Abstract
Damaged mitochondria are eliminated by mitophagy, a selective form of autophagy whose dysfunction associates with neurodegenerative diseases. PINK1, PARKIN and p62/SQTMS1 have been shown to regulate mitophagy, leaving hitherto ill-defined the contribution by key players in ‘general' autophagy. In basal conditions, a pool of AMBRA1 – an upstream autophagy regulator and a PARKIN interactor – is present at the mitochondria, where its pro-autophagic activity is inhibited by Bcl-2. Here we show that, upon mitophagy induction, AMBRA1 binds the autophagosome adapter LC3 through a LIR (LC3 interacting region) motif, this interaction being crucial for regulating both canonical PARKIN-dependent and -independent mitochondrial clearance. Moreover, forcing AMBRA1 localization to the outer mitochondrial membrane unleashes a massive PARKIN- and p62-independent but LC3-dependent mitophagy. These results highlight a novel role for AMBRA1 as a powerful mitophagy regulator, through both canonical or noncanonical pathways.
Autophagy is an important eukaryotic process involved in the lysosomal degradation of cytosolic components in both physiological and pathological conditions. During autophagy, the autophagosomes − specific double-membraned vesicles − engulf a number of different cargoes and then fuse with the lysosomes for subsequent recycling of their content. Several key proteins are involved in autophagosome formation, such as BECLIN 1 and its positive regulator AMBRA1;1, 2 a pool of AMBRA1 is localized at the mitochondria, where its pro-autophagic activity is inhibited by mitochondrial resident Bcl-2.3 Interestingly, mitochondria have been described as a source for autophagosome biogenesis;4 they play a key role in the cross-talk between autophagy and apoptosis regulation and they are involved in the cell death versus survival decision (reviewed in Strappazzon et al.3).
Another mechanistic link exists between autophagy and mitochondria in mammals. Indeed, mitochondria damaged by the uncoupler CCCP (carbonyl cyanide m-chlorophenyl hydrazone) − because of a loss of their mitochondrial membrane potential (ΔΨm) − are subjected to a form of selective autophagy, termed mitophagy.5, 6, 7 During this process, depolarized mitochondria are ubiquitylated; they then recruit p62 (a protein involved in linking polyubiquitinated protein aggregates to the autophagic machinery) and next they are transported along microtubules to the perinuclear region, where they form rough aggregate structures termed ‘mito-aggresomes',8, 9, 10 a step preceding their lysosomal degradation.
Although mitophagy has been described in a number of tissues and in various physiological or pathological conditions (reviewed in Andreux et al.11), very few are the known molecular mechanisms that regulate mitophagy; this is despite the fact that its manipulation may represent a forefront strategy in several human diseases. Thus, rather scarse is yet the availability of chemicals and drug candidates to modulate the process. The autophagy receptor NIX and the kinase Ulk1 mediate developmental removal of mitochondria during retyculocyte differentiation.6, 12, 13 Smurf1 has been defined as a new recognized mediator of both viral autophagy and mitophagy.14 In contrast, the E3 ubiquitin ligase PARKIN and the Ser/Thr kinase PINK1, both found to be mutated in autosomal recessive forms of Parkinson's disease (PD), regulate mitophagy after mitochondrial damage.5 In more detail, PINK1 recruits PARKIN to depolarized mitochondria in order to remove damaged mitochondria. This mitochondrial quality control, driven by PINK1/PARKIN proteins, has recently been better characterized by RNAi screens.15 In fact, new proteins such as HSPA1L, BAG4 and SIAH3 have been found to modulate translocation of PARKIN to damaged mitochondria, whereas TOMM7 stabilizes PINK1 on the mitochondria. Interestingly, it has been demonstrated that after mitochondrial depolarization, the cytosolic pool of AMBRA1 interacts with PARKIN to enhance mitochondrial clearance.16
In this study, we investigate the molecular mechanism(s) responsible for the AMBRA1-dependent enhancement of PARKIN-mediated mitophagy. We describe for the first time AMBRA1 as a new LIR (LC3 interacting region)-containing protein, and we demonstrate that this motif is essential for the binding between AMBRA1 and LC3, following mitophagy induction. Furthermore, we show that this interaction is crucial in a number of cell systems in order to both amplify PARKIN-mediated mitochondrial clearance and regulate PARKIN-independent mitophagy. In addition, to better understand the role of AMBRA1 at the mitochondria and as AMBRA1 does not possess a clear mitochondrial targeting sequence, we generated and expressed an organelle-targeted mutant of AMBRA1 in two different cell systems. Our data indicate that mitochondrial AMBRA1 induces (1) relocalization of the mitochondrial network around the nucleus, (2) depolarization and ubiquitylation of mitochondria and (3) recruitment of the molecular platform necessary to induce functional mitophagy through a PARKIN/p62-independent pathway.
This work thus places AMBRA1 as a central player of mitophagy: we suggest that this molecule facilitates mitochondrial clearance by bringing damaged mitochondria onto autophagosomes via its LIR-mediated LC3 interaction. In addition, we show that high levels of mitochondrial AMBRA1 trigger mitophagy, a finding that could herald new therapies to fight important human disorders, ranging from muscle dystrophy to neurodegeneration.
Results
FCCP induces AMBRA1 binding to LC3 through a previously undisclosed LIR motif, this enhancing PARKIN-mediated mitophagy
As AMBRA1 can localize at the mitochondria,3 and as this organelle has been described as a source for autophagosome biogenesis,4 we checked whether AMBRA1 could bind LC3, an autophagosome cargo adapter, on the mitochondria, so as to enhance autophagosome production following mitochondrial damage.
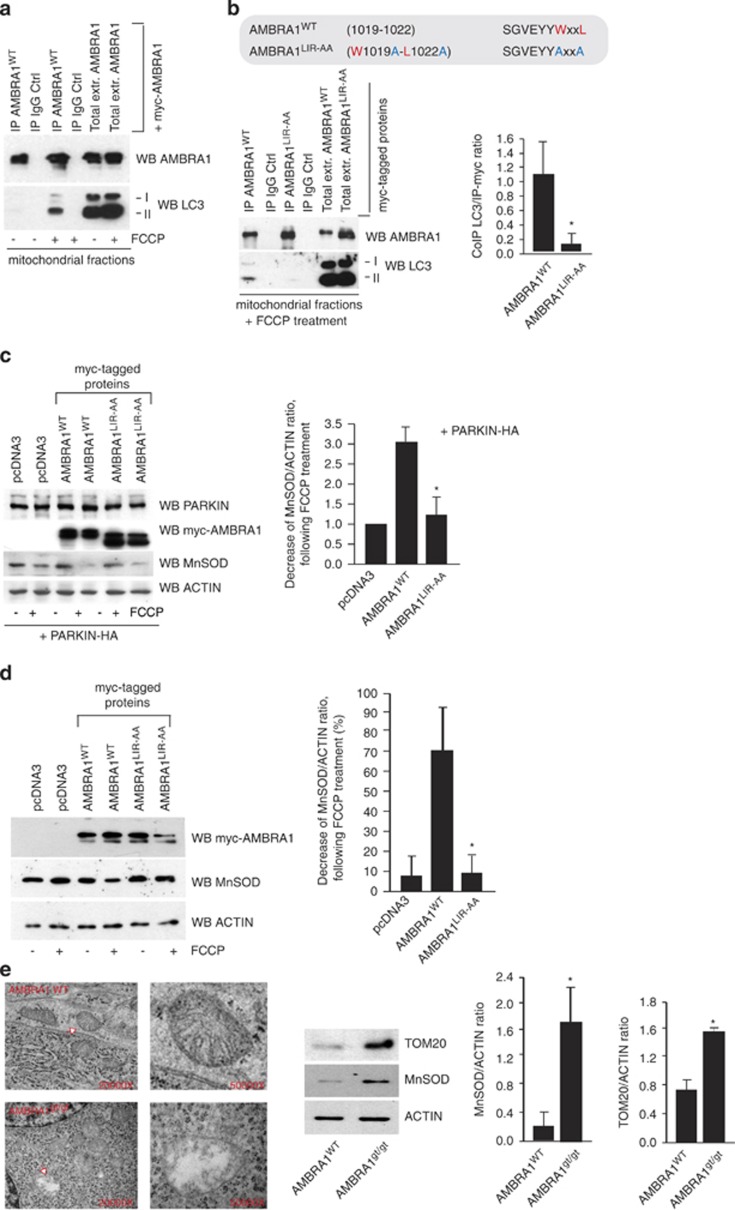
The known consensus sequence for the core LIR motif is W/F/Y-x-x-L/I/V.17 Therefore, first of all, we examined in silico the AMBRA1 sequence and found that AMBRA1 possesses, indeed, a putative LIR motif in its C-terminal region (1012SGVEYYWxxL1023). Primed by the hypothesis that the two factors could interact only locally at a subcellular level, we enriched our samples for mitochondrial proteins, by performing a mitochondrial fractioning assay in HEK293 cells transiently overexpressing myc-AMBRA1WT and after treatment with FCCP (trifluorocarbonylcyanide phenylhydrazone, 1 h, 10 μM), a routinely adopted inducer of mitochondrial depolarization in in vitro assays. We then precipitated myc- AMBRA1WT, and we checked for the presence of LC3-I and -II (hallmarks of autophagy). Interestingly, myc- AMBRA1WT can be found associated with LC3-I and -II forms following FCCP treatment in both endogenous and overexpression conditions (Figure 1a and Supplementary Figure 1). It is noteworthy that no binding between AMBRA1 and LC3 could be found in total extracts of HEK293 cells following autophagy induction (EBSS treatment, Supplementary Figure 1b).
