Abstract
The diffusion and drift motion of λ DNA molecules on Au coated membrane surface near nanopores prior to their translocation through solid-state nanopores are investigated using fluorescence microscopy. With the capability of controlling electric potential at the Au surface as a gate voltage, Vgate, the motions of DNA molecules vary dramatically near the nanopores in our observations, presumably generated by electrokinetic flow. We carefully investigate theses DNA motions with different values of Vgate in order to alter the densities and polarities of counterions; which are expected to change the flow speed or direction, respectively. Depending on Vgate, our observations have revealed the critical distance from a nanopore for DNA molecules to be attracted or to be repelled, DNA’s anisotropic and unsteady drifting motions and accumulations of DNA molecules near the nanopore entrance. Further finite element method (FEM) numerical simulations indicate that the electrokinetic flow could explain these unusual DNA motions near metal collated gated nanopores qualitatively. Finally, we demonstrate the possibility to control the speed and direction of DNA motion near or through a nanopore, for example, recapturing a single DNA molecule multiple times with AC voltages on the Vgate.
Keywords: biosensors, DNA, finite element analysis, nanofluidics, nanopores
1. Introduction
Because of the capability of single biomolecule detection and analysis, nanopore has gained much attention since its invention [1–3]. Using nanopore based devices, remarkable results have been published recently on the characterization of double- and single-stranded DNA, RNA, and proteins and their complex [4–7]. Briefly, the principle of single biomolecule detecting by a nanopore device is to measure the ionic current through a nanopore in a membrane. When a long chainlike biomolecule is translocating through a pore driven by an electric field, the presence of the molecule physically blocks a certain percentage of ion transportation and reduces the ionic conduction though the pore, which is often called ionic current blockade. The ionic current blockades generated by biomolecule translocation through a nanopore provide the information of the identification of the translocation molecule, as well as its structure [8–10].
The capture mechanism of biomolecules, especially DNA molecules toward and into a nanopore has been investigated both experimentally and theoretically to comprehend the capture rate and dynamics of DNA molecules approaching a pore [9, 11–15]. Numerous studies have shown that DNA molecules capture toward a nanopore is executed by electrophoretic force generated by an applied bias voltage across the membrane [11–14]. Other phenomena can also affect DNA motion and capturing process, for example, the influence of the electroosmotic flow via a nanopore generated by the drift motion of the anisotropically distributed ions near the nanopore wall induced by surface charge at the wall has been predicted theoretically [15]. In fact, the phenomenon such as the recirculating solution flow induced by electroosmotic flow has been optically observed in 2D micro or nano fluidics [16–18]. These fluid flow phenomena in nanofluidic channels have been intensively studied experimentally and numerically in order to develop reliable high-throughput and cost-effective “lab-on-a-chip” [19–25]. As a result, the profile of the solution flow can be quantitatively predicted numerically. Analogous with these studies, it has been predicted that the occurrence of the complex flows near a nanopore is resulting from an electrokinetic transport phenomenon as moleculer dynamics, numerical simulations and analytical calculations have indicated that the capture rate and the speed of DNA molecules passing through nanopores are strongly influenced by the electroosmotic flow [12, 26–29]. Exmerimentally, the electroosmotic flow in insulating nanopores by the surface charges on the nanopore surface or recently by the presence of the charged DNA molecules were confirmed by measuring the force acting on the DNA molecules [30, 31]. One may want to optically observe such complex flows near a nanopore as observed in the 2D micro or nano fluidics [16–18]. In fact, a few groups including our group have already successfully imaged the capture and translocations of fluorescently labeled DNA molecules optically by fluorescence microscopy [9, 13, 32]. These direct observations have confirmed the DNA translocations into a nanopore and add the insight of the electrophoretic capture mechanism near the nanopore. In fact, we have measured the DNA velocity prior to entering a nanopore under microscope resolution using near 100 nm diameter nanopores [13]. Our results showed that the electric field was approximately spherically symmetric around a nanopore. In addition, DNA molecules stuck in nanopores, called pore clogging, were observed, and counter-intuitively our observations showed that higher bias voltage increased the clogging probability [13]. However, the motion of DNA molecules under the influence of electroosmotic flow near a nanopore has not been directly observed likely because of micrometer-scale spatial resolution of an optical microscope with the limited amount of condensing counter-ions, which are responsible to generate the flow, near the pore wall. To increase the counter-ion density near nanopore wall, metallic nanopore having an externally controllable electric potential is preferred choice. It is because that the metallic nanopore is expected not only to enhance the electroosmotic flow via the pore but also to enable to alter the direction of the flow by simply reversing the polarity of the potential difference between the pore and buffer solution.
This concept, electrically conductive materials, such as metals, for nanopores, is analogous to field effect transistor (FET) in the semiconductor industry, recently it is called as ionic field effect transistor or just nanopore transistor [33–35]. This nanopore transistor, comparing with highly insulating silicon-based materials, such as Si3N4 or SiO2 [2, 36–39] made nanopore, can modify the chemical of the nanopores and their surfaces, resulting in one of recent trends for nanopore devise researches [40–44]. Some of nanopore transistors had demonstrated improved signal to noise ratio in current measurements resulting in the detection of the formation of molecular complexes or aggregates of DNA, RNA and proteins [45–47]. Recently, conductive materials have also been used to fabricate nano electrode gap across a nanopore entrance and attempted to detect novel signals, especially for the purpose to distinguish between DNA bases toward ultrafast and inexpensive DNA sequencing by tunneling current measurements [48–50].
As gate materials, chemically inert Au is attractive for low contact barrier electrodes in aqueous solutions. Au metallization of nanopore has been attempted and successfully coated not only on membrane surface but also hollow of inside of nanopores [51–53]. Interesting current voltage behavior, ion current rectification has already been observed with conical shape nanopores because of Cl ion passivation on Au surface [52]. A different group has demonstrated DNA translocation though Au metallized nanopores and found slow translocation speed suggested by the presence of a strong attractive interaction between DNA and Au surface of nanopore [26, 49, 53–55].
In this article, we will show that modulating the gate potential on nanopore membrane surface strongly influence on the motion of DNA molecules induced by the liquid flow generated by the electrokinetic behavior. For example, accumulation of DNA molecules which has been shown as a result of the electroosmotic flow in the 2D nanofluidic system was observed [55]. Surprisingly, anisotropic and unsteady DNA migrations near nanopores were also observed. Additionally, we will demonstrate the recapture of DNA molecules by applying alternate voltages on the gate electrodes [14].
To understand our observation of DNA motions near electrostatically gated nanopores and their Au surfaces, FEM based numerical simulations were used to estimate both the advection velocities of ionic solutions caused by electroosmosis and the electrophoretic velocities of DNA molecules generated by electric fields to calculate the speed of DNA molecules quantitatively [15, 26, 27, 56, 57]. Our simulation results will successfully reveal that the characteristic DNA motions can be explained by both electrophoresis and advection generated by electric field and electroosmosis, respectively, although further efforts will be necessary to describe all of phenomena observed in this context.
