Abstract
It is controversial whether cells truly die via autophagy or whether — in dying cells — autophagy is merely an innocent bystander or a well-intentioned ‘Good Samaritan' trying to prevent inevitable cellular demise. However, there is increasing evidence that the genetic machinery of autophagy may be essential for cell death in certain settings. We recently identified a novel form of autophagy gene-dependent cell death, termed autosis, which is mediated by the Na+,K+-ATPase pump and has unique morphological features. High levels of cellular autophagy, as occurs with treatment with autophagy-inducing peptides, starvation, or in vivo during certain types of ischemia, can trigger autosis. These findings provide insights into the mechanisms and strategies for prevention of cell death during extreme stress conditions.
Facts
Although physiological levels of autophagy are essential for the maintenance of cellular homeostasis during various stress conditions, excessive or uncontrolled levels of autophagy are able to induce autophagy-dependent cell death.
Autosis is an autophagy-dependent non-apoptotic form of cell death, characterized by enhanced cell substrate adhesion, focal ballooning of the perinuclear space, and dilation and fragmentation of endoplasmic reticulum.
Autosis is triggered by autophagy-inducing peptides, starvation, and neonatal cerebral hypoxia-ischemia.
Pharmacological inhibition or genetic inactivation of Na+,K+-ATPase blocks autosis in vitro and in vivo.
Open Questions
How is autosis initiated and executed?
What are the biomarker(s) of autosis?
Is autosis a subtype of autophagic cell death or does all autophagic cell death occur by autosis?
Does autosis occur under other pathophysiological conditions?
What are the biological and clinical implications of autosis?
Programmed cell death is a highly regulated cellular response in metazoans to control cell fate following various cellular stresses and/or extrinsic stimuli. Historically, three types of cell death have been identified based on morphological criteria, including Type I (apoptosis), Type II (autophagic cell death), and Type III (necrosis).1,2 Autophagic cell death was originally defined as a type of cell death accompanied by large-scale autophagic vacuolization of the cytoplasm and resultant vacuolated appearance. On the basis of these ultrastructural criteria, autophagic cell death was described during animal development, tissue homeostasis, and in diseased tissues, as well as in cultured mammalian cells treated with chemotherapeutic agents or other toxic compounds.3 Using more stringent genetic criteria to define a causative role of autophagy in cell death, several studies in the past decade have shown that autophagy-dependent cell death occurs under certain experimental conditions.4 Recently, we identified a novel form of autophagy gene-dependent, Na+,K+-ATPase-regulated, non-apoptotic cell death, termed ‘autosis', which is induced by autophagy-inducing peptides, starvation, and hypoxia-ischemia, and characterized by the disappearance of endoplasmic reticulum and focal swelling of the perinuclear space.5 In this review, we summarize our current understanding of autophagy-dependent cell death and autosis in mammalian systems.
Historical Perspective of Autophagy and Autophagic Cell Death
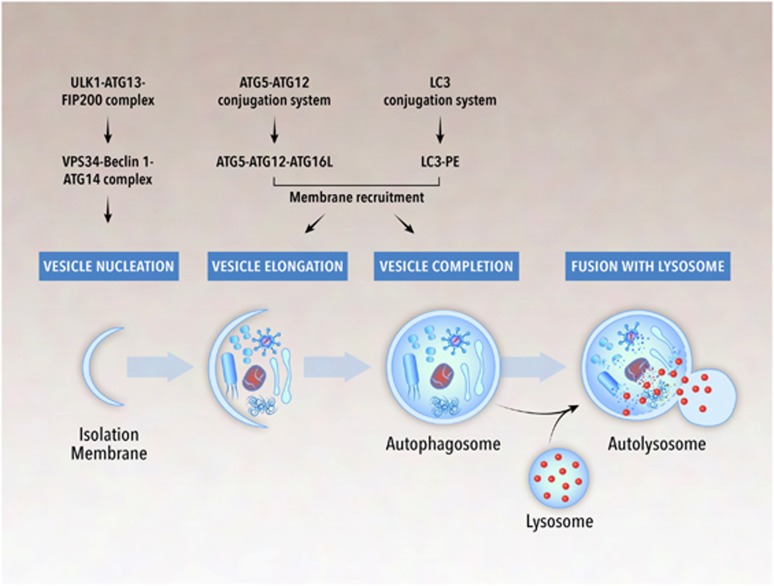
Physiological levels of autophagy promote cellular survival in response to a variety of stress conditions, including starvation, hypoxia, mitochondrial damage, and pathogen infection.6, 7, 8 A set of evolutionarily conserved proteins, the autophagy-related (Atg) proteins, mediate the homeostasis function of autophagy through the formation of a double-membrane bound structure termed the autophagosome (Figure 1). In the initial stage (vesicle nucleation), the ULK/Atg1 complex activates the class III phosphatidylinositol 3 kinase complex, which recruits a series of Atg proteins to the isolation membrane (phagophore) that forms either from preexisting organelles6 or via de novo lipid synthesis.9 In the subsequent stages of vesicle elongation and completion, two ubiquitin-like conjugation systems (Atg12-conjugation and microtubule-associated protein 1A/1B light chain 3 (LC3)-conjugation system) govern the covalent conjugation of Atg5 to Atg12 and the conversion of LC3-I to its phosphatidylethanolamine-conjugated form (LC3-II).10 The outer membrane of the mature autophagosome then fuses with the lysosome to form an autolysosome, in which the sequestered cytoplasmic material is degraded.
Figure 1.
Schematic overview of the autophagy pathway. Autophagy is a catabolic process in which damaged or redundant cellular organelles or exogenous pathogens are degraded by autophagosomes. The formation of the autophagosome includes vesicle nucleation, vesicle elongation, and vesicle completion, which are tightly regulated by various autophagy-related proteins, some of which are listed in the figure. The mature autophagosome then fuses with the lysosome to form an autolysosome. Inside the autolysosome, the sequestered contents are degraded
Excessive levels of autophagy have been observed in association with various forms of cell death and the term ‘autophagic cell death' was originally generated to describe cell death associated with autophagy.11 However, the evidence linking autophagy to cell death in these early reports was largely circumstantial, as the morphological criteria for ‘autophagic cell death' indicated only that autophagy was present in dying cells but could not be used to determine whether autophagy had a causative role in the death process.