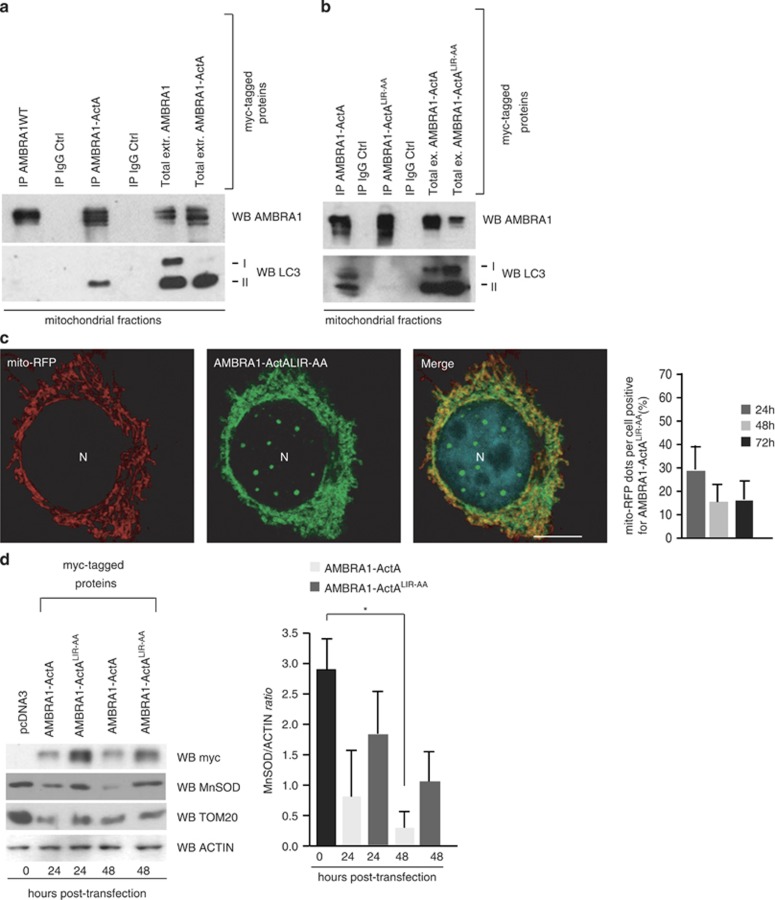
Figure 1.
FCCP induces AMBRA1 binding to LC3 through a previously undisclosed LIR motif, enhancing PARKIN-mediated mitophagy. (a) HEK293 cells transfected with a vector encoding myc-AMBRA1WT. At 24 h after transfection, cells were treated with DMSO (ctrl) or with FCCP for 1 h (10 μM). Mitochondrial extracts were immunoprecipitated using an anti-myc antibody or with IgG control. Purified complexes and corresponding total extracts were analysed by western blot (WB) using anti-AMBRA1 and anti-LC3 antibodies. (b) Identification of a LIR motif within the AMBRA1 sequence. Point mutations of both an aromatic residue and the conserved hydrophobic residue are shown (AMBRA1-LIRAA (W1019A-L1022A SGVEYYAxxA)). HEK293 cells were transfected with a vector encoding myc-AMBRA1WTor myc-AMBRA1-LIRAA. At 24 h after transfection, cells were treated with DMSO (ctrl) or FCCP for 1 h (10 μM). Mitochondrial extracts were immunoprecipitated using an anti-myc antibody or with IgG control. Purified complexes and corresponding total extracts were analysed by WB using anti-AMBRA1 and anti-LC3 antibodies. The band density ratio of immunoprecipitated AMBRA1 relative to immunoprecipitated LC3 is analysed in three independent experiments; each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05) versus AMBRA1. (c) HEK293 cells were transfected with a vector encoding myc-AMBRA1WT or myc-AMBRA1-LIRAA. At 24 h after transfection, cells were treated with DMSO (ctrl) or FCCP for 9 h (1 μM). Protein extracts were analysed using anti-MnSOD and anti-ACTIN (loading control) antibodies. The graph illustrates the MnSOD/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05) versus AMBRA1+PARKIN. (d) ETNA cells were transfected with a vector encoding myc-AMBRA1WTor myc-AMBRA1LIR-AA. At 24 h after transfection, cells were treated with DMSO (ctrl) or FCCP for 5 h (30 μM). Protein extracts were analysed using anti-MnSOD (mitochondria) and anti-ACTIN (loading control) antibodies. The graph illustrates the MnSOD/ACTIN ratio decrease following FCCP treatment (%). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05) versus AMBRA1WT. (e) Ultrastructural analysis of brain from wild-type and Ambra1gt/gt embryos. The arrow (in the Ambra1gt/gt panel) indicates alterations of the cristae and the overall structure of the organelle in damaged mitochondria. Protein extracts from embryo heads (E13.5) were analysed by WB using anti-MnSOD, anti-TOM20 and anti-ACTIN (loading control) antibodies. Magnification is indicated ( × ). Graphs represent MnSOD/ACTIN or TOM20/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed by Student's test (*P<0.05)
It is noteworthy that point mutations of both an aromatic residue and the conserved hydrophobic residue of the putative LIR motif (AMBRA1LIR-AA (W1019A-L1022A) SGVEYYAxxA) almost abolished the interaction between myc-AMBRA1WT and endogenous LC3 after mitophagy induction (Figure 1b). Next, as AMBRA1 overexpression has been shown to enhance PARKIN-mediated mitochondrial clearance,16 we set out to verify whether AMBRA1–LC3 interaction was required in this context. To this aim, we transiently coexpressed a vector encoding PARKIN and AMBRA1WT or AMBRA1LIR-AA in HEK293 cells. We then treated cells with FCCP in order to induce mitophagy, and then measured the expression of MnSOD, a mitochondrial marker. As shown in Figure 1c, overexpression of AMBRA1WTpotentiates PARKIN-mediated mitophagy, confirming previous results.16 In contrast, coexpression of PARKIN with AMBRA1LIR-AA did not increase mitochondrial clearance to the same extent as AMBRA1WT.
Next, we confirmed our results in a murine proneural cell line. We transiently coexpressed vectors encoding pcDNA3, AMBRA1WTor AMBRA1LIR-AA in embryonic telencephalic naive cells (ETNA).18 We then treated cells with FCCP, as in Figure 1c, and obtained even more striking results (Figure 1d).
Taken overall, these results indicate that, in different cell types, the LIR motif of AMBRA1 required for LC3 binding is responsible for the described enhancement of PARKIN-mediated mitochondrial clearance.16
To investigate the mitophagic role of AMBRA1 in vivo in physiological conditions, we looked at the mitochondrial content of embryonic forebrains (prosencephalon) from wild-type or AMBRA1-deficient embryos (Ambra1gt/gt). As revealed by ultrastructural analysis using electron microscope (EM), brains from Ambra1gt/gt embryos accumulated damaged mitochondria when compared with wild-type tissues. In addition, we observed a stronger expression of the two mitochondrial markers MnSOD and TOM20 in Ambra1gt/gt embryonic brains (Figure 1e). These results support a crucial role, in vivo, for AMBRA1 in mitochondrial homeostasis.