2. Experimental Section
2.1 Fabrication of nanopore transistors
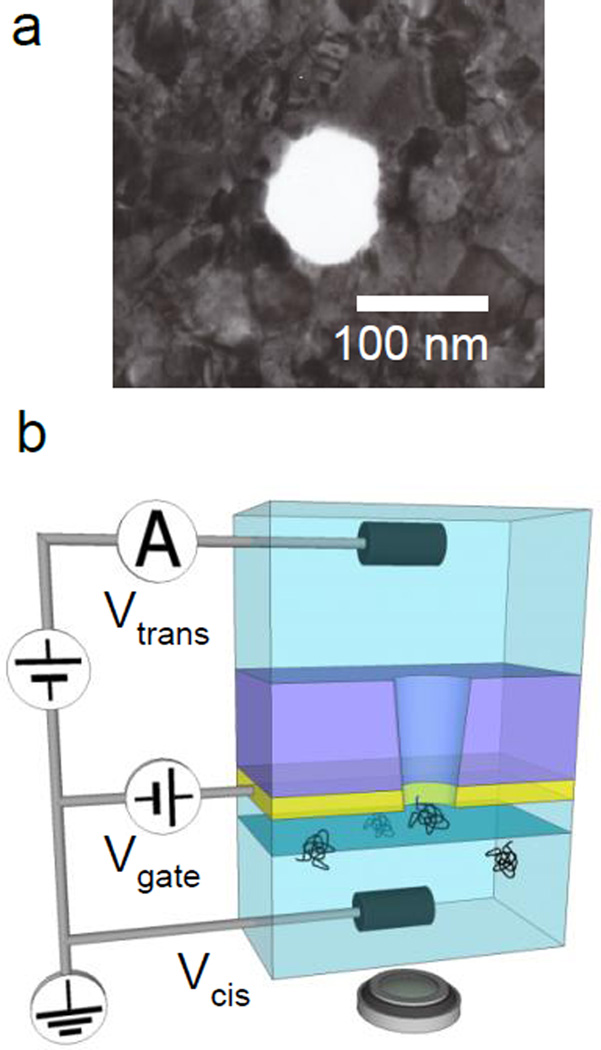
The fabrication procedure of our nanopore transistors is following. A 40 µm × 40 µm freestanding membrane of 200 nm thick silicon nitride supported on 500 µm thick silicon (100) substrate was created by conventional optical photolithography followed by anisotropic wet etching of silicon in KOH solution [1, 10]. A 50 nm thick gold film with 5 nm Cr film as adhesion layer was deposited by thermal evaporation on the silicon nitride membrane as the conductive gate. A nanopore was then milled through both the silicon nitride membrane and the Au/Cr films using focused ion beam (FIB) from the opposite side of Au/Cr evaporation, producing a truncated cone shape pore with near 5° vertex estimated by transmission electron microscopy (TEM) analysis. In this work, 100 nm and 200 nm diameter pores are used and a typical TEM image of the nanopore is shown in Figure 1a. The high contrast variations of the polycrystalline gold films are clearly displayed in the Figure 1a [58]. There is difference in fabrication processes resulting in the structures of nanopores between our Au/Cr/SiN nanopore and Au-metallized nanopore reported previously by Wei et al. Wei’s pore is that pore’s inner wall is entirely coated by thin Au film while our’s are not as shown schematically in Figure 1b. The amplitude of ionic current flowing through nanopores allows us to confirm the diameters of nanopores after completely wetting inside the nanopores. Only the pores producing the same current voltage characteristics for each 100 nm or 200 nm diameter pore were selected and used. As an illustration shows in Figure 1b, Bias voltages, Vcis (= 0 V) and Vtrans are applied via Ag/AgCl electrodes inserted into cis or trans chambers and a gate voltage, Vgate is applied on Au film via an Au wire. During experiments, the voltage differences between ionic solution in cis chamber, Vcis and Au film, Vgate were kept less than 0.5 V since these are directly facing each other to avoid electrochemical etching of the Au film. It has been reported that the electrochemical etching of Au rarely occur at the voltage differences less than 1.0 V [59] as I–V curves through conical Au nanochanels have been measured between ±1 V without etching the Au [52]. For example, higher than 1.4 V’s differences are necessary to etch Au substrate in HCl solution to fabricate a Au tip [60]. Details of this experimental set up containing wiring to the Au film are described in SI.
Figure 1.
(a) a TEM image of Au coated SiN nanopore. High contrast variations of the polycrystalline gold films is clearly displayed. (b) Schematic illustration of the experimental setup featuring the 50 nm thick Au film under the 200 nm thick SiN membrane. A cis chamber below the nanopore where DNA molecules are introduced faces to Au surface. The nanopore’s truncated cone angle is near 5° and the narrower end of the cone is on the cis side. Bias voltages, Vcis (= 0 V) and Vtrans are applied on the AgCl electrodes inserted into cis or trans chambers, respectively. As a gate voltage, Vgate is applied on Au film. The focus plane (a blue plane) and location of a microscope are schematically illustrated. More details of this experimental setup containing the wiring method on the Au film are described in SI.
2.2 Imaging of DNA Molecules
YOYO-1 dye (Molecular Probes), a fluorescent intercalating dye is used to visualize lambda phage DNA molecules. The amount of the dye was for the dye to base pair ratio near 1:10. The final DNA concentration was 1 ng/mL in 0.01 M KCl solution containing, 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. This DNA concentration is low enough not to observe the DNA- DNA interaction in aqueous solution. The estimated ionic strengths are 16 mM for 0.01 M KCl for this buffer solution based on the linear I–V characteristic plots along with the estimated pore diameter by TEM observation. These values are close to those found in previous studies by Tang et al [61]. The mechanical and electrical property changes by attaching YOYO-1 dyes are negligible for our study [62, 63].
2.3 Imaging and Analysis
Sequential images at 70 ms time intervals were taken using an intensified charge coupled device camera (ORCA-ER Hamamatsu Photonics) to trace the individual DNA motion. In this paper, focusing plane for an optical microscope is set at 0.5 µm above Au membrane surface where the microscope produces sharp images of DNA molecules on the surface likely because the radius of gyration of a DNA molecule is near 0.6 µm in solutions with high salt concentrations [61]. At this focusing condition, the location of DNA molecules in the range from surface to 1.5 µm above the surface are able to be extracted including z axis assuming that the center of mass is the location of a unfocused “DNA blob.” More details of the identification of the DNA location can be found in Supporting Information. The conformation of the DNA molecule is ignored and the location of a DNA molecule is determined as the geometric mean of equally weighted pixel positions within the DNA’s outlines. We have developed a MATLAB code for finding the locations of DNA molecules and estimating their velocities between two sequential frames automatically.
3. Results and Discussion
To classify the effects of the controlled gate potential on Au membrane surface on the cis side where DNA molecules are added, the applied gate voltages, Vgate are chosen in three ranges, 3.1. Vgate = Vcis (= 0 V), 3.2. Vgate < Vcis, and 3.3. Vgate > Vcis, as well as 3.4. AC voltage on Vgate. The counter ions to screen the gate potentials are K+ and Cl− for range 3.2. and 3.3., respectively. With the electric field generated by the voltage difference across the nanopores, these ions with different sign can alter the direction of the electroosmotic flow through a nanopore which can be shown by our numerical simulations in this work.
3.1. Vgate = Vcis (= 0 V), Vtrans = 0.3 V
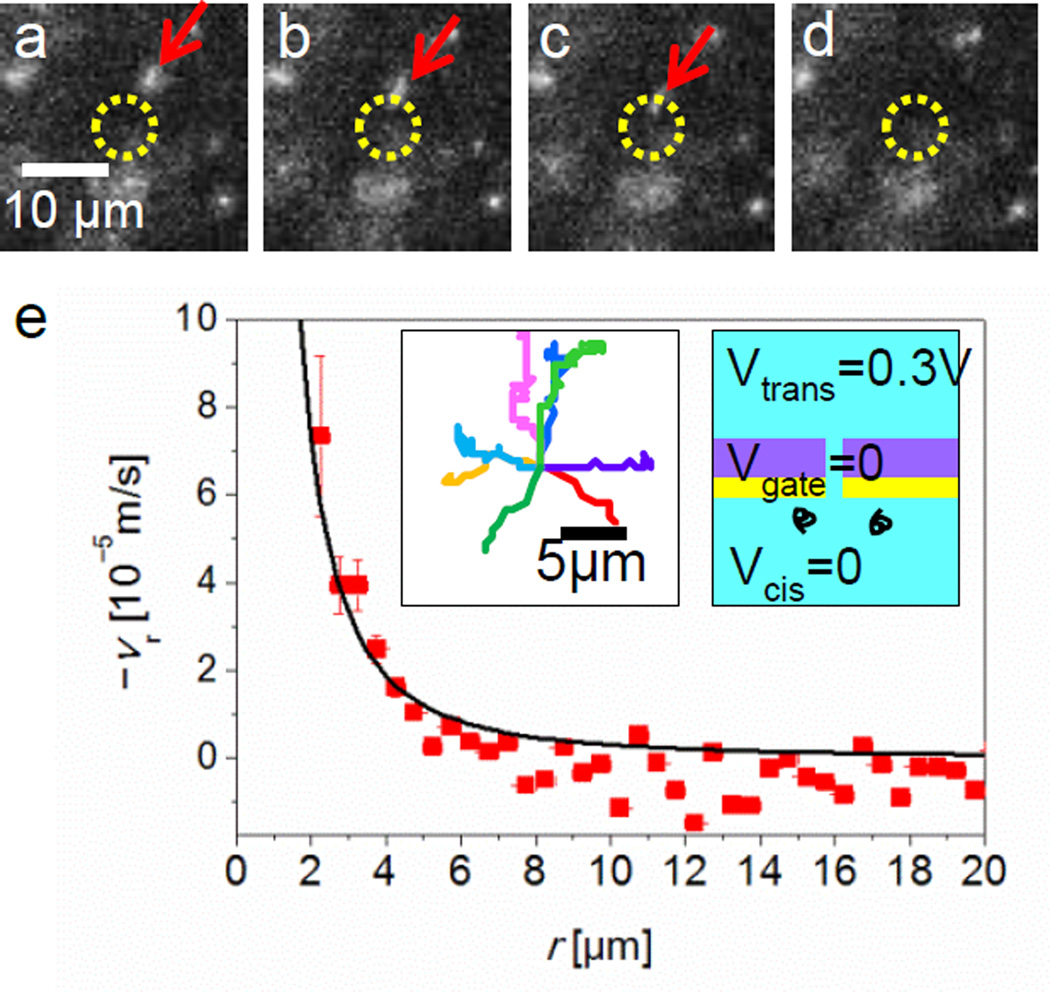
Figure 2a–c shows three examples of fluorescence microscope images of DNA molecules with fluorescent dyes (see Supporting Information Supplementary Movie S1). These excerpts from sequential images taken at a 14 Hz frame rate show the motion of DNA molecules as red arrows. Each image depicts a DNA molecule moving toward a 100 nm diameter pore that is marked by a yellow circle except in Figure 2d the molecule disappeared at the pore presumably as a consequence of DNA translocation. Since Vgate = Vcis, where the ion’s concentration distribution is likely uniform near the gate surface, no liquid flow by electroosmosis is expected. In this case, the drafting behavior of DNA molecules similar to our previous observation: the electric field near a nanopore generates the electrophoretic force to drive this DNA motions. Then, the average values of DNA velocities at various r’s can be compared with the velocity calculated by the linear relation between the velocity and the electric field in equation (1). The electric field is estimated by the Ohm’s law in equation (2) using the experimentally measured value of ionic current I though a nanopore. Reasonably, spherically symmetric field and no fluidic motions at this voltage relation, Vgate = Vcis, is assumed [14].