To avoid confusion, the Nomenclature Committee on Cell Death (NCCD) reintroduced the term ‘autophagic cell death' to describe cell death that is suppressed by inhibition of the autophagy pathway.4 As none of the presently available inhibitors of autophagy function exclusively in autophagy regulation, it became necessary to include genetic inactivation of essential autophagic proteins to establish the role of autophagy in cell death. Accordingly, the most recent guidelines from the NCCD specifies that autophagic cell death should be identified only if the process is blocked by genetic interventions targeting at least two components of the molecular machinery of autophagy.4 This requirement recognizes that many autophagy proteins have multiple autophagy-independent functions. Another requirement is that clonogenic survival assays should be used to demonstrate long-term protection against cell death with genetic inhibition of autophagy, to avoid misleading conclusions based solely on alterations of cell death kinetics.
Using these more stringent criteria to define a causative role of autophagy in cell death, several studies in the past decade have shown that autophagy can be mechanistically involved in cell death, including cell death in invertebrate development (e.g., Drosophila midgut degradation and salivary gland destruction) and hypersensitive cell death in plants.12,13 In cultured mammalian cells, autophagy can contribute to cell death under certain experimental conditions characterized by the absence of intact apoptosis pathways; these include etoposide- and staurosporine-induced death in Bax−/−; Bak−/− murine embryonic fibroblasts,14,15 caspase inhibition in mouse fibroblasts,16,17 expression of a Beclin 1 mutant that escapes Bcl-2 regulation in breast cancer cells that have inactive caspase 3,18 and caspase 10 inhibition in myeloma cells.19 In cells competent for apoptosis, high levels of autophagy can also lead to autophagy gene-dependent and caspase-independent cell death, including in cells expressing a short isoform of p19ARF,20 in human ovarian epithelial cells expressing oncogenic H-RasV12,21 and in cells exposed to a variety of environmental stresses and toxic agents.22,23
Although such studies provide genetic support for autophagy as a bona fide mode of cell death, the nature of autophagic cell death that occurs in mammalian cells and tissues in response to pathophysiological stimuli (such as starvation) has (until recently) remained poorly defined. Except for an accumulation of autophagosomes/autolysosomes, it has been unclear whether cells that die ‘by autophagy' have any unique morphological features or exclusive death machinery to distinguish autophagic cell death from apoptosis and necrosis. The only morphological feature that has been linked to autophagic cell death — autophagic vacuolization — is also commonly observed in cells undergoing apoptotic or necrotic cell death, and no proteins aside from the core autophagy proteins have been shown to be required for autophagic cell death.
Links between Autophagy and other Death Machineries
Perhaps the best-characterized example of molecular overlap between regulation of autophagy and apoptosis is the interaction of the BH3 domain of the Beclin 1 autophagy protein with the anti-apoptotic proteins, Bcl-2 and Bcl-XL.24 In nutrition-rich conditions, the autophagy activity of Beclin 1 is inhibited by endoplasmic reticulum (ER)-localized Bcl-2.18 Cells possess multiple different mechanisms to positively and negatively regulate this interaction, and thereby either inhibit or stimulate autophagy. For example, the complex is stabilized by Mst1 kinase phosphorylation of the Thr108 residue in the BH3 domain of Beclin 1,25 and the complex dissociates after JNK1-mediated Bcl-2 phosphorylation (during nutrient starvation),26 DAPK-mediated phosphorylation of the Thr119 residue in the BH3 domain of Beclin 1,27 or overexpression of BH3-only proteins or treatment with BH3 peptidomimetic drugs.28 Moreover, disruption of the Bcl-2/Beclin 1 complex is required for starvation- and exercise-induced autophagy in vivo.7,18 The dual regulation of apoptosis and autophagy of Bcl-2 family members by binding to BH3 domains of pro-apoptotic proteins and pro-autophagy proteins most likely reflects the need for cells to coordinately regulate these two pathways; however, to date, there is no evidence that full-length Beclin 1 participates in apoptosis, and the practical implications of this dual regulation are not understood.
The apoptotic machinery may inhibit autophagy (presumably as a means of shutting off a pro-survival pathway at a time when it is clearly futile) by triggering caspase-mediated cleavage of Atg proteins, including Beclin 1,29 Atg4D,30 and Atg16L.31 In addition, the pro-apoptotic molecule, Bim, binds to and sequesters Beclin 1 on microtubules, which thereby inhibits autophagy.32 Conversely, the autophagy machinery may modulate apoptosis by autophagic degradation of apoptotic proteins.33 In TRAIL-resistant tumor cells, autophagy-mediated degradation of active caspase 8 prevents TRAIL-mediated apoptosis.34 In addition, some autophagy proteins may function directly as a pro-apoptotic factor to initiate apoptosis, such as a calpain cleavage product of Atg535 and the unconjugated form of Atg12.36
Autophagy has recently been demonstrated to have a role in necroptosis. The combination of an mTOR and lysosomal inhibitor results in RIPK1- and oxidative stress-dependent necroptosis in human renal carcinoma cell lines,37 suggesting that autophagy may inhibit necroptosis via the degradation of RIPK1 and decreasing reactive oxygen species (ROS). Although autophagy also has a cytoprotective role in TCR stimulation-induced necroptosis in c-FLIP-deficient T cells,38 it contributes to rapamycin/obatoclax- and dexamethasone-induced necroptosis.39 Autophagy induction by obatoclax results in FADD/RIPK1/RIPK3 recruitment to the autophagosomal membranes by interaction with Atg5,40 suggesting that autophagy may promote necroptosis via the assembly of the necrosome on autophagosomes.
In addition to apoptosis and necroptosis, autophagy also regulates other death pathways, including immunogenic cell death, entosis, and pyroptosis. Suppression of autophagy results in diminished release of ATP from dying tumor cells, indicating an essential role of autophagy in immunogenic cell death.41,42 Downstream (e.g., Vps34, Atg5, and Atg7), but not upstream (e.g., ULK1, Atg13 and FIP200), autophagy components are involved in LC3 recruitment to single-membrane entotic vacuoles,43 suggesting that entosis may utilize autophagy proteins to facilitate the clearance of internalized cells by lysosomes. During Shigella infection, autophagy may have a protective role against pyroptosis as treatment with the autophagy inhibitor 3-MA enhances caspase-1-dependent cell death in infected macrophages.44
Autosis, a New Form of Non-Apoptotic Cell Death
To investigate cell death in the setting of very high levels of autophagy, we evaluated the effects of an autophagy-inducing peptide, Tat-Beclin 1, on autophagy-dependent cell death. Tat-Beclin 1 is derived from 18 amino acids of the evolutionarily conserved domain of Beclin 1 fused to 11 amino acids from the HIV Tat protein transduction domain to facilitate cellular peptide entry. This peptide induces robust levels of autophagy through a mechanism thought to involve disruption of Beclin 1/GAPR-1 binding in the Golgi complex.45 Doses that have no apparent cytotoxic effects have a variety of beneficial effects in vitro and in vivo in mouse models; Tat-Beclin 1 inhibits the replication of HIV, arboviruses, and Listeria monocytogenes in vitro; enhances the clearance of mutant huntingtin protein aggregates; and protects mice against lethal chikungunya and West Nile virus infection.45 In fact, the brains of West Nile virus-infected mice treated with Tat-Beclin 1 have a marked reduction in neuronal cell death, as measured by TUNEL staining, presumably as a consequence of reduced central nervous system (CNS) viral titers and/or cytoprotective effects of the peptide.