High levels of mitochondria-targeted AMBRA1 specifically induce a perinuclear distribution of mitochondria
To manipulate the dosage of the AMBRA1 mitochondrial pool, we generated a construct encoding myc-AMBRA1 fused to an ‘insertion sequence' that can target the molecule to the outer mitochondrial membrane. To this end, we used a DNA sequence from the Listeria monocytogenes Actin assembly-inducing protein (ActA).19
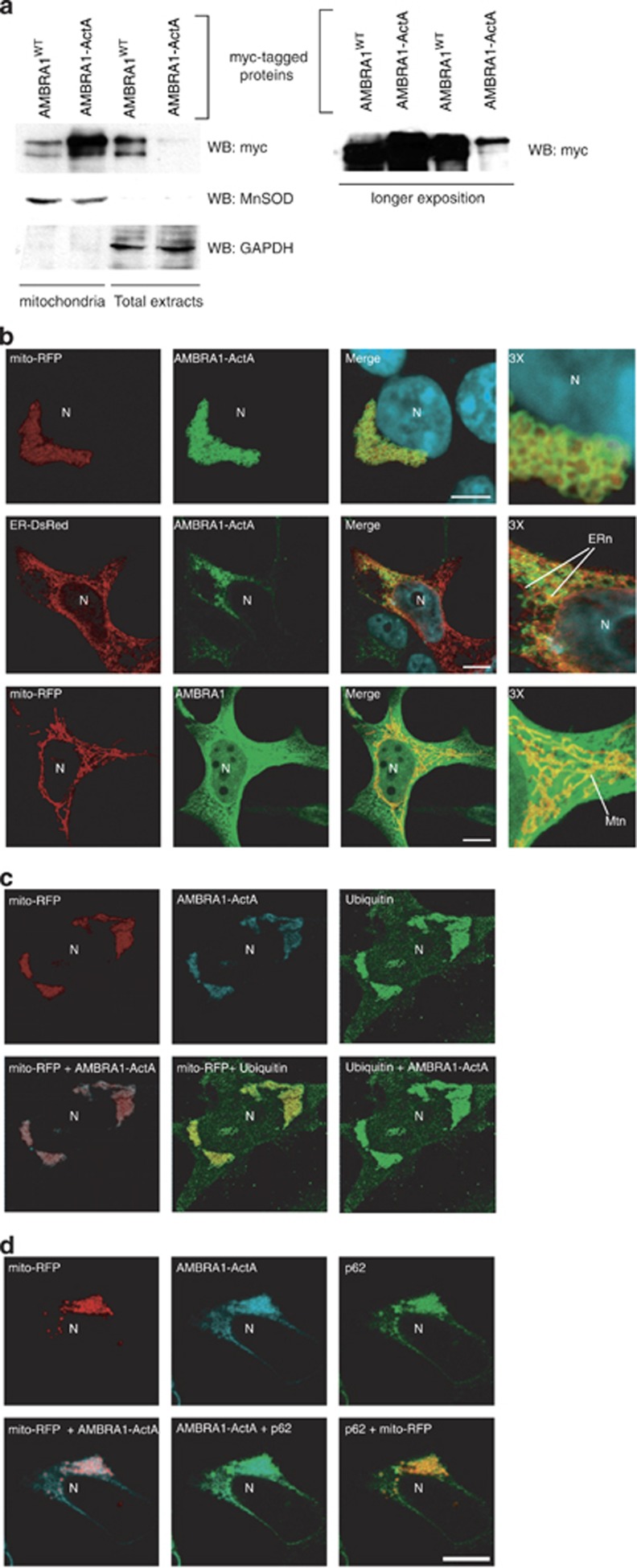
First, we checked for the localization of the AMBRA1–ActA protein. As expected, AMBRA1–ActA is enriched in the mitochondrial fraction of HEK293 cells (Figure 2a).
Figure 2.
AMBRA1–ActA colocalizes with mitochondria and induces the formation of ubiquitin-and p62-positive mito-aggresomes in HEK293 cells. (a) HEK293 cells were transfected with a vector encoding myc-AMBRA1–ActA. At 24 h after transfection, cells were lysed and mitochondrial extracts were analysed by western blot using an anti-myc antibody. The fraction purity was verified by means of antibodies against MnSOD (mitochondria) or GAPDH (cytosol). The longer exposure in the right panel shows the low levels of Ambra1–ActA outside mitochondria. (b) HEK293 cells were co-transfected with vectors encoding myc-AMBRA1–ActA (green) and mito-RFP (red) or ER-DsRed (red). As a control, HEK293 cells were co-transfected with vectors encoding myc-AMBRA1WT and mito-RFP. At 24 h after transfection, cells were fixed and stained with an anti-myc antibody (green). Merge of the fluorescence signals is shown in the right panel, together with a higher magnification image ( × 3). N, nucleus; ERn, ER network; Mtn, mitochondrial network. Scale bar, 6 μm. (c) HEK293 cells were co-transfected with vectors encoding myc-AMBRA1-ActA and mito-RFP (red). Twenty-four hours after transfection, cells were fixed and stained with anti-myc (blue) and anti-Ubiquitin (green) antibodies. Merged images of the two fluorescence signals are shown in the right panels. (d) HEK293 cells were co-transfected with vectors encoding myc-AMBRA1-ActA and mito-RFP (red). Twenty-four hours after transfection, cells were fixed and stained with anti-myc (blue) and anti-p62 (green) antibodies. Merged images of the two fluorescence signals are shown in the right panels. N, nucleus. Scale bar, 6 μm.
To confirm these results we performed a confocal microscopy analysis in HEK293 cells co-transfected with a vector encoding fluorescent mito-RFP, in order to stain the mitochondrial network, and a tagged vector encoding AMBRA1–ActA. Indeed, AMBRA1–ActA surrounds swollen and aggregated mitochondria (Figure 2b). In contrast, only a slight colocalization of AMBRA1–ActA was found with the ER network (Figure 2b, upper and mid panels). AMBRA1WT only partially colocalizes with mitochondria (Figure 2b, lower panels).
Most importantly, we observed that AMBRA1–ActA overexpression induces per se a strong relocalization of the mitochondrial network around the perinuclear envelope, leading to the formation of structures similar to those previously described as ‘mito-aggresomes'8, 10 (Figure 2b, upper panels). These structures cannot be found after overexpression of Bcl-2–ActA (Supplementary Figure 2), confirming that they represent a specific effect of AMBRA1 mitochondrial overexpression.
To summarize, AMBRA1 localization at the outer mitochondrial membrane potently induces the formation of structures similar to mito-aggresomes.
Mitochondria-targeted AMBRA1 induces massive mitophagy in PARKIN-competent cells
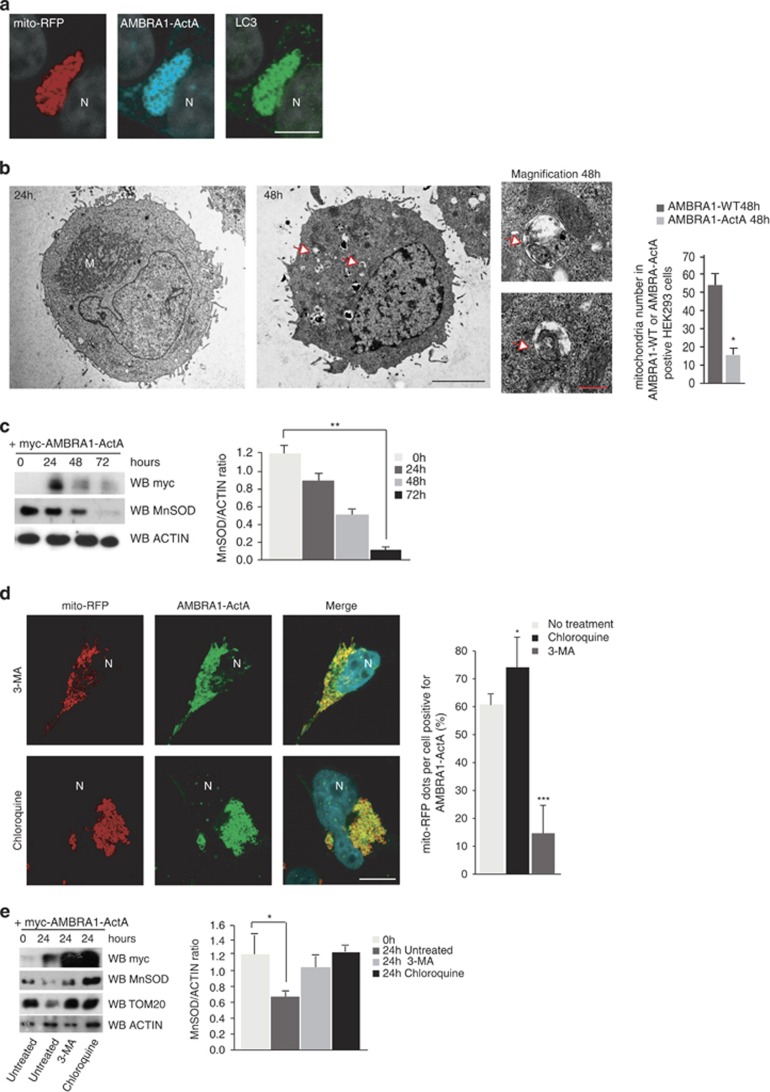
We next studied the dynamics of the mitochondria shape changes induced by AMBRA1. As shown in Supplementary Figure 3, at 8 h after transfection, AMBRA1–ActA staining significantly overlaps with the mitochondrial network that seems to be normally well organized. However, at 10 h after transfection, AMBRA1–ActA staining in the majority of transfected cells (≅75%) surrounds all mitochondria, forming a sort of ring. Finally, at 18 h after transfection, the mitochondrial network is completely redistributed around the perinuclear area and forms mito-aggresome-like structures.