| (1) |
where r is the radial coordinate in a spherical coordinate systems originated at the nanopore. µ is the electrophoretic mobility of the λ DNA molecule [64, 65].
| (2) |
where σ is the electrical conductivity of the ionic solution [61, 66]. By using the equations (1) and (2), vr can be estimated and plotted as a solid curve into the Figure 2e where experimentally measured the magnitude of the average DNA velocity are also plotted by tracing more than 3 hundreds of DNA molecules nearby a nanopore. The solid curve fits our measured velocity values quantitatively well. This result confirms that the above assumption, spherically symmetric field with no fluidic motions near a nanopore at this voltage relation, Vgate = Vcis.
Figure 2.
DNA motions at Vgate = Vcis = 0 and Vtrans = 0.3 V in 0.01 M KCl. (a) – (d) Time resolved fluorescence images focused near membrane surface showing the motions of DNA molecules. Images are extracted at t = 0.00, 0.14, 0.29 and 0.43 sec from a sequence of 1000 frames recorded at 14 Hz. A DNA molecule (red arrow) shows a drifting motion toward a nanopore located at the center in the images, which is shown in yellow circles. (e) The magnitude of the vr, as a function of r on the nanopore membrane surface. A solid curve, calculated value of vr plotted in (e), is based on the simple theoretical model by Ohm’s law with the experimentally measured ionic currents through nanopore. Inset images show typical trajectories of 8 DNA molecules into a nanopore before their translocations, and applied voltages on the trans, cis and gate electrodes.
3.2. Vgate (= − 0.4V) < Vcis (= 0 V), Vtrans = 0.3 V
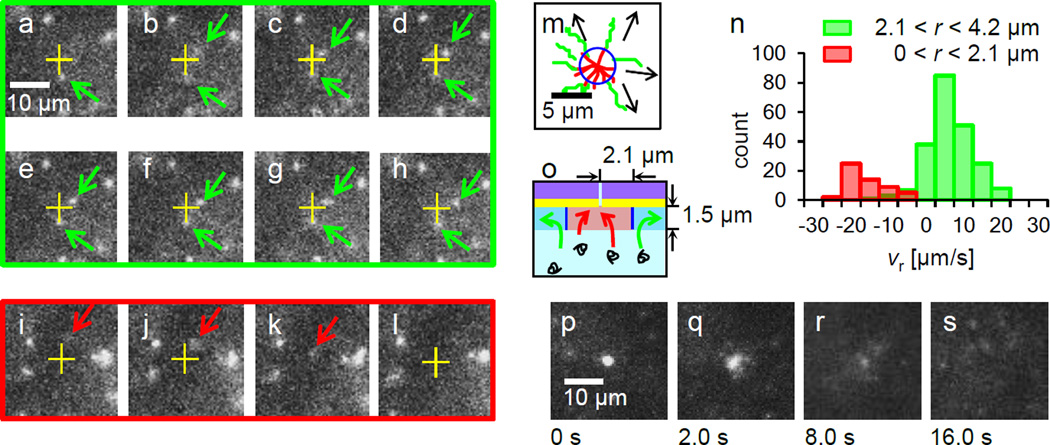
Remarkably, at parameters in the above voltage range, recorded movies (see also Supporting Information Supplementary Movie S2) revealed λ DNA’s motion of both drifting away from and toward the location of a 100 nm diameter nanopore, resulting in translocation as illustrated in Figure 3. Figure 3a–h display 8 sequential fluorescence images showing examples of the motion of two typical DNA molecules at Vgate = − 0.4 V, pointed by green arrows. The two green arrowed DNA molecules observed here did not enter the pore although their locations are r < 3 µm where almost all DNA molecules are expected to enter the pore within a few sequential frame period according to the calculations described in section 3.1.. Instead, one green arrowed DNA molecule above was drifting away from the nanopore by Figure 3h. Meanwhile, a DNA molecule entering the nanopore for its translocation was also observed frequently as a typical example pointed by a red arrow shown in Figure 3i–l. To examine the difference between these two cases, whether DNA molecules move toward and enter the nanopore or not, the examples of DNA’s trajectories near nanopore were plotted in Figure 3m. The green trajectories indicate the DNA molecules drifting away from the nanopore while the red trajectories correspond to entering into the pore. Typically, the speed of drifting away for single DNA molecules is near 7 µm/sec along the green trajectories in Figure 3m while the speed exceeds 20 µm/sec to enter the pore. These trajectories suggest a circular boundary near r = 2 µm as schematically summarized the motions of the DNA molecules near a nanopore in Figure 3o. As a remark, the appearance of the unfocused DNA molecules, ex., Figure 3i and j, corresponds to their Z height above the focusing plane, which is defined by the optics of microscope.
Figure 3.
DNA motions at Vgate = −0.4 V, Vcis = 0 and Vtrans = 0.3 V in 0.01 M KCl. (a)–(h) Images extracted from a sequential 14 frames per second movie showing typical behavior of DNA molecules. One green arrowed DNA below a nanopore (a yellow cross) is nearly at rest while the other green arrowed above is gradually drifting away. From (i) to (l), images are extracted at t = 0.00, 0.14, 0.29 and 0.43 sec. A red arrowed DNA is landing and entering into the nanopore. (m) A drawing of typical DNA trajectories in the vicinity of a nanopore reveals two types of DNA motions. 8 red trajectories present DNA molecules for translocations into the nanopore while 7 green trajectories show the existence of DNA molecules drifting away radially from the nanopore. Arrows indicate the direction of the green trajectories. A blue circle is likely the boundary between these two categories of motions. (n) A histogram of radial velocity of DNA molecules drifting in the ranges 0 <r ≤ 2.1 µm and 2.1 <r ≤ 4.2 µm. Apparently the distribution shows the anisotropic DNA motions in the radial direction. (o) A schematic of the extrapolated two different DNA motions, red and green arrows in the r – z plane near nanopore with the boundary is drawn in blue lines. The highlighted area corresponds to the imaging range by our optical microscope in the z axis. (p)–(s) DNA aggregations near a nanopore at Vgate= −0.4 V, Vcis = 0 and Vtrans = 0.3 V in 0.01 M KCl. (p) Over 20 DNA molecules are aggregated near the nanopore in 120 sec. (q)–(s) The images are taken 2, 8, 16 sec after turning all electrode’s voltages to 0 V at (p) for eliminating electric fields in solution. Free diffusion of DNA molecules from the aggregated DNA is observed in this image sequence. No DNA molecule is left at the nanopore in (s).