Although these results suggest that autophagy upregulation may be beneficial and pro-survival in certain settings, we also found that — at least in vitro — Tat-Beclin 1-induced time- and dose-dependent cell death.5 This death was blocked by pharmacological or genetic inhibition of autophagy, but not of apoptosis or necroptosis; it did not overlap genetically with apoptosis or necroptosis; and it displayed unique morphological features. This autophagy-dependent type of cell death, termed ‘autosis' (auto, autophagic; tosis, death), is characterized by enhanced cell-substrate adherence, dilated and fragmented ER (early) and ER disappearance (late), and nuclear membrane convolution (early) and focal swelling of the perinuclear space (late; Table 1). We noted that another published autophagy-inducing peptide, Tat-vFLIP α2, which acts by releasing ATG3 from cellular FLIP,46 also induced autosis,5 indicating that autosis may be the underlying mechanism of cell death in previous reports of autophagic cell death.
Table 1. Comparison of the morphological features of autosis, ‘classical' type II autophagic cell death, apoptosis, and necrosis.
|
Cell death |
Autosis |
‘Classical' Type II Autophagic cell death |
Apoptosis |
Necrosis |
| Plasma membrane | Focal plasma membrane rupture | Plasma membrane rupture; Blebbing sometimes observed | Blebbing of the plasma membrane with preserved integrity | Plasma membrane rupture |
| Nucleus | Nuclear membrane convolution and shrinkage; focal concavity of the nuclear surface; focal ballooning of perinuclear space | Minor changes | Nuclear compaction and fragmentation | Dilatation of the nuclear membrane |
| Chromatin | Mild-moderate chromatin condensation | Minor changes | Marked chromatin condensation | Mild-moderate chromatin condensation |
| Autophagic structures | Numerous autophagosomes and autolysosomes (early-mid stages) with disappearance in final stages | Numerous autophagosomes and autolysosomes | Varies | Varies |
| Other cytoplasmic organelles | Electron-dense mitochondria with abnormal internal structure (early); swollen mitochondria (late); dilated and fragmented ER (early) and ER disappearance (late) | Enlargement of Golgi, mitochondria, and ER sometimes observed; depletion of cytoplasmic organelles sometimes observed | Minor changes | Swelling |
| Other unique features | Enhanced cell-substrate adhesion; membrane-bound densities in perinuclear space | Rounding up of cells and detachment from substrate; formation of apoptotic bodies | Cell swelling |
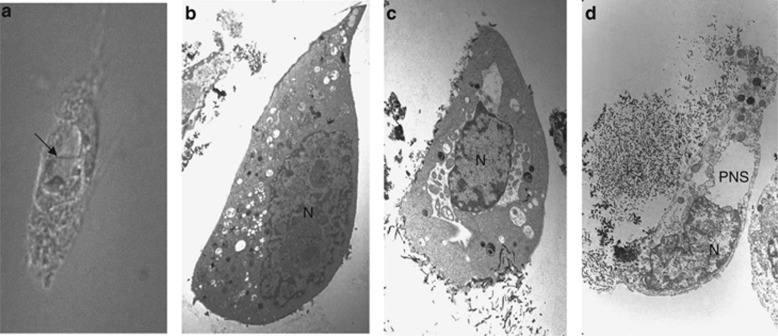
Autotic death can be identified by several criteria, including: (1) the absence of morphological, biochemical, and genetic evidence for other cell death pathways, (2) unique morphological changes, and (3) a unique dependence on Na+,K+-ATPase. While autosis is accompanied by several stereotypic morphological features (some of which have already been described in ‘classical' type II autophagic cell death; Table 1), the ‘sine qua non' of autosis is the nuclear membrane changes (Figure 2). At the light microscopic level, there is nuclear shrinkage with a focal concave portion adjacent to a round, vacuole-like entity (with a ‘balloon' appearance; Figure 2a); at the ultrastructural level, this balloon appearance corresponds to focal separation of the inner and outer nuclear membranes (Figures 2c and d). Although both autosis and apoptosis demonstrate chromatin condensation, it is very mild in autosis (Figure 2b) compared with apoptosis and neither DNA laddering nor TUNEL-positive staining occurs in autosis.5 Another unique feature of autosis compared with other forms of cell death is the increased substrate adherence of dying cells.
Figure 2.
Morphological features of autosis. (a) Representative light microscopic image of an autotic HeLa cell during starvation. Arrow shows area of focal nuclear concavity with adjacent focal ballooning of perinuclear space. (b–d) Representative electron microscopic images of different stages of autotic cell death; including (b) a cell in an early stage of autosis (referred to as phase 1a) with nuclear membrane convolution, mild chromatin condensation, numerous autophagosomes, and autolyososomes, dilated and fragmented ER, and electron-dense mitochondria; (c) a cell in a mid-stage of autosis (referred to as phase 1b) with separation of the inner and outer nuclear membrane, the presence of membrane-bound densities in the perinuclear space; and (d) a cell in the final stage of autosis (referred to as phase 2) with focal nuclear concavity, focal ballooning of the perinuclear space (which is empty) and disappearance of cellular organelles such as ER, autophagosomes, and autolysosomes. N, nucleus; PNS, perinuclear space
In addition to the unique nuclear membrane changes and increased substrate adherence, autosis demonstrates a distinct biochemical mechanism of regulation. Autotic cell death, but not apoptosis or necrosis, is inhibited by pharmacological or genetic inhibition of Na+,K+-ATPase.5 Conversely, treatment with caspase or RIP kinase inhibitors or genetic deletion of pro-apoptotic Bax and Bak or pro-necroptotic RIPK1 and RIPK3 does not protect cells against autotic cell death.5 Unlike necrosis or necroptosis, ROS is not a mediator in autosis.5 Thus, autosis utilizes an exclusive death machinery to initiate or execute cell death that is distinct from that of previously described forms of cell death. However, no biochemical markers are currently available to identify cells dying by autosis.