Next, we characterized these structures and found that at 24 h following AMBRA1–ActA overexpression, mitochondria are stained for ubiquitin and p62 (Figure 2). This observation suggests that AMBRA1–ActA induces canonical mito-aggresomes, decorated by ubiquitin and p62.9, 20 The appearance of mito-aggresomes led us to hypothesize that AMBRA1–ActA overexpression was able to induce mitophagy. Thus, we decided to check the occurrence of this event by the presence of LC3, a well-known marker of autophagosome formation.21 As shown in Figure 5a, a perfect colocalization between AMBRA1–ActA, mitochondria and LC3 can be observed. Cells overexpressing AMBRA1–ActA showed a colocalization of ≅82% with ubiquitin, ≅69% with p62 and ≅89% with LC3 (see Supplementary Figure 4).
Furthermore, in order to better characterize the mito-aggresomes as regions with high density of mitochondria-containing autophagosomes, we performed an ultrastructural analysis by EM on cells overexpressing AMBRA1–ActA. The EM image at 24 h indicates the presence of mitochondria redistributed on one side of the nucleus. At 48 h after transfection, fewer mitochondria can be observed in whole cells and engulfment events of mitochondria by autophagosomes can be found (Figure 3b, 48 h+ magnification panels). In contrast, after transfection of a vector encoding AMBRA1WT, no changes in the mitochondrial distribution are observed (Supplementary Figure 5). To ascertain that mitophagy was induced by AMBRA1–ActA, we evaluated, by electronic microscopy, the mitochondria number 48 h after transfection of AMBRA1WT or AMBRA1–ActA. As shown in the graph (Figure 3b), a third of mitochondria disappeared upon transfection of AMBRA1–ActA versus AMBRA1WT. To confirm these data, we checked for mitochondrial proteins at different time points after AMBRA1–ActA overexpression. As shown in the graph in Figure 3c, at 48 h after AMBRA1–ActA overexpression, a third of mitochondria have disappeared, whereas almost all mitochondria have been degraded at 72 h after transfection. We can thus conclude that massive mitophagy is induced by AMBRA1–ActA.
Figure 3.
AMBRA1–ActA induces mitophagy in HEK293 cells. (a) HEK293 cells were co-transfected with vectors encoding myc-AMBRA1–ActA and mito-RFP (red). At 24 h after transfection, cells were fixed and stained with anti-myc (blue) and anti-LC3 (green) antibodies. Merge of the two fluorescence signals is shown in the right panel. Scale bar, 5 μm. (b) An ultrastructural analysis by EM on whole cells. HEK293 cells were co-transfected with vectors encoding myc-AMBRA1–ActA and GFP mitochondria. At 24 or 48 h after transfection, GFP-positive cells were sorted by using FACS and fixed in glutaraldehyde. Morphological analysis was performed. In the left panel, mitochondria are already redistributed around the nucleus (24 h following transfection). M, mitochondria. The center panel (48 h following transfection) shows a whole cell containing less mitochondria than at 24 h. Arrows indicate fragmented mitochondria into autophagosomes. Scale bar, 5 μm. The upper and lower panels on the right are two magnifications of the autophagosomes observed in the whole cell (see arrows in the middle panel, 48 h). Scale bar, 500 nm. The graph shows quantification of mitochondria number per cells positive for AMBRA1WT and AMBRA1–ActA at 48 h post transfection. Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05) (c) HEK293 cells were transfected with a vector encoding myc-AMBRA1–ActA. At 24, 48 and 72 h after transfection, proteins extracts were analysed by western blot using the following antibodies: anti-MnSOD, anti-myc (to control AMBRA1–ActA expression) and anti-ACTIN (loading control). The graph represents the MnSOD/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.01; 24 h versus 72 h). (d) 3-MA treatment partially inhibits AMBRA1–ActA/mito-DsRed vacuole formation. HEK293 cells were co-transfected with vectors encoding AMBRA1–ActA and mito-RFP and treated directly with 10 mM 3-MA. After 24 h, cells were fixed and stained with an anti-myc antibody (green). Nuclei were stained with 1 μg/μl DAPI for 20 min. Merge of the three fluorescence signals is shown in the right panel. Scale bar, 6 μm. Chloroquine treatment increases the percentage of AMBRA1–ActA/mito-RFP colocalizations on mito-aggresomes per cell. HEK293 cells were co-transfected with a vector encoding AMBRA1–ActA and a vector encoding mito-RFP. After 24 h, cells were treated with chloroquine for 1 h, and cells were fixed and stained with an anti-myc antibody (green). Nuclei were stained with 1 μg/μl DAPI for 20 min. Merge of the three fluorescence signals is shown in the right panel. Scale bar, 6 μm. The graph shows the quantification of AMBRA1–ActA/mito-RFP colocalization on mito-aggresomes per cell (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05, no treatment versus Chloroquine; and *P<0.001, no treatment versus 3-MA). N, nucleus. (e) HEK293 cells were co-transfected with vectors encoding AMBRA1–ActA and mito-RFP and treated directly with 10 mM 3-MA. After 24 h, proteins extracts treated with Chloroquine (1 h) or left untreated were analysed by western blot using the following antibodies: anti-MnSOD, anti-TOM20, anti-myc (to control AMBRA1–ActA expression) and anti-ACTIN (loading control). The graph represents the MnSOD/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05)
To determine the role of PI3K on mitophagy induced by AMBRA1–ActA overexpression, HEK293 cells co-transfected with AMBRA1–ActA and mito-RFP were treated with 10 mM 3-MA (3-methyladenine) or 75 nM Wortmannin (two inhibitors of PI3K22, 23). Aggregation of the mitochondrial network was reduced by treating the cells with 3-MA or Wortmannin (Figure 3d and Supplementary Figure 6a). Thus, AMBRA1–ActA-induced mitophagic sequestration seems to depend, at least in part, on the classical upstream PI3K pathway. We also examined the on/off rate of autophagy upon AMBRA1–ActA overexpression in the presence of a lysosomal-inhibiting agent, chloroquine. This treatment resulted in a significantly higher level of mito-aggresomes accumulation in AMBRA1–ActA-positive cells, hence indicating that autophagosomes induced by AMBRA1–ActA overexpression are normally degraded in the lysosome (Figure 3d). In addition, LAMP1/LC3 colocalization is present in AMBRA1–ActA-overexpressing cells, indicating a canonical lysosomal destination for the observed mito-aggresomes (Supplementary Figure 6b). We next confirmed the involvement of the PI3K pathway in AMBRA1–ActA-induced mitophagy by a more biochemical approach, that is, by analysing the expression of MnSOD and TOM20 following AMBRA1–ActA transfection in cells treated or not with 3-MA and Chloroquine. As shown in Figure 3e, both 3-MA and chloroquine treatments block MnSOD and TOM20 decrease following AMBRA1–ActA overexpression. Moreover, downregulation of BECLIN 1, an activator of the PI3K complex, is able to delay AMBRA1–ActA-induced mitophagy (Supplementary Figures 6c and d). It is noteworthy that AMBRA1–ActA overexpression is also sufficient to induce a massive mitochondrial clearance in the ETNA proneural cell line (Supplementary Figure 7).
Taken together, these observations demonstrate that AMBRA1–ActA transfection per se triggers a fully functional and massive mitophagy in different cell types.