To further verify the existence of this r ~ 2 µm boundary of altering the direction of DNA’s drift motions, a histogram of the radial velocity, vr of DNA molecules located in 0 < r ≤ 2.1µm and 2.1 < r ≤ 4.2 µm above z = 1.0 µm is plotted in Figure 3n. Apparently vr in 0 < r ≤ 2.1 µm is mostly inward as the majority of the vr is negative while vr in 2.1 < r ≤ 4.2 µm is appeared to be outward. Based on these experimental observations, there must be equilibrium in DNA motions near r = 2.1 µm where forces to drive DNA molecules are balanced. This behavior has been observed on all pores with both diameters of 100 nm and 200 nm. For the 200 nm diameter pore, the boundary of the radius to alter the directions of the DNA drift motions is increased to near 3.4 µm (see Supporting Information Figure S5).
As described later with numerical simulations in section 3.5, the outward flow near a nanopore membrane is possibly be generated initially by electroosmotic flow inside the nanopore toward the trans chamber (see Supporting Information Figure S4a). This flow will force DNA molecules to drift outward, which is opposite to the inward electrophoretic force via the nanopore.
The observation of DNA aggregation, shown in Figure 3p supports the existence of the counter flow by the electroosmosis as the DNA’s aggregation near the entrance of a 2D fluidic nanochannel was observed with similar experimental parameters to ours where the electrophoretic force acting on DNA molecules and the electroosmotic advection flow were in opposite directions [55]. The DNA aggregation on the top of a nanopore in the Figure 3p can be seen while applying voltages, Vgate ≤ − 0.4 V, Vcis = 0 V and Vtrans = 0.3 V in 0.01 M KCl for 120 sec. This aggregation typically occurs after one DNA molecule was stuck at a nanopore, and then other DNA molecules keep accumulating on the top of the nanopore as one aggregation, near 20 of DNA molecules can be seen in Figure 3p. Since the presence of a negatively charged DNA molecules further enhances the electroosmotic flow in opposite directions to the DNA translocation by electrophoresis [30], it likely generates the area where the speeds of DNA molecules by the advection flow and the electrophoresis are balanced near a nanopore thus generates the DNA stagnation.
This appearance of DNA aggregation is very similar to the heavily DNA clogged nanopore, meaning all DNA molecules are stuck inside of the pore which is frequently observed when the potential difference between Vcis and Vtrans exceeds 0.5 V [13]. To distinguish whether the clogging or aggregating DNA molecules at or near a nanopore, we demonstrate free diffusion by turning to 0 V for all applied trans, cis and gate voltages to eliminate electric fields. As shown in the images extracted at t = 2.0, 8.0 and 16.0 sec from an image sequence in Figure 3q–s after eliminating the fields at t = 0 in Figure 3p, no clogged DNA molecules are observed at the nanopore but all DNA molecules diffuse away in free solution by t = 16.0 of Figure 3s (see also Supporting Information Supplementary Movie S3). In contrast, if the DNA molecules are clogged in a nanopore initially, almost all of the DNA molecules stayed at the nanopore continuously as clogged after eliminating electric fields [13]. Based on previous studies by other research groups, the mean diffusion distances a 48.5 kbp lambda DNA molecule in a 2 D plane are 2.3, 4.5 and 6.4 µm for 2.0, 8.0 and 16.0 sec respectively [64, 67]. Widening the aggregation sizes in Figure 3 is comparable with the average travel distances of the free diffusion above.
3.3. Vgate (= 0.5V) > Vcis (= 0 V), Vtrans = 0.3 V
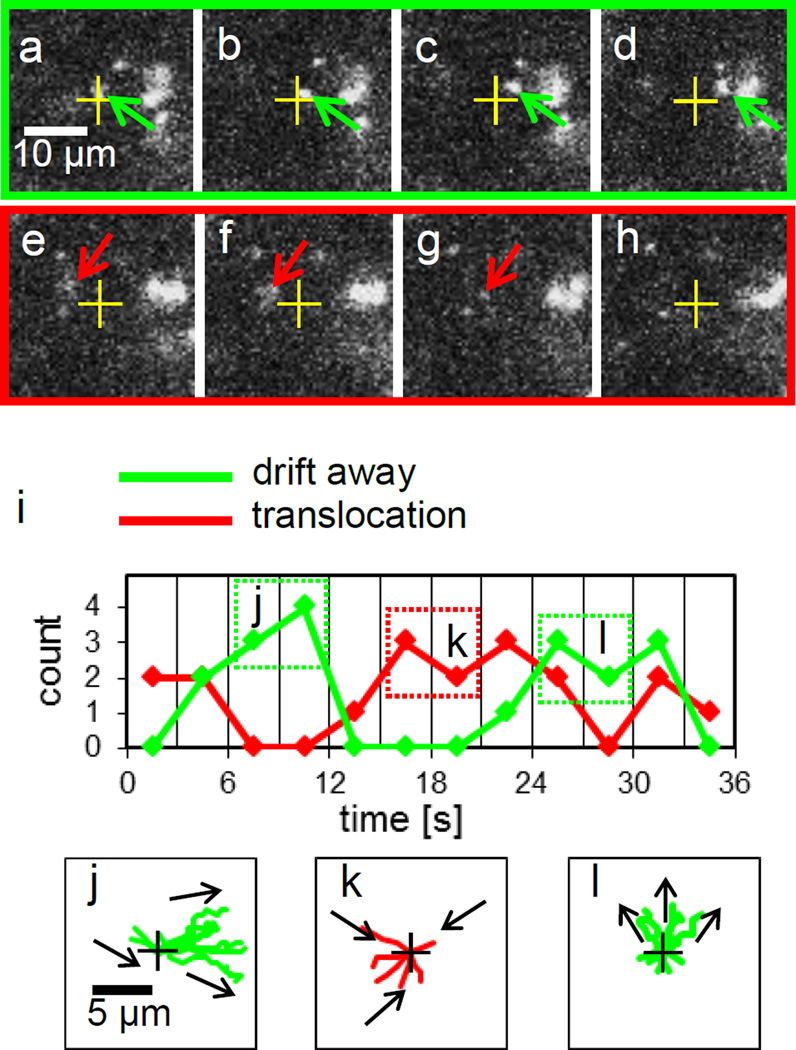
At this Vgate range, the motion of DNA molecules was not steady in time because with the exactly same applied voltages, both the DNA motions moving into and passing by a 100 nm diameter nanopore in the vicinity of the nanopore are observed at different times. Furthermore, the DNA molecules passing by a nanopore eventually drift away from the nanopore, and their directions of the drift are anisotropic, i.e. accumulated DNA molecules are present on the right sides in Figure 4 (see also Supporting Information Supplementary Movie S4). To present these notable DNA motions, sequential snap shots of the typical motions, drifting away shown in Figure 4a–d and toward a nanopore in Figure 4e–h are extracted from a movie. The time intervals between the images presented here are all 0.14 sec. As an example, a DNA molecule (green arrow) appeared at r = 1.6 µm in Figure 4a moving toward right and away from a nanopore, presumably against electrophoretic force, are shown in Figure 4b–d. By contrast, a DNA molecule toward a nanopore for its translocation can be seen in an image sequence of Figure 4e–h. To demonstrate this unsteady motion better, we have counted the number of DNA molecules both going into and drifting away in the vicinity of the nanopore after the DNA molecules entered within the spatial range of r < 2.1 µm, and plotted the number in every 3 seconds in Figure 4i. Apparently, two states of the DNA motions, going into the nanopore and drifting away toward a particular direction exist and switch alternatively near every 10 seconds.
Figure 4.
DNA motions near a nanopore at Vgate = 0.5 V, Vcis = 0 and Vtrans = 0.3 V in 0.01 M KCl. (a) – (h) Images extracted every other frame from a 14 frames per second movie. Time intervals between the sequential images are 0.14 sec. (a) The image near t = 6 sec. One green arrow points a DNA molecule moving away from a nanopore depicted as a cross in the images from (a) to (d). (e) The image near t =15 sec. One red arrow points a DNA molecule moving toward a nanopore in the images from (e) to (g) and then entering the pore before the image (h). (i) Count the number of DNA molecules entering within r < 2.1 µm either translocations (red) or drift away (green) in every 3 seconds. For example, from t = 6 to 12 sec, no DNA molecules enter the nanopore, they all drift away toward right as shown by the 7 DNA trajectories, depicted in the green dotted box j in (i). Instead, all 6 DNA molecules in the red dotted box k on (i), entered the nanopore from t = 15 to 21 sec as their trajectories are plotted in (k). After t = 24, as the green dotted box l depicts, DNA molecules drifting away again. The anisotropic DNA drift motions are shown in their trajectories plotted in (l).