Although autosis was initially identified in the context of high levels of autophagy triggered by an autophagy-inducing peptide, this death pathway also occurs naturally during certain stress conditions. In vitro, autosis occurs in a subpopulation of cells that undergo the highest levels of autophagy and become substrate-adherent during nutrient starvation.5 Moreover, in vivo, autotic cell death occurs in hippocampal CA3 region neurons of neonatal rats subjected to cerebral hypoxia-ischemia.5 The unique features of autosis may have been missed in other tissues with ischemia-induced autophagic cell death that lack features of apoptosis and necrosis. More broadly, it remains to be determined whether autosis represents a subtype of autophagic cell death or whether all bona fide cell death by autophagy occurs through an autotic route.
How Do Cells Die in Autosis and Autophagic Cell Death?
In a chemical screen of ~5000 compounds with known biological targets, cardiac glycosides, antagonists of Na+,K+-ATPase, were the only identified inhibitors of autosis.5 Cardiac glycosides rescue clonogenic survival in vitro of cells that die by autophagy-inducing peptide or starvation-induced autosis, and they also blocks hippocampal autotic death and decrease cerebral infarct size following neonatal rat carotid artery ligation. In addition, knockdown of the α-subunit of Na+,K+-ATPase protects cells against autosis,5 confirming the specificity of the target of cardiac glycosides. The magnitude of autotic death rescue with cardiac glycosides or Na+,K+-ATPase knockdown is greater than with single autophagy gene knockdown.5 Although this may reflect incomplete ablation of the autophagy pathway by autophagy gene knockdown, it is also possible that autophagy proteins do not function in the core death machinery of autosis. Rather, excessive levels of autophagy may act upstream as a primer to ignite the death machinery that occurs in autosis.
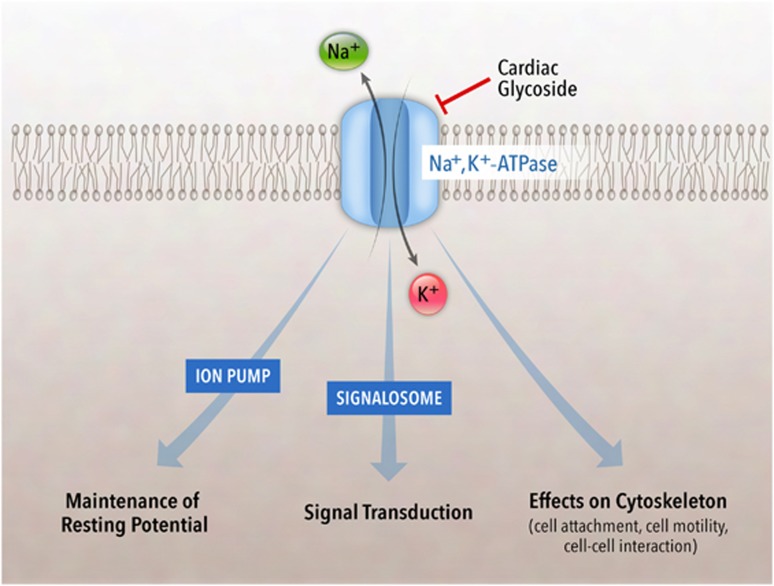
Na+,K+-ATPase is a well-characterized membrane pump that generates Na+ and K+ gradients across plasma membranes47 and also functions as a versatile transducer to mediate various cellular signaling pathways48 (Figure 3). In addition, Na+,K+-ATPase has a role in cell-substrate adhesion,49,50 which may explain the enhanced substrate adherence of cells undergoing autotic death. Na+,K+-ATPase localizes to the inner nuclear membrane51 as well as to the plasma membrane; nuclear envelope-associated Na+,K+-ATPase activity may alter membrane ionic transport and osmolarity, and thereby, contribute to the ER and perinuclear space expansion observed in autotic cells.
Figure 3.
Multiple functions of Na+,K+-ATPase. Na+,K+-ATPase is a ubiquitous plasma membrane protein complex (consisting of α- and β-subunits) which functions in ion homeostasis by pumping Na+ out of cells and pumping K+ into cells. The ion pump function of Na+,K+-ATPase maintains the cell membrane potential, which is specifically inhibited by cardiac glycosides. In addition to its ion transporter function, Na+,K+-ATPase functions as a signalosome, recruiting diverse signaling molecules to initiate a series of signaling pathways. Na+,K+-ATPase also regulates the cytoskeleton, tight junctions, cell motility, and cell polarity
The distinct cellular morphological features of autosis may provide clues regarding the mechanisms involved in autotic death. The most striking morphological feature of autosis, separation of inner and outer nuclear membrane and resultant focal expansion of the perinuclear space, is rescued by cardiac glycosides or Na+,K+-ATPase knockdown. Before the final stage of cellular demise (when this perinuclear space is empty), there are several ~100-nm-sized membrane-bound densities in between the inner and outer nuclear membrane (Figure 2c); the origin of these structures is not known, but their presence is suggestive of dynamic membrane mobilization of the ER/outer nuclear membrane during autosis. Although similar expansion of the perinuclear space and dilation of the ER has not been previously reported in mammalian cell death, it has been observed in cells expressing disease-associated inner nuclear membrane lamin B receptor mutants.52 The molecular mechanism of this cellular phenotype was not defined, but it is noteworthy that expression of mutant sterol reductases localized to the ER and outer nuclear membrane, TM7SF2 and DHCR1, result in a similar phenotype as that of disease-associated lamin B mutants.52 This suggests that disruption of cholesterol metabolism in the ER/outer nuclear membrane is sufficient to produce the phenotype of ER and focal perinuclear space expansion. This phenotype may be caused by alterations of ER membrane properties including transport or channel conductance, which would result in osmotic changes and disruption of signaling through the nuclear envelope. In fact, members of the LINC complex, which links the outer and inner nuclear membranes to establish nucleocytoplasmic communication, are lost from the perinuclear expansions in cells expressing abnormal levels of sterol reductase.52 Interestingly, in plant wounds, there is an accumulation of phytoecdysteroids (e.g., 20-hydroxyecdysone),53 which is associated with cell death that has rapid nuclear collapse and nuclear envelope separation.54 In the Drosophila salivary gland, the steroid hormone 20-hydroxyecdysone induces autophagy-dependent cell death.55 Together, these observations suggest that disturbances in cholesterol metabolism and ER membrane homeostasis may contribute to autosis.