AMBRA1–ActA induces mitophagy independently of PARKIN and p62 recruitment
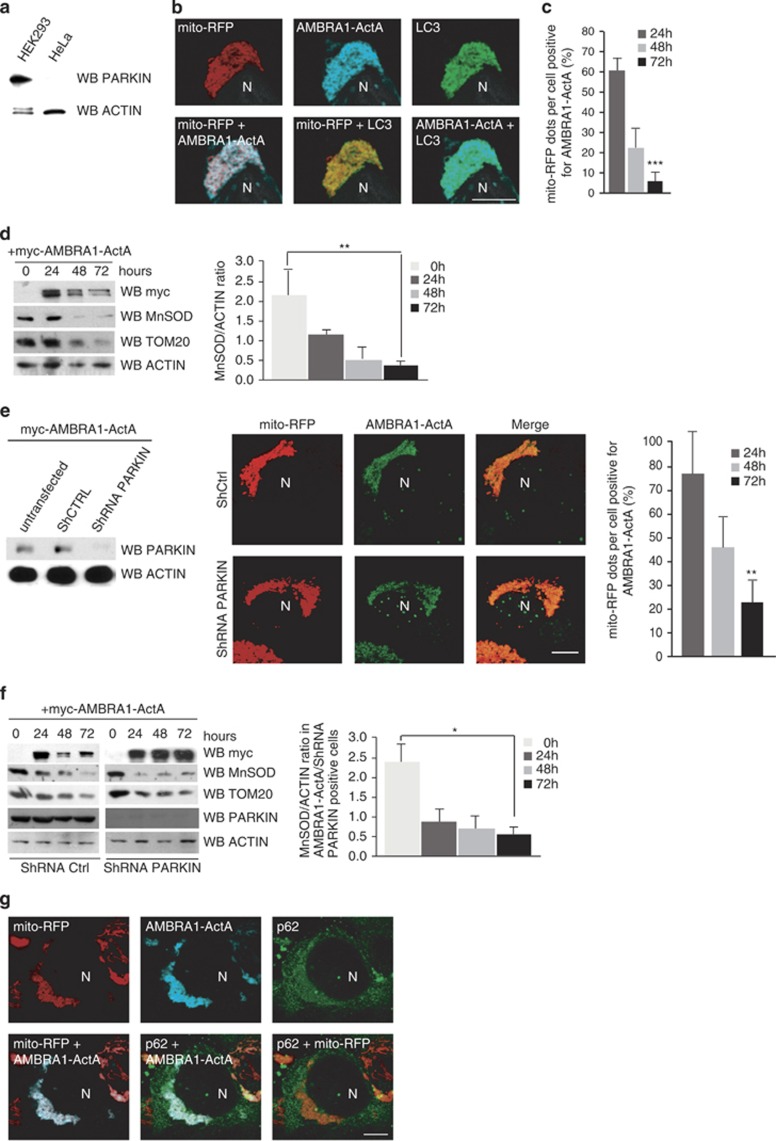
To identify the specific mitophagy pathway induced by AMBRA1–ActA, we decided to test whether our construct was able to induce mitophagy in a PARKIN-free system. We used HeLa cells as a cell line containing no PARKIN (Figure 4a), where AMBRA1–ActA induces mito-aggresomes (Figure 4b). Then we measured mitochondria degradation following AMBRA1–ActA overexpression. As shown in Figure 4c, at 48 h after transfection, 50% of mito-aggresomes disappear, when compared with the 24 h time point. At 72 h after transfection, it is also difficult to find cells with mito-aggresomes, and mitochondrial degradation is almost completed. This finding was also confirmed by western blot analysis, as revealed by the decrease of MnSOD and Tom20, two mitochondrial markers, following AMBRA1–ActA overexpression. Quantification of the MnSOD/ACTIN ratio indicates that 72 h after AMBRA1–ActA overexpression, almost all mitochondria are lost (Figure 4d). These results strongly suggest that AMBRA1–ActA is also able to induce mitophagy in a PARKIN-deficient cell line.
Figure 4.
AMBRA1–ActA induces mitophagy in PARKIN-deficient cell lines. (a) HeLa and HEK293 cells were grown in normal media. After extraction of proteins, we performed a western blot analysis by using antibodies against PARKIN and against ACTIN (as a loading control). (b) Colocalization between AMBRA1–ActA, mito-RFP and LC3 protein. HeLa cells were co-transfected with vectors encoding AMBRA1–ActA and mito-RFP, and grown in normal media. Cells were then stained using antibodies anti-myc (AMBRA1, blue) and anti-LC3 (green). Nuclei were stained with 1 μg/μl DAPI for 20 min. Merge of the different fluorescence signals is illustrated. Scale bar, 8 μm. (c) Quantification in HeLa cells of AMBRA1–ActA/mito-RFP mito-colocalizations at 24, 48 and 72 h after transfection per cell (±S.D.). Each time point value is the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (***P<0.001; 24 h versus 72 h). (d) HeLa cells were transfected with a vector encoding myc-AMBRA1–ActA. At 24, 48 and 72 h after transfection, proteins extracts were analysed by western blot using the following antibodies: anti-MnSOD, anti-TOM20, anti-myc (to control AMBRA1–ActA expression) and anti-ACTIN (loading control). The graph represents the MnSOD/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (**P<0.01; 24 h versus 72 h). (e) HEK293 cells were co-transfected with vectors encoding ShPARKIN or control ShRNA and myc-AMBRA1-ActA. At 24 h after transfection, expression of PARKIN was controlled by western blot analysis by means of anti-PARKIN and anti-ACTIN (loading control) antibodies. Cells were fixed and stained with an anti-myc antibody (green). The merge of the fluorescence signals is shown in the right panels. Scale bar, 4 μm. The graph illustrates the quantification of AMBRA1–ActA/mito-RFP colocalizations on mito-aggresomes per cell (±S.D.). Each point value represents the mean±S.D. from two independent experiments. Statistical analysis was performed using Student's test (*P<0.01; 24 h versus 72 h). (f) HEK293 cells were co-transfected with vectors encoding ShPARKIN or control ShRNA, and myc-AMBRA1–ActA. At 24 h after transfection, expression of PARKIN was checked by western blot analysis by means of anti-PARKIN and anti-ACTIN (loading control) antibodies. At 24, 48 and 72 h after transfection, protein extracts were analysed by western blot using the following antibodies: anti-MnSOD, anti-TOM20, anti-myc (to control AMBRA1–ActA expression) and anti-ACTIN (loading control). The graph represents the MnSOD/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed using Student's test (*P<0.05; 24 h versus 72 h). (g) HeLa cells were co-transfected with vectors encoding myc-AMBRA1–ActA and mito-RFP (red). At 24 h after transfection, cells were fixed and stained with anti-myc (AMBRA1, blue) and anti-p62 antibodies (green). Merge of the fluorescence signals are shown. N, nucleus. Scale bar, 4 μm
To confirm these results, we analysed HEK293 cells in which PARKIN-mediated autophagy normally takes place and where PARKIN recruitment to mitochondria also occurs upon AMBRA1–ActA transfection (Supplementary Figure 8). We downregulated PARKIN (ShPARKIN, see Figure 4e) and, in the same cells, we again coexpressed AMBRA1–ActA and mito-RFP. As shown in Figure 4e, AMBRA1–ActA staining surrounds mitochondria in cells overexpressing either ShPARKIN or control ShRNA. Our measurement of mitochondria degradation at 24, 48 and 72 h post transfection was also consistent (see graph in Figure 4e). These data were also confirmed by western blot analysis, by analysing the expression of MnSOD and TOM20 (Figure 4f).
Next, we decided to look at the formation of mito-aggresomes following AMBRA1–ActA overexpression in HeLa cells. Surprisingly, as shown in Figure 4e, we found that even though mitochondria are positive for ubiquitin (Supplementary Figure 9), p62 staining is almost negligible (at variance with what was observed in HEK293 cells, see Figure 4b). In more detail, cells overexpressing AMBRA1–ActA showed a colocalization of ≅80% with ubiquitin; ≅15% with p62 and ≅87% with LC3 (Supplementary Figure 9).
Altogether, these data strongly suggest that high levels of AMBRA1 at the mitochondria can also drive this process in a PARKIN- or p62-independent manner.