To disclose the anisotropic drift motions in each state, the trajectories of the DNA motions are plotted from t = 6 to 12 sec and 24 to 30 for drifting away and from t = 15 to 21 sec for entering a nanopore for translocation in Figure 4j, k and l, respectively. The trajectories in Figure 4j reveal that all 7 DNA molecules drift toward the right in the plot. The direction of the anisotropic drift DNA motions changes in time during the same experiment, this can be seen as the direction changed toward the top in the plot after t = 24 depicted in Figure 4l. Interestingly, the typical speed of these single DNA molecules to drift away from the nanopore is near 18 µm/sec, which is much faster than the speed of near 5 µm/sec for the DNA’s drifting away motions observed in previous section.
On the other hand, the motion toward a nanopore is appeared to be isotropic in radial directions as depicted in Figure 4k. Apparently the physical conditions at the vicinity of the nanopore in Figure 4j and k are different, likely due to fluidic motions as we will discuss with the results of the numerical simulations. Such asymmetric drift motions of DNA molecules stop instantaneously when the voltage is changed to Vgate = Vcis (= 0) and then all the DNA molecules move toward a nanopore isotropically. This suggests that the asymmetric drift motions are not caused by background flow.
These unsteady and anisotropic drift DNA motions with the two states, entering the nanopore or drifting away have been observed for both 100 nm and 200 nm diameter pores at this voltage relation. However, it tends to “damp” the distinct oscillations between the two states near 200 nm diameter pores (see Supporting Information Figure S6). After switching between the states a few times, DNA molecules mostly enter the 200 nm diameter pores while drifting away occasionally. Further experiments and analyses will be necessary to quantify the unsteady motions by extend the experimental parameters including pore shape, for example, ellipse pores.
3.4. AC on Vgate relative to Vcis (= 0), Vtrans = 0.3 V
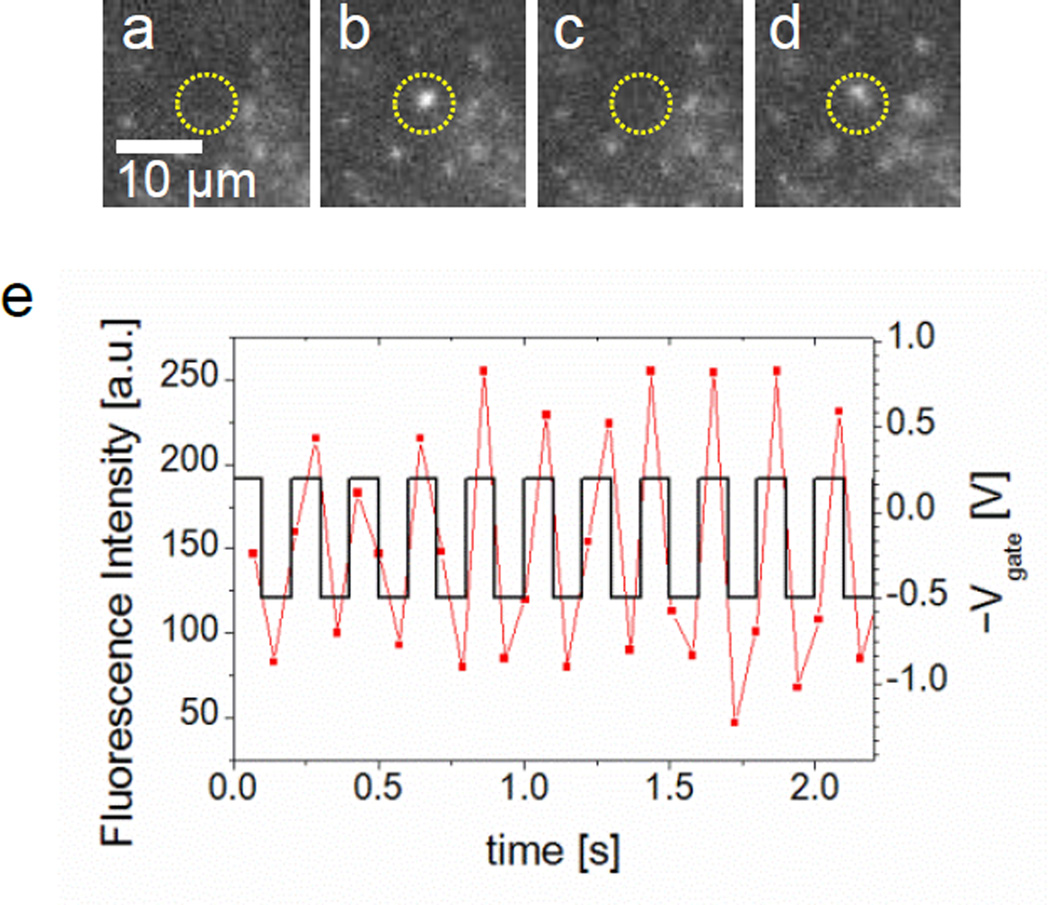
AC voltages on Vgate relative to Vcis were applied with their frequencies from 0.5 to 10 Hz. By optimizing the amplitudes and frequency for the AC voltage, oscillations of single DNA molecules at a nanopore were observed. A yellow circle in each image from Figure 5a–d shows the location of a nanopore and a fluorescence light from a DNA molecule blinks there between the images by applying a 5 Hz square wave with 0.7 Vpp and 0.15 V offset on Vgate resulting in Vgate = −0.2 or 0.5 V, where the number of blinking is maximized (see Supporting Information Supplementary Movie S5).
Figure 5.
A single DNA molecule oscillating in a nanopore by applying a 5 Hz square wave with 0.7 Vpp and 0.15 V offset on Vgate relative to Vcis in 0.01 M KCl. (a) – (d) Image frames with a nanopore marked in a yellow circle are selected at t = 0.79, 0.86, 0.93 and 1.08 sec. A DNA molecule blinking at the nanopore is detected as a DNA appears in (b) and (d) while not in (a) and (c). (e) Fluorescence intensity of the DNA with the square wave is plotted in time. Synchronization between them is identified. Blue circled points are the intensity values for the presented images from (a) to (d) above.
Considering the low concentration of DNA molecules near the nanopore, we believe this blinking is caused by repeatedly oscillating a single DNA molecule between the trans and cis side of chambers via the nanopore synchronized with this optimized AC square wave on the Vgate. Although difficulties to adjust the phase between the DNA oscillation and the AC on the Vgate in time in our experimental setup, one can easily recognized synchronicity by plotting both the fluorescence intensity of a recaptured DNA molecule and the AC voltage vs. time on a single graph as shown in Figure 5e. Typically, a DNA molecule can oscillate more than 10 times between the cis and trans chambers via a nanopore corresponding to 20 times of the translocation by the optimized square wave. This DNA oscillation through a nanopore, named DNA recapturing has been observed by measuring ionic current blockages applying AC between Vcis and Vtrans by Gershow et al [14]. Applying AC voltages on Vgate in our DNA oscillation experiment appears to be some advantages to increase the returning probability. The typical number, 20 times of the consecutive recapturing is nearly equal to the maximum recapturing number in their observation while the frequency, 5 Hz of the DNA oscillation is nearly 10 times slower. However, this 5 Hz is limited by the image recording frequency at 14 Hz for reasonably observation of the DNA oscillations in our observation scheme. Therefore, the recapturing number may increase by applying higher frequency of AC on Vgate. Nevertheless, the slowing down the translocation related process may have some advantages to increase signal to noise ratio for scrutinizing the information of a single DNA molecule.
The driving force to return a DNA molecule from trans to cis side of the chamber via a nanopore during AC voltage applying to the Vgate is likely due to the electric field instantaneously generated by the Au gate electrode on the nanopore membrane. As we have described the DNA motions near a nanopore at various DC Vgate’s relative to Vcis, there are no apparent voltage parameters which can drive a DNA molecule toward the cis side as observed here with the AC voltage on the Vgate. In contrast, DNA molecules can be likely pulled from the trans side of the chamber to the cis side when Vgate > Vtrans.