Given the crucial role of the ER in autophagosomal biogenesis,56 we speculate that stimulation of very high levels of autophagy in certain contexts may perturb the normal homeostatic mechanisms of the ER, leading to similar expansions of the ER lumen and perinuclear space as that seen with overexpression of mutant sterol reductases. Alternatively, the outer nuclear membrane may serve as a key site of initiation of the autosis program. The outer nuclear membrane is physically contiguous with the ER, but under physiological conditions, the autophagy machinery selectively avoids using the outer nuclear membrane as a membrane source to form autophagosomes. In the setting of extremely high levels of autophagy, however, the ER membrane may become exhausted as a source of autophagosomes, triggering the pathological use of the outer nuclear membrane (and perhaps some as-of-yet undefined signal that the cell should self-destruct). This hypothesis is consistent with the depletion of the ER at late stages of autosis and the presence of membrane-bound structures in the perinuclear space.
Although ROS may contribute to other forms of autophagy-dependent cell death,13,16,17,57 they do not appear to contribute to autosis (our unpublished data). Furthermore, other reported factors in autophagic cell death, including JNK,14,15,58 AMPK,59 MAPK,60 BNIP3,61 Noxa,21 cathepsin L,62 and BCLAF119 have been demonstrated to participate in apoptosis or other cellular stress responses, suggesting that the crosstalk between apoptosis and autophagy has important regulatory functions in autophagic cell death. However, it is not clear whether these mechanisms induce or mediate autosis. Moreover, it is not known which cellular alterations function directly as inducers and executors of autosis and other forms of autophagic cell death; the possibilities include alterations in lipid or membrane dynamics, protein functions, ion homeostasis, ROS or other factors.
A Na+,K+-ATPase-mediated function is an essential upstream component of autosis; however, it is not known how this ion pump leads to the downstream morphological changes and demise of the cell. Na+,K+-ATPase may also function upstream of other death pathways, as neonatal rat treatment with cardiac glycosides blocks not only autotic death in the CA3 region of the hippocampus, but also other forms of cell death that occur in other regions of the brain.5 Despite recent evidence that autophagy, in the absence of other death pathways, can lead to cell death, it remains unknown whether the ultimate demise of the cell is a function of uncontrolled autophagy (i.e., eating oneself to death) or a byproduct of other cellular processes that occur as a consequence of autophagy. In autosis, the morphological features, including the late disappearance of the ER, are consistent with a scenario in which cells deplete organelles essential for cell survival through excessive autophagy. However, the dependence of this death process on the Na+,K+-ATPase pump favors a model in which autophagy and Na+,K+-ATPase activity cooperate to trigger an as-of-yet undefined death execution pathway.
Indeed, the concept that distinct signals from those involved in autophagy may be crucial in determining whether autophagy results in cell death has been previously demonstrated in Dictyostelium discoideum.63 The differentiation factor, DIF-1, triggers the death of starved cells undergoing autophagy, but not of non-starved cells, and conversely, starvation and autophagy, in the absence of DIF-1, do not result in cell death. An intriguing new frontier in autophagy and cell death research will be to more precisely define the spectrum of ‘second signals' for autophagic cell death, and how such signals interface with the autophagic machinery to result in cellular demise.
Cardiac Glycosides and Na+K+-ATPase
Cardiac glycosides, a large family of naturally derived steroidal compounds, were first described for the treatment of heart failure in 1785,64 and ~50 years ago, the cellular target of cardiac glycosides was identified as Na+,K+-ATPase (Figure 3). Cardiac glycosides bind to the extracellular site of the α-subunit of Na+,K+-ATPase, which results in a conformational change of Na+,K+-ATPase and stabilizes the enzyme in a specific enzymatic conformation (E2-P).65 Inhibition of the pumping function of Na+,K+-ATPase by cardiac glycosides increases intracellular sodium concentrations, which inactivates Na+/Ca2+ exchange and causes an increase of intracellular calcium concentrations, and thereby a positive inotropic effect in cardiac muscle. Independently of this action, cardiac glycosides directly activate the Na+,K+-ATPase-mediated signalosome complex,48,64,66 which is subsequently transduced by the MAPK cascade and IP3 receptor to generate ROS, Ca2+ oscillations, and other cellular stresses. Na+,K+-ATPase also modulates cell polarity, cell motility, the cytoskeleton, and cell–cell interactions.67,68 It is unclear which of these actions of Na+,K+-ATPase are important in autosis.
The Na+,K+-ATPase is a heterodimeric protein consisting of a large α and a heavily glycosylated β-subunit.69 The α-subunit has four isoforms; α1 is ubiquitously expressed, whereas α2, α3, and α4, exhibit tissue-specific expression. However, it should be noted that, although all human α-isoforms are sensitive to cardiac glycosides, the α1 isoform in rodents is resistant to cardiac glycosides.70 Consistent with this species difference, autosis in mouse fibroblasts (which express predominantly the α1 isoform) is inhibited by siRNA knockdown of the Na+,K+-ATPase α1 subunit but not by treatment with the cardiac glycosides.5 However, as rodent brain expresses cardiac glycoside-sensitive α2 and α3 isoforms, treatment with the CNS-penetrating cardiac glycoside, neriifolin, prevents autotic neuronal cell death in rat cerebral hypoxic-ischemic injury.5
Cardiac glycosides modulate autophagy in different experimental settings. In a high-content screen for autophagy inducers, cardiac glycosides were found to induce autophagy under nutrition-rich conditions.71 In addition, cardiac glycosides induce autophagy in human non-small cell lung cancer cells through mTOR and ERK1/2 signaling pathways.72 However, in these studies, there was no genetic evidence that the autophagy-inducing effect of cardiac glycosides were due to Na+,K+-ATPase inhibition. In contrast, cardiac glycosides reduce levels of autophagy under autosis-inducing conditions, including treatment with the autophagy-inducing peptides, starvation, and cerebral hypoxia-ischemia.5 These data are consistent with dual roles of cardiac glycosides to maintain levels of autophagy within a physiological range.