AMBRA1–ActA surrounds depolarized mitochondria
A large body of evidence indicate that mitochondria depolarization is a ‘call-out' signal to start mitophagy.24 Therefore, we set out to establish whether or not AMBRA1–ActA overexpression was localized around depolarized mitochondria. We used Tetramethylrhodamine, methyl ester (TMRM).25 As shown in Supplementary Figure 10A, AMBRA1–ActA is, indeed, localized around depolarized mitochondria, with AMBRA1–ActA-overexpressing cells showing an ≅60% reduction in their mitochondrial potential (see graph), similar to the effect obtained by FCCP treatment (≅80%). Overexpression of a vector coding for GFP-mito alone does not affect mitochondrial potential (Supplementary Figure 10B).
These data suggest that AMBRA1–ActA-mediated clearance of mitochondria is summoned into action by depolarized organelles.
The LIR motif of AMBRA1 is required for AMBRA1–ActA-induced mitophagy
Given our initial results regarding the importance of AMBRA1 LIR domain in mitophagy (see Figures 1c and d), we decided to check the occurrence of AMBRA1–ActA/LC3 interaction through this domain. We thus performed an immunoprecipitation between AMBRA1–ActA and LC3 on mitochondrial fractions. We found that, in the absence of any known mitophagy inducers, AMBRA1–ActA interacts with LC3, whereas no interaction can be found between AMBRA1WT and LC3 (Figure 5a). Point mutations of both the aromatic residue and the conserved hydrophobic residue of the LIR motif (AMBRA1LIR-AA-ActA (W1019A-L1022A SGVEYYAxxA)) suppress the interaction between AMBRA1–ActA and endogenous LC3 (Figure 5b). In addition, we observed that AMBRA1LIR-AA–ActA was able to colocalize with the mitochondrial network, but not, in contrast to AMBRA1–ActA, with mito-aggresomes (Figure 5c).
Figure 5.
AMBRA1–ActA interacts with LC3, with this interaction being crucial for AMBRA1–ActA-induced mitophagy. (a) HEK293 cells were transfected with a vector encoding myc-AMBRA1–ActA. At 24 h after transfection, mitochondrial extracts were immunoprecipitated using an anti-myc antibody. Purified complexes and corresponding total extracts were analysed by western blot (WB), using anti-AMBRA1 and anti-LC3 antibodies. (b) HEK293 cells were transfected with a vector encoding myc-AMBRA1–ActA or myc-AMBRA1–ActA-LIRAA. At 24 h after transfection, mitochondrial extracts were immunoprecipitated using an anti-myc antibody. Purified complexes and corresponding total extracts were analysed by WB using anti-AMBRA1 and anti-LC3 antibodies. (c) HEK293 cells were transfected with a vector encoding myc-AMBRA1–ActA-LIRAA and mito-RFP. At 24 h hours after transfection, cells were fixed and stained with an anti-myc antibody (green). Merge of the fluorescence signals is shown in the right panels. Scale bar, 8 μm. The graph reports the quantification of mitochondrial clearance in HEK293 cells overexpressing AMBRA1–ActALIR-AA/mito-RFP at 24, 48 and 72 h after transfection per cell (±S.D.). Each time point value is the mean±S.D. from three independent experiments. (d) HEK293 cells were transfected with a vector encoding myc-AMBRA1–ActA-LIRAA. At 0, 24 and 48 h after transfection, protein extracts were analysed using anti-MnSOD, anti-myc (AMBRA1) and anti-ACTIN (loading control) antibodies. The graph represents the MnSOD/ACTIN ratio (±S.D.). Each point value represents the mean±S.D. from three independent experiments. Statistical analysis was performed by Student's test (*P<0.05; 0 h versus 48 h)
Next, by measuring mitochondria degradation by western blot analysis, we found that the mitochondrial markers MnSOD and TOM20 were more stable after transfection of AMBRA1LIR-AA–ActA when compared with cells overexpressing AMBRA1–ActA (Figure 5d).
Our data indicate that the LIRAA mutation on AMBRA1–ActA delays mitochondrial clearance. We can thus conclude that AMBRA1–ActA-driven PARKIN-independent mitophagy also absolutely requires LC3 binding via the AMBRA1 LIR domain.
AMBRA1 mediates PINK1–PARKIN-independent mitophagy in different cell systems
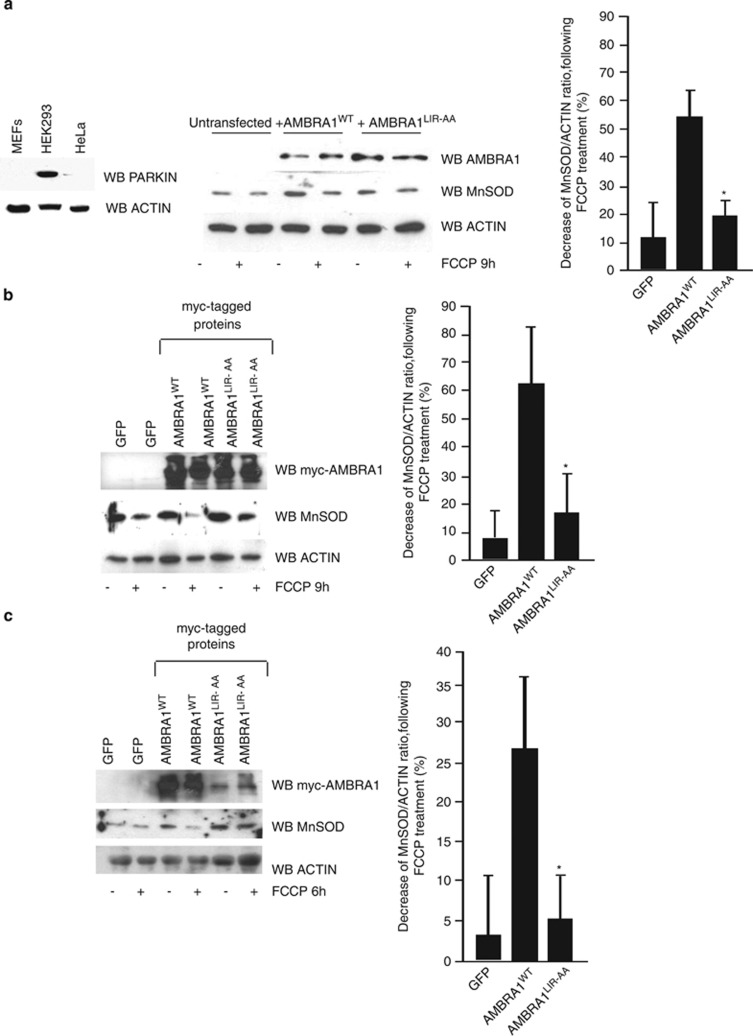
Finally, to extend our analysis to the PINK1–PARKIN regulatory axis and to get an in vivo corroboration of our results, we decided to investigate whether AMBRA1WTwas able to induce mitochondrial clearance in several systems in which the PINK1–PARKIN pathway was deficient or absent. To this aim, we reconstituted Parkin-Ambra1 double-deficient MEFs (Ambra1gt/gt) with AMBRA1WT or AMBRA1LIR-AA and treated these cells with 30 μM FCCP for 9 h, in order to induce mitophagy. As shown in Figure 6a, AMBRA1WT is able to induce mitochondrial clearance, at variance with AMBRA1LIR-AA, that is not able to promote mitophagy. We next decided to use mouse embryonic fibroblasts (MEFs) from PINK1−/− mice in order to ascertain that the PINK1–PARKIN pathway was abolished. We overexpressed AMBRA1WT or AMBRA1LIR-AA and we treated cells with FCCP for 9 h (30 μM). Again, as shown in Figure 6b, overexpression of AMBRA1WT induces mitochondrial clearance, whereas overexpression of AMBRA1LIR-AA did not increase mitochondrial clearance to the same extent. To complete our analysis, we next used primary human cells from a PD patient with PINK1 loss of function (fibroblasts from Biobank of Genova, Italy). We introduced lentivirus expressing AMBRA1WTor AMBRA1LIR-AA in those cells and then treated them with FCCP for 5 h (40 μM). As shown in Figure 6c, overexpression of AMBRA1WT induces mitochondrial clearance. At variance, overexpression of AMBRA1LIR-AA did not increase mitochondrial clearance to the same extent as AMBRA1WT.
Figure 6.
See next page for Figure caption
These results indicate that AMBRA1 expression can partially rescue mitochondrial clearance in different ex vivo models, including cells from a PD patient. In sum, AMBRA1 can induce mitophagy through its LIR domain, sinergically with FCCP and in the absence of the PINK1–PARKIN pathway activity.