3.5. Numerical simulations of electrokinetic theory for nanopores
To better understand our experimental observations, here we use a theoretical model that consists of coupled three partial differential equations: the Poisson equation, the Nernst–Planck equation, and the Navier-Stokes equation to describe the ion transport near nanopores [15, 25–27, 30, 55, 68, 69]. The Poisson equation estimates the distribution of the electric potential in the vicinity of the nanopore and the Nernst–Planck equation describes the electric field-driven ion transports through the nanopore. The convective flux term corresponding to the advection is added on this Nernst–Planck equation and the flux speed can be estimated by the Navier-Stokes equation which represents a conservation of momentum in a differential form. Previously, ion transportation via a nanopore with its typical geometry, for example, its diameter less than 10 nm, had been investigated using the finite element analysis (COMSOL Multiphysics) and had successfully revealed the electrokinetics near a nanopore [27, 68, 69]. For our purpose here to observe DNA motions directly under the influence of the electrokinetic phenomena, we used much wider diameter nanopores, e.g. 100 nm and 200 nm, with Au gate electrodes to enhance the electrokinetic effects. Our geometry of the nanopores with Au gate electrodes and the parameters, for example ionic solutions, was accurately specified to do the simulations. As observed previously, the DNA velocity at the vicinity of the 2D channel outlet can be estimated successfully by adding two velocities of electrophoresis and advection [55]. We also estimate the DNA velocity in such a way for the three representative voltage relations 3.1., 3.2. and 3.3. shown in Figure 6 as an example of the estimated velocities of electrophoresis, advection and the addition of both velocities for the Figure 6b is plotted in Figure S4a, b and c, respectively. Details of the parameter values, the nanopore geometry and the boundary conditions can be also found in Supporting Information.
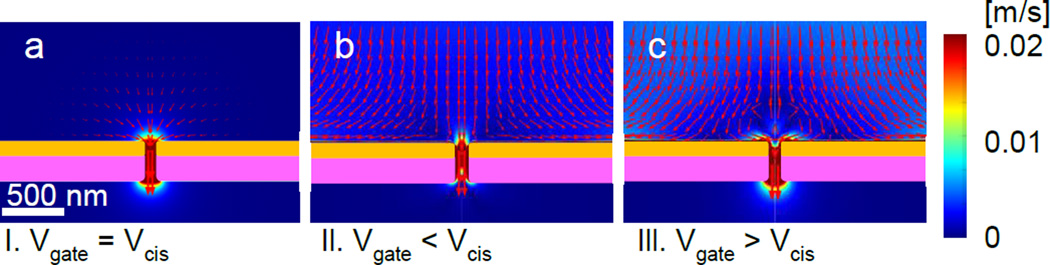
Figure 6.
DNA transport simulations under the voltage relations of our experiments, Vcis= 0 and Vtrans = 0.3 V with various Vgate voltages. Red arrows indicate the direction and the logarithmic scale of the magnitude of the predicted velocities for DNA molecules described in text. (a) 3.1.Vgate = Vcis = 0. Spherically symmetric motions of DNA molecules toward a nanopore can be expected. (b) 3.2. Vgate(= − 0.4 V) < Vcis. The arrows indicated that DNA molecules on the top of the nanopore opening entrance move toward nanopore while DNA molecules near nanopore membrane surface move away from the nanopore. (c) 3.3. Vgate (= 0.5 V) > Vcis. The arrows reveal circulating vortex motions above nanopore opening.
For the condition 3.1. Vgate = Vcis, where no counterions are presumably accumulated in the vicinity of a nanopore wall and membrane surface, our numerical simulations indicate that the DNA motions are predominantly generated by electrophoresis as expected. The ionic current values via nanopore and the DNA velocities near nanopore membrane surface fit to our experimental measured values within the error of our experiments are already presented in Figure 2. Figure 6a shows the direction in red arrows and the magnitude in color scale of the DNA velocity, which depicts the spherically symmetric velocity fields generated by the spherically symmetric potential distribution around the nanopore as its center. This experimental condition 3.1. is appropriate to validate our numerical simulations for the DNA motions when the advection can be ignored.
When 3.2. Vgate < Vcis, interestingly our experimental observation showed both DNA molecules move toward and away from a nanopore whether the DNA molecules are r < 2.1 µm or r > 2.1 µm. Numerical simulations at this voltage relation 3.2. indicate that the direction of the volumetric flow from the nanopore induced by electrokinetic advection by electroosmosis is opposite to the direction by the electrophoresis induced DNA motion as these flow field and DNA velocity field were shown in Figure S4a and b, respectively. The arrow’s lengths corresponding to the speeds in logarithmic scale are multiplied by the same scale factor in order to compare between the magnitudes of the velocities in both Figures. It can be seen that the magnitude of the velocity induced by the electrophoresis exceeds the electrokinetic advection speed inside the nanopore, as calculated by adding both velocities as shown in Figure S4c. As a result, the DNA molecules likely can enter and translocate toward the trans side of the chambers although the DNA speed is slowing down inside of the nanopore. In contrast, the directions of arrows point away from the nanopore in the region z ~ 500 nm, r > 1 µm in Figure 6b where single DNA molecules are imaged in focus indicating that DNA molecules can possibly be observed by moving either toward for their translocations or away from a nanopore depending on r. These results are consistent with our experimental observations qualitatively. Although quantitatively the estimated boundary values, r = 2 µm from the experimental observations and r = 1 µm from the numerical calculations do not agree exactly, the drift away motion of a DNA molecule near a nanopore can be attributed to the electroosmosis. Similar DNA’s drift motions are identified in both numerical and experimental results near a 200 nm diameter nanopore although the boundary values, 3.4 µm by experimental observations and 1 µm by numerical calculations, do not match exactly. Further investigations are necessary to explain this qualitative inconsistency.
The dynamics of DNA molecules near a nanopore in the voltage relation 3.3. where Vgate > Vcis disclosed the unsteady flow motions of DNA molecules moving toward or away from a nanopore alternatively with near 10 seconds. The direction dependent drift motions away from the nanopore were also observed in our experiments. The complex unsteady motions of DNA molecules near nanopore entrance at this voltage relation 3.3. in our experimental observations cannot be explained by turbulence because of the low Reynolds number in nano-micro fluidics that does not generate the turbulence but a predictable laminar flow such as circulating vortex flow [70]. Our simulations are steady and angle independent in cylindrical coordinates because the assumption of geometrical symmetry of the nanopore which would not reproduce our experimental observations from the beginning. However, it may provide us a clue to find the origin that generates this complex fluidic motion. Our simulation results for this voltage relation reveal the creation of a circulating vortex flow above a nanopore hole which is analogous to the previous findings from numerical simulation on hydrodynamic flow near a nanopore with charged surface [27]. This circulating vortex flow near pore entrance has been well described as microvortics in 2D nanofluidics system [17, 18]. However, about direction dependent diffusions of DNA molecules, one possible explanation is that non-circular nanopore or tilted its channel geometry may induce the direction dependent volume flux via nanopore generated by the electroosmotic flow. Further investigations and considerations which contain these effects into the numerical simulations will be necessary. In addition, the presence of the DNA molecules is likely taken into account for the simulations to predict the translocation of DNA molecules in a nanopore channel would generate an osmotic flow found experimentally with DNA molecules inside the pore [30].
The observation of DNA oscillations across a nanopore at AC on Vgate in section 3.4. cannot be explained by our above numerical simulations. Again further simulations will be necessary especially in time dependent manner for this AC cases. However, our numerical simulation indicates the presence of the advection by electro-osmosis and this advection with electrophoresis successfully reproduces our experimentally observed DNA motions at the vicinity of a nanopore. The DNA motions near a nanopore under the 4 different gate voltages presented from section 3.1 to 3.4. are summarized in Table 1.
Table 1.