Of note, animals increase circulating levels of endogenous inhibitors of Na+,K+-ATPase inhibitors (for example, ouabain-like factors) following various physiological and pathological stresses. It is intriguing to speculate that these endogenous Na+,K+-ATPase inhibitors allow animals to maintain proper physiological levels of autophagy by restraining excessive levels that may maladaptively promote autosis.
Clinical Implications
Although the underlying mechanisms of the Na+,K+-ATPase-mediated autosis remain to be determined, the discovery of this unique form of cell death may have important clinical implications. Ischemic stroke, the destruction of cerebral tissue following deprivation of oxygen- and nutrient-rich blood flow, is an important cause of neurological deficits.73 Because the CNS cannot regenerate, the protection of neurons against ischemia-induced cell death is a key target in stroke therapy.
Multiple forms of cell death have been observed in neonatal ischemic brain damage,74 but neurons in CA3 regions of the hippocampus have a preference for an exclusive form of ischemia-induced cell death, which is characterized by early autophagic features in the absence of any signs of apoptosis or necrosis.75 Inhibition of autophagy by a pharmacological inhibitor, 3-methyladenine,76,77 or genetic inactivation of the autophagy genes, Atg7 or beclin 1,78, 79, 80 significantly protects against ischemia-induced neuronal death, indicating that autophagy contributes to cell death and infarct size in neonatal ischemic brain injury. Furthermore, the cardiac glycoside, neriifolin, protects against neuronal injury in a mouse brain slide-based model81 and whole-animal studies.5,81, 82, 83, 84 These observations suggest that CNS-penetrating cardiac glycosides may warrant investigation in neonatal cerebral hypoxia-ischemia.
Autosis may contribute to ischemic injury in other organs, such as the heart and kidney. The induction of autophagy and cardiac injury during ischemia reperfusion is significantly attenuated in beclin 1+/− mice,85 suggesting that autophagy may function as a pro-death factor in this setting. In mouse renal ischemia/reperfusion injury, Atg5 deletion sensitizes the kidneys to ischemic injury,86, 87, 88 but autophagy has been shown to be detrimental in prolonged ischemia.89, 90, 91, 92, 93, 94 However, it is not clear that reperfusion injury is an important mechanism of cardiac dysfunction in the clinical setting, and there is no clinical evidence that cardiac glycosides are beneficial in preventing the consequences of cardiac or renal ischemia.
Although autophagy has a crucial role in degrading aggregate-prone proteins, in a mouse model of familial amyotrophic lateral sclerosis (ALS), autophagy induction by rapamycin increases motor neuron degeneration95 and beclin 1 haploinsufficiency prolongs life span of mutant SOD1 transgenic mice,96 suggesting that autophagy may contribute to neurodegeneration in certain settings. If autotic cell death mediates neuronal cell death in mouse models of ALS, clinical trials of cardiac glycosides in patients with ALS may be warranted.
Summary
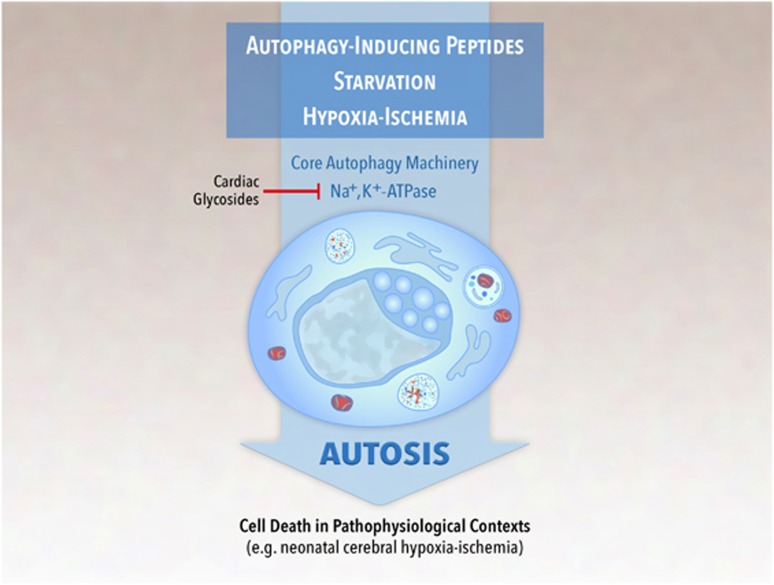
We identified a novel form of non-apoptotic autophagy-dependent cell death, termed autosis, which has unique morphological features; depends on the cellular Na+,K+-ATPase; and occurs during starvation, autophagy-inducing peptide treatment, and in vivo during cerebral hypoxia-ischemia (Figure 4). The discovery of autosis should stimulate exploration of the molecular mechanisms of this form of cell death; will pave the road to understanding the role of autophagic cell death under various physiological and pathophysiological conditions; may define a role for autosis in human disease; and is likely to provide novel targets for therapeutic strategies. Thus, vis-à-vis our understanding of autosis, it seems that death is only the beginning.
Figure 4.
Summary of autosis inducers, mediators, and known pathophysiological contexts. Autosis is a novel form of autophagy-dependent, non-apoptotic cell death, which is induced by autophagy-inducing peptides, starvation, and hypoxia-ischemia. This process requires the core autophagy machinery and Na+,K+-ATPase. Thus far, it is known to contribute to cerebral infarct size in rodent neonatal cerebral hypoxia-ischemia. Cardiac glycosides, antagonists of Na+,K+-ATPase, inhibit autosis induced by peptides, starvation, and hypoxia-ischemia, and reduce cerebral infarct size in rodent neonatal cerebral hypoxia-ischemia
Acknowledgments
We thank Haley Harrington for help with manuscript preparation and Angela Diehl for help with medical illustration. The work in the authors' laboratory was supported by CPRIT grant RP120718-P1 (BL); NIH grants R01CA84254 (BL), R01CA109618 (BL), U19AI109725 (BL), and an American Heart Fellowship 14POST20040022 (YL).
Glossary
- Atg
autophagy related
- LC3
microtubule-associated protein 1A/1B light chain 3
- NCCD
Nomenclature Committee on Cell Death
- ER
endoplasmic reticulum
- PNS
perinuclear space
- ROS
reactive oxygen species
- CNS
central nervous system
- ALS
amyotrophic lateral sclerosis
The authors declare no conflict of interest.
