minimum degree ordering algorithm. ACM Transactions on Mathematical Software (TOMS) 2004;30(3):381–388. [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen D-H, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. Journal of structural biology. 2012;177(3):589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter WT, Grassucci RA, Gao H, Frank J. Determination of signalto-noise ratios and spectral SNRs in cryo-EM low-dose imaging of molecules. Journal of structural biology. 2009;166(2):126–132. doi: 10.1016/j.jsb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella G, Berger RL. Statistical Inference. Duxbury Press; 2001. [Google Scholar]
- Fischer N, Konevega A, Wintermeyer W, Rodnina M, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466(7304):329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-dimensional electron microscopy of macromolecular assemblies: visualization of biological molecules in their native state. Oxford University Press; USA: 2006. [Google Scholar]
- Frank J. The ribosome comes alive. Israel journal of chemistry. 2010;50(1):95–98. doi: 10.1002/ijch.201000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski KM, Hivon E, Banday A, Wandelt BD, Hansen FK, Reinecke M, Bartelmann M. HEALPix: a framework for high-resolution discretization and fast analysis of data distributed on the sphere. The Astrophysical Journal. 2005;622(2):759. [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, Lyumkis D, Potter CS, Carragher B. Appion: An integrated, database-driven pipeline to facilitate EM image processing. Journal of Structural Biology. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois R, Pallesen J, Ash JT, Ho DN, Rubinstein JL, Frank J. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. Journal of Structural Biology. 2014;186(1):1–7. doi: 10.1016/j.jsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. Journal of structural biology. 2013;183(3):377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D, et al. Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Milazzo A-C, Cheng A, Moeller A, Lyumkis D, Jacovetty E, Polukas J, Ellisman MH, Xuong N-H, Carragher B, Potter CS. Initial evaluation of a direct detection device detector for single particle cryo-electron microscopy. Journal of Structural Biology. 2011;176(3):404–408. doi: 10.1016/j.jsb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012a;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. A Bayesian view on cryo-EM structure determination. Journal of molecular biology. 2012b;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo J-M. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature Methods. 2007;4(1):27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Grob P, Nogales E, Carazo J-M. Maximum likelihood refinement of electron microscopy data with normalization errors. Journal of structural biology. 2009;166(2):234–240. doi: 10.1016/j.jsb.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Nuñez R, Sorzano CO, Marabini R, Herman GT, Carazo J-M. Maximum-likelihood multi-reference refinement for electron microscopy images. Journal of molecular biology. 2005;348(1):139–149. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Trujillo R, LeBarron JS, Baxter WT, Frank J. Particle verification for single-particle, reference-based reconstruction using multivariate data analysis and classification. Journal of structural biology. 2008;164(1):41. doi: 10.1016/j.jsb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Chen B, Liao H, Frank J. Quantitative analysis in iterative classification schemes for cryo-EM application. In: Herman GT, Frank J, editors. Computational Methods for Three-Dimensional Microscopy Reconstruction. Applied and Numerical Harmonic Analysis. Springer; New York: 2014. pp. 67–95. [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Yin Z, Zheng Y, Doerschuk PC, Natarajan P, Johnson JE. A statistical approach to computer processing of cryo-electron microscope images: virion classification and 3-D reconstruction. Journal of structural biology. 2003;144(1-2):24. doi: 10.1016/j.jsb.2003.09.023. [DOI] [PubMed] [Google Scholar]