Summary of the voltage parameters, their relations and the observed DNA motions near nanopores.
| Voltage conditions Vcis = 0, Vtrans = 0.3 V |
Vgatez V |
Description of DNA motions | Notes |
|---|---|---|---|
| Vgate = Vcis | 0 V | Toward nanopore by electrophoretic force | Section 3.1. |
| Vgate<Vcis | − 0.4 V | r < 2.1 µm: Toward nanopore by electrophoretic force r > 2.1 µm: Away from nanopore by advection | Section 3.2. |
| Vgate>Vcis | 0.5 V | Unsteady and anisotropic drift motions: entering nanopore or drifting away | Section 3.3. |
| Vgate ~AC Voltage | 0.15 ± 3.5 V 0.5 ~ 10 Hz | Oscillations at nanopore synchronized with AC Vgate at 5 Hz | Section 3.4. |
We have observed and investigated the DNA motions at the vicinity of voltage gated Au coated nanopores. It will be interesting to extending our studies to modify the surface chemistry of Au including surface charges and hydrophobicity, such as long chain alkane thiols (SAM) in the future. These surface modifications will allow us to observe the interactions of the surface with DNA molecules directly as the non-observing preliminary work is mentioned in the Introduction
4. Conclusions
By using fluorescent microscopy, we have observed DNA motions at the vicinity of a nanopore with different gate voltage, Vgate, controlled on the pore membrane surface. To observe the effects caused by advection generated by electro-osmosis, the DNA motions with three voltage relations, 3.1. Vgate = Vcis, 3.2. Vgate < Vcis and 3.3. Vgate > Vcis are studied. At 3.1. Vgate = Vcis, DNA motions are essentially driven by electrophoresis as expected. The magnitude of velocity of DNA molecules toward the nanopore fits to the estimated velocity with simple Ohm’s law. When 3.2. Vgate < Vcis, where the directions to drive DNA motions by electro-osmosis and electrophoresis are opposite for each other, DNA molecules landing within r < 2.1 µm move toward nanopore for their translocations statistically, while outside of the area, DNA molecules tend to diffuse away from the nanopore. Our numerical simulation explains that the dominant velocity for DNA motion inside and above nanopore is by electrophoresis which, in turn let DNA enter the nanopore from the top of the nanopore. Our simulation also shows the DNA motion drifting away from the nanopore near membrane surface qualitatively. Accumulations of DNA molecules were observed where the magnitude of the DNA velocity by electrophoresis and electro-osmosis is balanced because of the enhanced advection by the presence of DNA molecules. When 3.3. Vgate > Vcis, the DNA motions were very complex, unsteady and asymmetric in angle around the nanopore hole. For example, the translocation toward cis side and trans side of the cambers are alternatively observed about every 10 seconds. This implies that the DNA motions under the influence of advection flow are sensitive to DNA molecules themselves and further numerical investigations by adding the DNA motion will be necessary. Finally, with AC voltages on Vgate (3.4.) to alter the direction of advection, synchronized DNA oscillation across a nanopore has been observed and the directions for DNA molecules to move are the same direction as the advection. To control the direction and the speed of DNA motions by advection as well as by electrophoresis is beneficial for the development of future nanopore based biomolecular sensor devices.
Supplementary Material
Acknowledgements
T.M. acknowledges support from Ministry of Education, Culture, Sports, Science and Technology - Supported Program for the Strategic Research Foundation at Private Universities, 2013–2017. J.L. acknowledges support from NHGRI R21HG004776. The authors are grateful to Dr. Ogata at KESCO Co. for valuable discussions and suggestions related to COMSOL simulations.
Referances
- 1.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 2.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 3.Stein D, Li J, Golovchenko JA. Ion-beam sculpting time scales. Phys. Rev. Lett. 2002;89:276106. doi: 10.1103/PhysRevLett.89.276106. [DOI] [PubMed] [Google Scholar]
- 4.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keyser UF. Controlling molecular transport through nanopores. J. R. Soc. Lond. Interface. 2011;8:1369–1378. doi: 10.1098/rsif.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 7.Wanunu M. Nanopores: A journey towards DNA sequencing. Phys. Life. Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storm AJ, Storm C, Chen J, Zandbergen H, Joanny JF, Dekker C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, Gu JJ, Brandin E, Kim YR, Wang Q, Branton D. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 2004;4:2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 11.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muthukumar M. Theory of capture rate in polymer translocation. J. Chem. Phys. 2010;132:195101. doi: 10.1063/1.3429882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ando G, Hyun C, Li J, Mitsui T. Directly observing the motion of DNA molecules near solid-state nanopores. ACS nano. 2012;6:10090–10097. doi: 10.1021/nn303816w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gershow M, Golovchenko JA. Recapturing and trapping single molecules with a solid-state nanopore. Nat. Nanotechnol. 2007;2:775–779. doi: 10.1038/nnano.2007.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong CT, Muthukumar M. Polymer capture by electro-osmotic flow of oppositely charged nanopores. J. Chem. Phys. 2007;126:164903. doi: 10.1063/1.2723088. [DOI] [PubMed] [Google Scholar]
- 16.Kim SJ, Wang YC, Lee JH, Jang H, Han J. Concentration polarization and nonlinear electrokinetic flow near a nanofluidic channel. Phys. Rev. Lett. 2007;99:044501. doi: 10.1103/PhysRevLett.99.044501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yossifon G, Frankel I, Miloh T. On electro-osmotic flows through microchannel junctions. Phys. Fluids. 2006;18:117108. [Google Scholar]
- 18.Chang HC, Yossifon G, Demekhin EA. Nanoscale Electrokinetics and Microvortices: How Microhydrodynamics Affects Nanofluidic Ion Flux. Annu. Rev. Fluid. Mech. 2012;44:401–426. [Google Scholar]
- 19.Nguyen NT, Huang XY, Chuan TK. MEMS-micropumps: A review. J. Fluid. Eng.-T. Asme. 2002;124:384–392. [Google Scholar]
- 20.Laser DJ, Santiago JG. A review of micropumps. J. Micromech. Microeng. 2004;14:R35–R64. [Google Scholar]
- 21.Urbanski JP, Thorsen T, Levitan JA, Bazant MZ. Fast AC electro-osmotic micropumps with nonplanar electrodes. Appl. Phys. Lett. 2006;89:143508. [Google Scholar]
- 22.Zangle TA, Mani A, Santiago JG. Theory and experiments of concentration polarization and ion focusing at microchannel and nanochannel interfaces. Chem. Soc. Rev. 2010;39:1014–1035. doi: 10.1039/b902074h. [DOI] [PubMed] [Google Scholar]
- 23.Craighead H. Future lab-on-a-chip technologies for interrogating individual molecules. Nature. 2006;442:387–393. doi: 10.1038/nature05061. [DOI] [PubMed] [Google Scholar]
- 24.Stone HA, Stroock AD, Ajdari A. Engineering flows in small devices: Microfluidics toward a lab-on-a-chip. Annu. Rev. Fluid. Mech. 2004;36:381–411. [Google Scholar]
- 25.Schoch RB, Han JY, Renaud P. Transport phenomena in nanofluidics. Rev. Mod. Phys. 2008;80:839–883. [Google Scholar]
- 26.He Y, Tsutsui M, Fan C, Taniguchi M, Kawai T. Controlling DNA translocation through gate modulation of nanopore wall surface charges. ACS nano. 2011;5:5509–5518. doi: 10.1021/nn201883b. [DOI] [PubMed] [Google Scholar]
- 27.Mao M, Ghosal S, Hu G. Hydrodynamic flow in the vicinity of a nanopore induced by an applied voltage. Nanotechnology. 2013;24:245202. doi: 10.1088/0957-4484/24/24/245202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luan B, Aksimentiev A. Electro-osmotic screening of the DNA charge in a nanopore. Physical review. E, Statistical, nonlinear, and soft matter physics. 2008;78:021912. doi: 10.1103/PhysRevE.78.021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan B, Aksimentiev A. Control and reversal of the electrophoretic force on DNA in a charged nanopore. Journal of physics. Condensed matter : an Institute of Physics journal. 2010;22:454123. doi: 10.1088/0953-8984/22/45/454123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laohakunakorn N, Ghosal S, Otto O, Misiunas K, Keyser UF. DNA interactions in crowded nanopores. Nano Lett. 2013;13:2798–2802. doi: 10.1021/nl401050m. [DOI] [PubMed] [Google Scholar]