References
- Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- Schweichel JU, Merker HJ. The morphology of various types of cell death in prenatal tissues. Teratology. 1973;7:253–266. doi: 10.1002/tera.1420070306. [DOI] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict. J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shoji-Kawata S, Sumpter RM, Jr, Wei Y, Ginet V, Zhang L, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Hackenberg T, Juul T, Auzina A, Gwizdz S, Malolepszy A, Van Der Kelen K, et al. Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell. 2013;25:4616–4626. doi: 10.1105/tpc.113.117192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Konishi A, Nishida Y, Mizuta T, Nishina H, Yamamoto A, et al. Involvement of JNK in the regulation of autophagic cell death. Oncogene. 2010;29:2070–2082. doi: 10.1038/onc.2009.487. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Lamy L, Ngo VN, Emre NC, Shaffer AL, 3rd, Yang Y, Tian E, et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23:435–449. doi: 10.1016/j.ccr.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Sharma K, Le N, Alotaibi M, Gewirtz DA. Cytotoxic autophagy in cancer therapy. Int J Mol Sci. 2014;15:10034–10051. doi: 10.3390/ijms150610034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz DA. When cytoprotective autophagy isn't... and even when it is. Autophagy. 2014;10:391–392. doi: 10.4161/auto.27719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–277. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell. 2012;47:359–370. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead. Protein Cell. 2012;3:17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One. 2012;7:e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He MX, He YW. A role for c-FLIP(L) in the regulation of apoptosis, autophagy, and necroptosis in T lymphocytes. Cell Death Differ. 2013;20:188–197. doi: 10.1038/cdd.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace L, Bornhauser BC, Schmitz M, Cario G, Ziegler U, Niggli FK, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 2010;120:1310–1323. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit F, Cristofanon S, Fulda S. Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 2013;20:1161–1173. doi: 10.1038/cdd.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- Ko A, Kanehisa A, Martins I, Senovilla L, Chargari C, Dugue D, et al. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell Death Differ. 2014;21:92–99. doi: 10.1038/cdd.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- Newman RA, Yang P, Pawlus AD, Block KI. Cardiac glycosides as novel cancer therapeutic agents. Mol Interv. 2008;8:36–49. doi: 10.1124/mi.8.1.8. [DOI] [PubMed] [Google Scholar]
- Contreras RG, Shoshani L, Flores-Maldonado C, Lazaro A, Cereijido M. Relationship between Na(+),K(+)-ATPase and cell attachment. J Cell Sci. 1999;112 (Pt 23:4223–4232. doi: 10.1242/jcs.112.23.4223. [DOI] [PubMed] [Google Scholar]
- Belusa R, Aizman O, Andersson RM, Aperia A. Changes in Na(+)-K(+)-ATPase activity influence cell attachment to fibronectin. Am J Physiol Cell Physiol. 2002;282:C302–C309. doi: 10.1152/ajpcell.00117.2001. [DOI] [PubMed] [Google Scholar]
- Galva C, Artigas P, Gatto C. Nuclear Na+/K+-ATPase plays an active role in nucleoplasmic Ca2+ homeostasis. J Cell Sci. 2012;125:6137–6147. doi: 10.1242/jcs.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Kolb T, Richter K, Karakesisoglou I, Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol Biol Cell. 2010;21:354–368. doi: 10.1091/mbc.E09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Grebenok RJ, Galbraith DW, Bowers WS. Damage-induced accumulation of phytoecdysteroids in spinach: a rapid root response involving the octadecanoic acid pathway. J Chem Ecol. 1998;24:339–360. [Google Scholar]
- Cutler SR, Somerville CR. Imaging plant cell death: GFP-Nit1 aggregation marks an early step of wound and herbicide induced cell death. BMC Plant Biol. 2005;5:4. doi: 10.1186/1471-2229-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Kim EH, Sohn S, Kwon HJ, Kim SU, Kim MJ, Lee SJ, et al. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res. 2007;67:6314–6324. doi: 10.1158/0008-5472.CAN-06-4217. [DOI] [PubMed] [Google Scholar]
- Chen SY, Chiu LY, Maa MC, Wang JS, Chien CL, Lin WW. zVAD-induced autophagic cell death requires c-Src-dependent ERK and JNK activation and reactive oxygen species generation. Autophagy. 2011;7:217–228. doi: 10.4161/auto.7.2.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZX, Liang J, Haridas V, Gaikwad A, Connolly FP, Mills GB, et al. A plant triterpenoid, avicin D, induces autophagy by activation of AMP-activated protein kinase. Cell Death Differ. 2007;14:1948–1957. doi: 10.1038/sj.cdd.4402207. [DOI] [PubMed] [Google Scholar]
- Guo WJ, Zhang YM, Zhang L, Huang B, Tao FF, Chen W, et al. Novel monofunctional platinum (II) complex Mono-Pt induces apoptosis-independent autophagic cell death in human ovarian carcinoma cells, distinct from cisplatin. Autophagy. 2013;9:996–1008. doi: 10.4161/auto.24407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma H, Gangadhar NM, Letso RR, Wolpaw AJ, Sriramaratnam R, Stockwell BR. Identification of a small molecule that induces ATG5-and-cathepsin-l-dependent cell death and modulates polyglutamine toxicity. Exp Cell Res. 2013;319:1759–1773. doi: 10.1016/j.yexcr.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti C, Luciani MF, Golstein P. A second signal for autophagic cell death. Autophagy. 2010;6:823–824. doi: 10.4161/auto.6.6.12750. [DOI] [PubMed] [Google Scholar]
- Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- Yatime L, Laursen M, Morth JP, Esmann M, Nissen P, Fedosova NU. Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase. J Struct Biol. 2011;174:296–306. doi: 10.1016/j.jsb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Xie Z, Cai T. Na+-K+—ATPase-mediated signal transduction: from protein interaction to cellular function. Mol Interv. 2003;3:157–168. doi: 10.1124/mi.3.3.157. [DOI] [PubMed] [Google Scholar]
- Geering K. Functional roles of Na,K-ATPase subunits. Curr Opin Nephrol Hypertens. 2008;17:526–532. doi: 10.1097/MNH.0b013e3283036cbf. [DOI] [PubMed] [Google Scholar]