Discussion
AMBRA1 binds LC3 following mitophagy induction
We show that AMBRA1 uses a LIR motif to interact with LC3 following mitophagy induction. This interaction takes place both upon enhanced levels of AMBRA1 at the mitochondria and after FCCP treatment joint to PARKIN endogenous expression or overexpression. Our results thus suggest that LC3–AMBRA1 interaction is functional to an efficient mitochondrial clearance, rather than to the capture of AMBRA1 within the autophagosome in a sort of negative feedback control of autophagy. However, we cannot exclude that AMBRA1 is degraded by the autophagosomes in other conditions. Indeed, several ATG proteins are located on the outer surface of the phagophores, and some are also located within the autophagosomes.26, 27, 28 The interaction with LC3 may imply that a binding of AMBRA1 with other ATG proteins on LC3-positive structures could be required for correct autophagosome maturation.
AMBRA1–ActA induces depolarization and ubiquitylation of mitochondria
By another set of experiments, we demonstrate that all mitochondria found to be positive for AMBRA1–ActA are depolarized. An open question is how high levels of mitochondrial AMBRA1 would induce mitochondrial depolarization. We previously demonstrated that AMBRA1 could bind mito-Bcl-2.3 Considering all our findings, we can now postulate that high levels of AMBRA1 at the mitochondria, by mimicking BH3-only proteins, could sequester the majority of mito-Bcl-2, thus inducing Bax/Bak activation and mitochondrial membrane depolarization. As for the fate of depolarized mitochondria, it is important to bear in mind that ubiquitin is a signal for selective autophagy in mammalian cells. In fact, in the PARKIN-induced mitophagy pathway, PARKIN, an E3-ubiquitin ligase, translocates from the cytosol to mitochondria in cells treated with CCCP.5, 7 After translocation to the depolarized mitochondria, PARKIN catalyzes polyubiquitylation of several substrates, calling into play the autophagy receptor p62/SQSTM1 that simultaneously binds ubiquitin and autophagy-specific ubiquitin-like modifiers (LC3/GABARAP proteins).9, 20 Here we found that all mitochondria are ubiquitylated after AMBRA1–ActA overexpression. As we also found mitochondrial ubiquitin in PARKIN-deficient cells, AMBRA1–ActA could serve, most likely, as an adaptor for other E3 ligases. Noteworthy, we have recently demonstrated that AMBRA1 can bind the E3 ubiquitin ligase TRAF6.29 It will be interesting to investigate whether the binding between AMBRA1 and TRAF6 is an important event for mitochondria ubiquitylation. However, other candidates can help AMBRA1 to ubiquitylate mitochondria. In fact, AMBRA1 has also been found to be associated in a complex with the CUL4–DDB1 complex.30, 31
AMBRA1 mediates PINK1–PARKIN-independent mitophagy
Another issue is the mechanism by which endogenous AMBRA1 participates in mitochondrial clearance. We confirm that AMBRA1 potentiates PARKIN-mediated mitophagy, in line with previous work.16 We show that AMBRA1–ActA promotes PARKIN translocation to depolarized mitochondria in HEK293 cells, where PARKIN is present. However, in PARKIN-deficient cells, AMBRA1–ActA alone can induce mitochondrial clearance, indicating that a parallel pathway for mitophagy may exist when PARKIN is not present. Of the highest importance, wild-type AMBRA1, in combination with FCCP treatment, can also induce mitochondrial clearance in a PARKIN-independent manner, as in Pink1- or Parkin-deficient mouse fibroblasts or in human fibroblasts in which a mutant inactive form of PARKIN is present.
In sum, it is certainly possible that different mitophagy pathways coexist, depending on the inducing conditions or the cell type. Moreover, we found a new mechanism accounting for the effect of AMBRA1 on PARKIN-mediated mitophagy. In fact, AMBRA1 binding to LC3 increases mitochondrial clearance. In the absence of PARKIN, we also found a binding between AMBRA1 and LC3 after AMBRA1–ActA overexpression. Therefore, in the absence of PARKIN, AMBRA1 could be proposed as a selective mitophagy receptor, similar to the factor Atg32 in yeast.32, 33
Interestingly, it has recently been demonstrated that BECLIN 1 interacts with PARKIN and regulates PARKIN translocation to mitochondria.34 As we show that downregulation of BECLIN 1 reduces AMBRA1–ActA-induced mitophagy, it would be interesting to investigate the role of BECLIN 1 in AMBRA1–ActA-induced mitophagy. We can hypothesize that BECLIN 1 is functional to bring a putative E3 Ubiquitin ligase (alternative to PARKIN) in order to ubiquitylate mitochondria in AMBRA1–ActA-induced mitophagy.
AMBRA1–ActA is a powerful inducer of mitophagy
Hitherto, in mammalian cells, overexpression of PARKIN in combination with CCCP treatment is the only known method to induce mitophagy. We have now been able to prove that targeting AMBRA1 to mitochondria is sufficient to induce massive mitochondrial clearance. The next step will be to check whether AMBRA1–ActA induces mitophagy in vivo, this being of the highest importance in biomedicine. Furthermore, given the fact that AMBRA1 has a LIR motif used for its binding to LC3 during mitophagy, it would be interesting to investigate whether AMBRA1 could be mutated in this domain in neurodegenerative conditions, such as in PD. Similar mutations may also be associated with the risk of developing PD. At the same time, it could also be important to check AMBRA1 levels in other human disorders in which mitophagy is deregulated, such as muscular dystrophies.35
Materials and Methods
Antibodies
The antibodies used were as follows: polyclonal anti-PARKIN (Santa Cruz Biotechnology, Santa Cruz, CA, USA, H-300- sc-30130, Abcam, Cambridge, UK, 15954), mouse monoclonal anti-ACTIN (Sigma-Aldrich, St. Louis, MO, USA), monoclonal anti-BCL-2 (Santa Cruz Biotechnology, sc-7382), polyclonal anti-MnSOD (Assay Designs, Danvers, MA, USA), polyclonal anti-AMBRA1 (Novus Biologicals, Littleton, CO, USA, 26190002), polyclonal anti-LC3 (Cell Signaling, Danvers, MA, USA), monoclonal anti-GAPDH and rabbit polyclonal anti-LAMP1 (Abcam 24170).
Cells
The human embryonic kidney HEK293 cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich) supplemented with 10% FCS, 2 mM L-glutamine, and 1% penicillin/streptomycin solution at 37 °C under 5% CO2. MEFs cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), 2 mM L-glutamine and 1% penicillin/streptomycin solution at 37 °C. Cells were incubated in the presence of 30 μM FCCP (Sigma-Aldrich) for 9 h. Human fibroblasts (code: FFF0592011, Biobank Genova, Laboratorio di diagnosi Prenatale, Ist. Gaslini, Genova, Italy, kind gift of Dr. Filocamo) were obtained from a man bearing a mutation in the PINK1 gene, who developed PD when he was 40 years old (early-onset form of the disease). Human fibroblasts were then cultured in RPMI-140 medium supplemented with 10% FBS (Sigma-Aldrich), 2 mM L-glutamine and 1% penicillin/streptomycin solution at 37 °C. Pink1−/− mice were jointly provided by Professor Jie Shen (Harvard Medical School, Boston MT, USA) and Professor Antonio Pisani (IRCCS, Fondazione Santa Lucia, Roma, Italy). MEFs derived from Pink1−/− embryos at day 13.5 p.c. were grown in DMEM supplemented with 10% FBS (Sigma-Aldrich), 2 mM L-glutamine and 1% penicillin/streptomycin solution at 37 °C.
Cloning and plasmids
To create AMBRA1–ActA fusion gene, we first amplified by PCR, with PfuTurbo DNA polymerase, the oligonucleotides corresponding to aa 613–639 of Listeria ActA, using a vector encoding Bcl-2-ActA (a kind gift of Dr. Beth Levine, Dallas, TX, USA). The AMBRA1 coding region has also been generated by PCR with PfuTurbo DNA Polymerase, using wild-type myc-AMBRA1. Next, we digested these two PCR products with two restriction enzymes ApaI and HindIII, respectively, and then they were cloned into PcDNA3 vector (Invitrogen, Carlsbad, CA, USA). Construct coding for AMBRA1WT was cloned in pLPCX vector (Clontech Laboratories, Mountain View, CA, USA).
Point mutations were generated using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and all plasmid constructs made in this study were verified by DNA sequencing (Eurofins, Rome, Italy). The oligonucleotides used for mutagenesis, PCR and DNA sequencing were purchased from Invitrogen.
ShRNA PARKIN
ShRNA PARKIN (PGIP2 449497) or shRNA Control (PGIP2 ) were transfected in HEK293 cells using Turbofect Transfection reagent (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA, R0531), according to the manufacturer's instructions. At 24 or 48 h after transfection, PARKIN expression was controlled by western blot analysis.