- 31.van Dorp S, Keyser UF, Dekker NH, Dekker C, Lemay SG. Origin of the electrophoretic force on DNA in solid-state nanopores. Nat. Phys. 2009;5:347–351. [Google Scholar]
- 32.Soni GV, Singer A, Yu Z, Sun Y, McNally B, Meller A. Synchronous optical and electrical detection of biomolecules traversing through solid-state nanopores. The Review of scientific instruments. 2010;81:014301. doi: 10.1063/1.3277116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Huber DE, Tabard-Cossa V, Dutton RW. Descreening of field effect in electrically gated nanopores. Appl. Phys. Lett. 2010;97:143109. [Google Scholar]
- 34.Jiang Z, Mihovilovic M, Chan J, Stein D. Fabrication of nanopores with embedded annular electrodes and transverse carbon nanotube electrodes. J. Phys.: Condens. Matter. 2010;22:454114. doi: 10.1088/0953-8984/22/45/454114. [DOI] [PubMed] [Google Scholar]
- 35.Albrecht T. How to understand and interpret current flow in nanopore/electrode devices. ACS nano. 2011;5:6714–6725. doi: 10.1021/nn202253z. [DOI] [PubMed] [Google Scholar]
- 36.Storm AJ, Storm C, Chen JH, Zandbergen H, Joanny JF, Dekker C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 37.Yemini M, Hadad B, Liebes Y, Goldner A, Ashkenasy N. The controlled fabrication of nanopores by focused electron-beam-induced etching. Nanotechnology. 2009;20:245302. doi: 10.1088/0957-4484/20/24/245302. [DOI] [PubMed] [Google Scholar]
- 38.Mitsui T, Stein D, Kim YR, Hoogerheide D, Golovchenko JA. Nanoscale volcanoes: accretion of matter at ion-sculpted nanopores. Phys. Rev. Lett. 2006;96:036102. doi: 10.1103/PhysRevLett.96.036102. [DOI] [PubMed] [Google Scholar]
- 39.Chen P, Mitsui T, Farmer DB, Golovchenko J, Gordon RG, Branton D. Atomic layer deposition to fine-tune the surface properties and diameters of fabricated nanopores. Nano Lett. 2004;4:1333–1337. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LM, Dekker C. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 42.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson AT, Drndic M. DNA translocation through graphene nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 43.Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae MH, Aluru NR, Pop E, Bashir R. Stacked graphene-Al2O3 nanopore sensors for sensitive detection of DNA and DNA-protein complexes. ACS nano. 2012;6:441–450. doi: 10.1021/nn203769e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, He J, Tang J, Liu H, Pang P, Cao D, Krstic P, Joseph S, Lindsay S, Nuckolls C. Translocation of single-stranded DNA through single-walled carbon nanotubes. Science. 2010;327:64–67. doi: 10.1126/science.1181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 2012;7:257–263. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- 46.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yusko EC, Prangkio P, Sept D, Rollings RC, Li J, Mayer M. Single-particle characterization of Abeta oligomers in solution. ACS nano. 2012;6:5909–5919. doi: 10.1021/nn300542q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhijun J, Mirna M, Jason C, Derek S. Fabrication of nanopores with embedded annular electrodes and transverse carbon nanotube electrodes. J. Phys.: Condens. Matter. 2010;22:454114. doi: 10.1088/0953-8984/22/45/454114. [DOI] [PubMed] [Google Scholar]
- 49.Xie P, Xiong QH, Fang Y, Qing Q, Lieber CM. Local electrical potential detection of DNA by nanowire-nanopore sensors. Nat. Nanotechnol. 2012;7:119–125. doi: 10.1038/nnano.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spinney PS, Collins SD, Howitt DG, Smith RL. Fabrication and characterization of a solid-state nanopore with self-aligned carbon nanoelectrodes for molecular detection. Nanotechnology. 2012;23:135501. doi: 10.1088/0957-4484/23/13/135501. [DOI] [PubMed] [Google Scholar]
- 51.Nishizawa M, Menon VP, Martin CR. Metal nanotubule membranes with electrochemically switchable ion-transport selectivity. Science. 1995;268:700–702. doi: 10.1126/science.268.5211.700. [DOI] [PubMed] [Google Scholar]
- 52.Siwy Z, Heins E, Harrell CC, Kohli P, Martin CR. Conical-nanotube ion-current rectifiers: the role of surface charge. J. Am. Chem. Soc. 2004;126:10850–10851. doi: 10.1021/ja047675c. [DOI] [PubMed] [Google Scholar]
- 53.Pedone D, Langecker M, Abstreiter G, Rant U. A pore-cavity-pore device to trap and investigate single nanoparticles and DNA molecules in a femtoliter compartment: confined diffusion and narrow escape. Nano Lett. 2011;11:1561–1567. doi: 10.1021/nl104359c. [DOI] [PubMed] [Google Scholar]
- 54.Menard LD, Ramsey JM. Electrokinetically-driven transport of DNA through focused ion beam milled nanofluidic channels. Anal. Chem. (Wash.) 2013;85:1146–1153. doi: 10.1021/ac303074f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stein D, Deurvorst Z, van der Heyden FH, Koopmans WJ, Gabel A, Dekker C. Electrokinetic concentration of DNA polymers in nanofluidic channels. Nano Lett. 2010;10:765–772. doi: 10.1021/nl902228p. [DOI] [PubMed] [Google Scholar]
- 56.Vlassiouk I, Smirnov S, Siwy Z. Ionic selectivity of single nanochannels. Nano Lett. 2008;8:1978–1985. doi: 10.1021/nl800949k. [DOI] [PubMed] [Google Scholar]
- 57.Grosberg AY, Rabin Y. DNA capture into a nanopore: interplay of diffusion and electrohydrodynamics. J. Chem. Phys. 2010;133:165102. doi: 10.1063/1.3495481. [DOI] [PubMed] [Google Scholar]
- 58.Wei R, Pedone D, Zurner A, Doblinger M, Rant U. Fabrication of metallized nanopores in silicon nitride membranes for single-molecule sensing. Small. 2010;6:1406–1414. doi: 10.1002/smll.201000253. [DOI] [PubMed] [Google Scholar]
- 59.Bard AJ, Parsons R, Jordan J. Standard potentials in aqueous solution. New York: M. Dekker; 1985. International Union of Pure and Applied Chemistry. [Google Scholar]
- 60.Ren B, Picardi G, Pettinger B. Preparation of gold tips suitable for tip-enhanced Raman spectroscopy and light emission by electrochemical etching. Rev. Sci. Instrum. 2004;75:837–841. [Google Scholar]
- 61.Tang J, Levy SL, Trahan DW, Jones JJ, Craighead HG, Doyle PS. Revisiting the Conformation and Dynamics of DNA in Slitlike Confinement. Macromolecules. 2010;43:7368–7377. [Google Scholar]
- 62.Zhang C, Zhang F, van Kan JA, van der Maarel JR. Effects of electrostatic screening on the conformation of single DNA molecules confined in a nanochannel. J. Chem. Phys. 2008;128:225109. doi: 10.1063/1.2937441. [DOI] [PubMed] [Google Scholar]
- 63.Gunther K, Mertig M, Seidel R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010;38:6526–6532. doi: 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nkodo AE, Garnier JM, Tinland B, Ren H, Desruisseaux C, McCormick LC, Drouin G, Slater GW. Diffusion coefficient of DNA molecules during free solution electrophoresis. Electrophoresis. 2001;22:2424–2432. doi: 10.1002/1522-2683(200107)22:12<2424::AID-ELPS2424>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 65.Stellwagen NC, Gelfi C, Righetti PG. The free solution mobility of DNA. Biopolymers. 1997;42:687–703. doi: 10.1002/(SICI)1097-0282(199711)42:6<687::AID-BIP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 66.Klein SD, Bates RG. Conductance of tris(hydroxymethyl)-aminomethane hydrochloride (Tris·HCl) in water at 25 and 37°C. J. Solution Chem. 1980;9:289–292. [Google Scholar]
- 67.Balducci A, Mao P, Han JY, Doyle PS. Double-stranded DNA diffusion in slit like nanochannels. Macromolecules. 2006;39:6273–6281. [Google Scholar]
- 68.Pardon G, van der Wijngaart W. Modeling and simulation of electrostatically gated nanochannels. Adv Colloid Interface Sci. 2013;199–200:78–94. doi: 10.1016/j.cis.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Constantin D, Siwy ZS. Poisson-Nernst-Planck model of ion current rectification through a nanofluidic diode. Phys. Rev. E: Stat. Phys Plasmas, Fluids. 2007;76:041202. doi: 10.1103/PhysRevE.76.041202. [DOI] [PubMed] [Google Scholar]
- 70.Squires TM, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005;77:977–1026. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.