- Rajasekaran SA, Barwe SP, Rajasekaran AK. Multiple functions of Na,K-ATPase in epithelial cells. Semin Nephrol. 2005;25:328–334. doi: 10.1016/j.semnephrol.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. Na+, K+-ATPase: functions in the nervous system and involvement in neurologic disease. Neurology. 2011;76:287–293. doi: 10.1212/WNL.0b013e3182074c2f. [DOI] [PubMed] [Google Scholar]
- Lingrel JB, Orlowski J, Shull MM, Price EM. Molecular genetics of Na,K-ATPase. Prog Nucleic Acid Res Mol Biol. 1990;38:37–89. doi: 10.1016/s0079-6603(08)60708-4. [DOI] [PubMed] [Google Scholar]
- Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qiu Q, Shen JJ, Li DD, Jiang XJ, Si SY, et al. Cardiac glycosides induce autophagy in human non-small cell lung cancer cells through regulation of dual signaling pathways. Int J Biochem Cell Biol. 2012;44:1813–1824. doi: 10.1016/j.biocel.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Apoptosis in neurodegenerative disorders. Nat Rev Mol Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- Puyal J, Ginet V, Clarke PG. Multiple interacting cell death mechanisms in the mediation of excitotoxicity and ischemic brain damage: a challenge for neuroprotection. Prog Neurobiol. 2013;105:24–48. doi: 10.1016/j.pneurobio.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Ginet V, Puyal J, Clarke PG, Truttmann AC. Enhancement of autophagic flux after neonatal cerebral hypoxia-ischemia and its region-specific relationship to apoptotic mechanisms. Am J Pathol. 2009;175:1962–1974. doi: 10.2353/ajpath.2009.090463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Sheng R, Zhang LS, Han R, Zhang X, Zhang XD, et al. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008;4:762–769. doi: 10.4161/auto.6412. [DOI] [PubMed] [Google Scholar]
- Puyal J, Vaslin A, Mottier V, Clarke PG. Postischemic treatment of neonatal cerebral ischemia should target autophagy. Ann Neurol. 2009;66:378–389. doi: 10.1002/ana.21714. [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, et al. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J Pathol. 2008;172:454–469. doi: 10.2353/ajpath.2008.070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Zhang Y, Li J, Zhang J, Li Y, Dang C, et al. Beclin 1 knockdown inhibits autophagic activation and prevents the secondary neurodegenerative damage in the ipsilateral thalamus following focal cerebral infarction. Autophagy. 2012;8:63–76. doi: 10.4161/auto.8.1.18217. [DOI] [PubMed] [Google Scholar]
- Ginet V, Spiehlmann A, Rummel C, Rudinskiy N, Grishchuk Y, Luthi-Carter R, et al. Involvement of autophagy in hypoxic-excitotoxic neuronal death. Autophagy. 2014;10:846–860. doi: 10.4161/auto.28264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JK, Portbury S, Thomas MB, Barney S, Ricca DJ, Morris DL, et al. Cardiac glycosides provide neuroprotection against ischemic stroke: discovery by a brain slice-based compound screening platform. Proc Natl Acad Sci USA. 2006;103:10461–10466. doi: 10.1073/pnas.0600930103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Rehni AK, Singh N, Jaggi AS. Studies on cerebral protection of digoxin against ischemia/reperfusion injury in mice. Yakugaku Zasshi. 2009;129:435–443. doi: 10.1248/yakushi.129.435. [DOI] [PubMed] [Google Scholar]
- Dunn DE, He DN, Yang P, Johansen M, Newman RA, Lo DC. In vitro and in vivo neuroprotective activity of the cardiac glycoside oleandrin from Nerium oleander in brain slice-based stroke models. J Neurochem. 2011;119:805–814. doi: 10.1111/j.1471-4159.2011.07439.x. [DOI] [PubMed] [Google Scholar]
- Van Kanegan MJ, He DN, Dunn DE, Yang P, Newman RA, West AE, et al. BDNF mediates neuroprotection against oxygen-glucose deprivation by the cardiac glycoside oleandrin. J Neurosci. 2014;34:963–968. doi: 10.1523/JNEUROSCI.2700-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- Liu S, Hartleben B, Kretz O, Wiech T, Igarashi P, Mizushima N, et al. Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy. 2012;8:826–837. doi: 10.4161/auto.19419. [DOI] [PubMed] [Google Scholar]
- Jiang M, Liu K, Luo J, Dong Z. Autophagy is a renoprotective mechanism during in vitro hypoxia and in vivo ischemia-reperfusion injury. Am J Pathol. 2010;176:1181–1192. doi: 10.2353/ajpath.2010.090594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011;22:902–913. doi: 10.1681/ASN.2010070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Nishihara K, Inui K, Masuda S. Involvement of autophagy in the pharmacological effects of the mTOR inhibitor everolimus in acute kidney injury. Eur J Pharmacol. 2012;696:143–154. doi: 10.1016/j.ejphar.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Chien CT, Shyue SK, Lai MK. Bcl-xL augmentation potentially reduces ischemia/reperfusion induced proximal and distal tubular apoptosis and autophagy. Transplantation. 2007;84:1183–1190. doi: 10.1097/01.tp.0000287334.38933.e3. [DOI] [PubMed] [Google Scholar]
- Decuypere JP, Pirenne J, Jochmans I. Autophagy in renal ischemia-reperfusion injury: friend or foe. Am J Transplant. 2014;14:1464–1465. doi: 10.1111/ajt.12717. [DOI] [PubMed] [Google Scholar]
- Yeh CH, Hsu SP, Yang CC, Chien CT, Wang NP. Hypoxic preconditioning reinforces HIF-alpha-dependent HSP70 signaling to reduce ischemic renal failure-induced renal tubular apoptosis and autophagy. Life Sci. 2010;86:115–123. doi: 10.1016/j.lfs.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Wu HH, Hsiao TY, Chien CT, Lai MK. Ischemic conditioning by short periods of reperfusion attenuates renal ischemia/reperfusion induced apoptosis and autophagy in the rat. J Biomed Sci. 2009;16:19. doi: 10.1186/1423-0127-16-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaka Y, Suzuki C, Abe T, Okumi M, Ichimaru N, Imamura R, et al. Bcl-2 protects tubular epithelial cells from ischemia/reperfusion injury by dual mechanisms. Transplant Proc. 2009;41:52–54. doi: 10.1016/j.transproceed.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li L, Chen S, Yang D, Wang Y, Wang Z, et al. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- Nassif M, Valenzuela V, Rojas-Rivera D, Vidal R, Matus S, Castillo K, et al. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014;10:1256–1271. doi: 10.4161/auto.28784. [DOI] [PMC free article] [PubMed] [Google Scholar]