Lentiviral production and infection
AMBRA1 cDNAs, wild-type and mutant, were subcloned from pLPCX, by cutting with NheI and NotI (blunted) restriction enzymes and transferred into the pRRL lentiviral plasmid digested with XbaI and SalI (blunted). For AMBRA1 ectopic expression in MEFs, lentiviral particles were produced by transfecting HEK293T cells with the lentiviral vector pRRLsinPPT.CMV.WPRE (Follenzi et al.36) encoding AMBRA1WT or AMBRA1-mutant (AMBRA1-LIRAA), together with the pMDLg/pRRE, pMD2.G and pRSV-Rev plasmids. Supernatants were collected 48 h post transfection. The lentiviral suspension was supplemented with polybrene (4 μg/ml, Sigma-Aldrich), filtered (PES 0.45 μM filters, Corning, Santa Clara, CA, USA) to remove cell debris and incubated with target cells for 8–12 h. To increase the transduction efficiency, the infection was repeated twice. Infected cells were then analysed starting from 48 h after infection.
Reagents
3-Methyladenine, Chloroquine and wortmannin were purchased from Sigma Chemical Co. (St. Louis, MO, USA). TMRM (cat. no. T668) was obtained from Invitrogen/Molecular Probes (Carlsbad, CA, USA). FCCP was from Sigma.
Western blot analysis
Cell extracts were centrifuged at 13 000 g for 10 min at 4 °C. Protein concentrations were determined with the Bio-Rad Protein Assay (Bio-rad Laboratories, Hercules, CA, USA). Cell extracts or immunoprecipitates were separated by SDS-PAGE and transferred onto nylon membranes (Immobilon P, Millipore, Bedford, MA, USA). Membranes were incubated with primary antibodies followed by horseradish peroxidase-conjugate secondary antibody (Jackson Laboratories, Ann Arbor, MI, USA) and visualized with ECL plus (Amersham Bioscience, Little Chalfont, UK).
Immunocytochemistry
Cells were washed in PBS and fixed with 4% paraformaldehyde in PBS for 15 min. After permeabilization with 0.4% Triton X-100 in PBS for 5 min, cells were blocked in 3% normal goat serum in PBS and incubated overnight at 4 °C with primary antibodies. We used the antibodies directed against AMBRA1, PARKIN and LC3. Cells were then washed in blocking buffer and incubated for 1 h with labelled anti-mouse (Alexa Fluor 488 or Alexa Fluor 555, Molecular Probes, Eugene, OR, USA) or anti-rabbit (FITC or Cy3, Jackson ImmunoResearch, West Grove, PA, USA) secondary antibodies. Nuclei were stained with 1 μg/ml DAPI and examined under a Zeiss LSM 700 100 × oil-immersion objective (Zeiss, Oberkoechen, Germany). We used ‘ZEN 2009 Light edition' software for image analysis. All measurements in this work were performed by a blind approach. All colocalization analyses were performed in nonsaturated single z-confocal planes.
Mitochondria purification
Mitochondria were isolated from HEK293 cells by standard differential centrifugation, and suspended in isolation buffer (IB; 0.2 M sucrose, 10 mM Tris-MOPS (pH 7.4), 0.1 mM EGTA-Tris and 0.1% delipidated BSA).
Immunoprecipitation
Cells were lysed in RIPA buffer (plus protease inhibitors). Equal amounts of protein (500 μg) were incubated with 2 μl of monoclonal anti-Myc antibody conjugated with protein A agarose beads (Clontech and Sigma, respectively) for 4 h followed by 60 min incubation with 30 μl of protein A Sepharose beads (Roche, Boulogne-Billancourt, France). The beads were collected by centrifugation and washed four times with the RIPA buffer. Proteins bounds to the beads were eluted with 30 μl of SDS-polyacrylamide gel electrophoresis sample buffer and heated to 95 °C for 10 min.
Electron microscopy
For conventional EM, HeLa cells previously transfected with AMBRA1–ActA and mito-GFP were sorted 24 or 48 h after transfection and fixed with 2.5% glutaraldehyde buffered with 0.1 M sodium phosphate, pH 7.4. Samples were postfixed with osmium tetroxide, then stained with uranyl acetate, dehydrated in ethanol and embedded in Epon resin (Fluka, Sigma-Aldrich). After sectioning, samples were collected on uncoated nickel grids and observed and photographed in a Technai 20 (FEI Company, Eindhoven, The Netherlands) EM.
Embryos lysis heads
Heads of embryos from stage E13.5 were subjected to mechanical lysis in 50 mM Tris HCl pH 7.5, 320 mM Sucrose, 50 mM NaCl, 1% Triton X-100 and protease inhibitors. Protein extracts were analysed by western blot analysis.
Ultrastructural analysis of brain from wild-type and Ambra1gt/gt embryos
The embryos (E13.5) were extracted from the deciduas and fixed by immersion in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 45 min at 4 °C, rinsed in buffer, postfixed in 1% OsO4 in 0.1 M cacodylate buffer, pH 7.4, dehydrated and embedded in Epon resin. Grids were thoroughly rinsed in distilled water, stained with aqueous 2% uranyl acetate for 20 min and photographed in a Zeiss EM 900 EM.
Acknowledgments
We thank Mrs M Acuña Villa and Dr M Bennett for secretarial and proofreading work and A Di Rita for research assistance. We are indebted to Professor B Levine (Dallas, TX, USA) for kindly providing us with the Bcl-2-ActA construct. We acknowledge Professor J Shen (Boston, MA, USA) who provided Pink1/mice and Dr. P Bonsi from Professor A. Pisani's lab (Rome, Italy) for her help in breeding these mice. The cell line from patients affected by Parkinson's disease were obtained from the ‘Cell Line and DNA Biobank from Patients affected by Genetic Diseases' (Istituto G. Gaslini) and the ‘Parkinson Institute Biobank' (Milan, http://www.parkinsonbiobank.com/), members of the Telethon Network of Genetic Biobanks ((http://www.biobanknetwork.org, project no. GTB12001), funded by Telethon Italy. This work was supported in part by grants from the Telethon Foundation (GGP10225), AIRC (IG2010 to FC and MyFAG 2009 to SC), FISM (2009), the Italian Ministry of University and Research (PRIN 2009 and FIRB Accordi di Programma 2011), the Italian Ministry of Health (Ricerca Finalizzata and Ricerca Corrente to FC, Ricerca Finalizzata–Progetto Giovani Ricercatori to SC) and Ricerca Finalizzata RF-OGR-2008-120-3614 to MP.
Glossary
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- ΔΨm
mitochondrial membrane potential
- LIR
LC3 interacting region
- FCCP
trifluorocarbonylcyanide phenylhydrazone
- ETNA
embryonic telencephalic naive cell
- ActA
Actin assembly-inducing protein
- EM
electron microscope
- 3-MA
3-methyladenine
- TMRM
Tetramethylrhodamine, methyl ester
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- PD
Parkinson's disease
- MEF
mouse embryonic fibroblast
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Kroemer
Supplementary Material
References
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;3:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Nardacci R, Corazzari M, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;4:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra N, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen S, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;6:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahi A, Sumpter R, Xiao G, Ng A, Zou Z, Tang Y, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson S, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, et al. High-content genome-wide RNAi screens identify regulators of parkin upstresam of mitophagy. Nat Lett. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosi. 2011;28:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M, Ferraro E, Ferri A, Rigamonti D, Quondamatteo F, Ding H, et al. Apoptosome inactivation rescues proneural and neural cells from neurodegeneration. Cell Death Differ. 2004;11:1179–1191. doi: 10.1038/sj.cdd.4401476. [DOI] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Diesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;9:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella M, Parker N, Tan CH, Hall AM, Duchen MR. IF(1):setting the pace of the F(1)F(o)-ATP synthase. Trends Biochem Sci. 2009;7:343–350. doi: 10.1016/j.tibs.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol. 2011;192:839–853. doi: 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitination, self-association and function via AMBRA1 and TRAF6. Nat Cell Biol. 2013;4:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1 interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;5:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Wade Harper J. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey V, Cagalinec M, Liiv J, Safiulina D, Hockey M, Kuum M, et al. BECN1 is involved in the initiation of mitophagy. It facilitates PARK2 translocation to mitochondria. Autophagy. 2014;10:1092–1106. doi: 10.4161/auto.28615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Berteggia E, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;11:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.