Abstract
Human pluripotent stem cells, under the right conditions, can be engineered to generate populations of any somatic cell type. Knowledge of what mechanisms govern differentiation towards a particular lineage is often quite useful for efficiently producing somatic cell populations from hPSCs. Here, we have outlined a strategy for deriving populations of simple epithelial cells, as well as more mature epidermal keratinocyte progenitors, from hPSCs by exploiting a mechanism previously shown to direct epithelial differentiation of hPSCs. Specifically, we describe how to direct epithelial differentiation of hPSCs using a Src family kinase inhibitor, SU6656, which has been shown to modulate β-catenin translocation to the cell membrane and thus promote epithelial differentiation. The differentiation platform outlined here produces cells with the ability to terminally differentiate to epidermal keratinocytes in culture through a stable simple epithelial cell intermediate that can be expanded in culture for numerous (>10) passages.
Keywords: Human pluripotent stem cells, differentiation, epithelial cells, Src kinase inhibitor, epidermal
1. Introduction
Human pluripotent stem cells (hPSCs) can be coerced into generating populations of any somatic cell type in the human body [1-3]. Given this ability, hPSCs can be used for in vitro models to study human development and disease, as well as for applications in regenerative medicine. To generate populations of somatic cells to be used for such applications, it is imperative to design differentiation systems that are robust and produce high purity populations of cells. While there are different strategies to obtain epithelial populations from hPSCs [4-8], a recent study demonstrated how epithelial differentiation can be modulated by β-catenin localization, providing insight as to what mechanisms are involved in governing the epithelial differentiation process [9]. Here, we describe a method to produce simple epithelial cells and, subsequently, epidermal keratinocyte progenitor populations by exploiting this mechanism using a Src family kinase inhibitor.
To efficiently derive populations of epithelial cells to be used for tissue engineering applications, it is optimal first to generate highly-enriched populations of simple epithelial cells. These cells can be characterized by high levels of cytokeratin 18 (K18), expressed by simple, or single-layered epithelial cells in vivo [8], and the lack of transcription factors such as Oct4 and Nanog, expressed in hPSCs and play critical roles in regulating pluripotency [10]. Upon further differentiation and epithelial maturation, simple epithelial cells lose K18 expression and acquire expression of cytokeratin 14 (K14), found in the basal layer of many epithelial tissues, including the epidermis [8,11]. In addition, the transcription factor, p63, which plays a role in the regenerative ability of many epithelial tissues, is expressed during and throughout epithelial differentiation [12-14]. Cells can be monitored using assays such as immunofluorescence and flow cytometry to detect these marker proteins representing cells at various stages of differentiation and to ensure that populations of cells generated from hPSCs are highly enriched in epithelial cells for future incorporation into tissue constructs for various clinical and research applications.
2. Materials
2.1 Cell growth and differentiation
hPSC growth medium: mTeSR1 (STEMCELL Technologies, Vancouver, Canada).
hPSC differentiation medium 1: Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (1:1) supplemented with 20% Knockout Serum Replacer (KSR), 1X non-essential amino acids (NEAA), 1 mM L-glutamine (all from Life Technologies, Carlsbad, CA), 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO), and 6 μM SU6656 (Sigma).
hPSC differentiation medium 2: Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (1:1) supplemented with 20% Knockout Serum Replacer (KSR), 1X non-essential amino acids (NEAA), 1 mM L-glutamine (all from Life Technologies), 0.1 mM β-mercaptoethanol (Sigma), 1 μM retinoic acid (RA, Sigma), and 10 ng/ml bone morphogenetic protein 4 (BMP4, Life Technologies).
Matrigel (BD, Biosciences, San Jose, CA). Store at −80°C in single use aliquots. Thaw at 4°C. All manipulations must be conducted on ice using chilled pipette tips to avoid gelation of Matrigel solution. To coat a 6-well plate with Matrigel, dissolve 0.5 mg of Matrigel (solution) in 6 ml of DMEM/F12 and coat each well with 1 ml of solution. Allow Matrigel to gel at 37°C for at least 1 hour prior to plating cells.
Dispase (Life Technologies). Reconstituted in DMEM/F12 at 2 mg/ml. Store aliquots at −20°C.
Gelatin powder (Sigma) dissolved in water at 0.1% (w/v). To coat a 6-well plate with gelatin, coat each well with 1 ml of gelatin solution and store at 37°C for at least 4 hours prior to plating cells.
Defined keratinocyte serum-free medium (K-DSFM) and supplement (Life Technologies).
Epithelial cell expansion medium: K-DSFM supplemented with 5% fetal bovine serum (both from Life Technologies).
ROCK inhibitor Y27632 (Sigma). Add to culture medium for a final concentration of 10 μM.
Trypsin (0.05%)-ethylenediamine tetraacetic acid (EDTA, 1 mM, Life Technologies).
Accutase (Life Technologies).
Versene (Life Technologies).
2.2 Immunofluorescent staining
IF fixation buffer:16% (w/v) paraformaldehyde (PFA, Sigma) diluted to 4% (v/v) in PBS.
Blocking buffer: PBS with 5% milk or chick serum (Sigma) and 0.4% (v/v) Triton X-100 (Fisher, Pittsburgh, PA) added.
Primary antibodies (recommended dilution): rabbit anti-Nanog polyclonal antibody (1:800, Cell Signaling Technology, Danvers, MA), rabbit anti-Oct4 polyclonal antibody (1:100), mouse anti-p63 monoclonal antibody (1:25, both from Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-K14 polyclonal antibody (1:100), mouse anti-K18 monoclonal antibody (1:100), mouse anti-K10 monoclonal antibody (1:100, all from Lab Vision, Fremont, CA), rabbit anti-Nanog polyclonal antibody (1:800, Cell Signaling Technology), mouse anti-K3 monoclonal antibody (1:100, Millipore, Bedford, MA).
Secondary antibodies: goat anti-mouse IgG1 AlexaFluor 488 conjugated antibody, goat anti-mouse IgG2 AlexaFluor 488 conjugated antibody, donkey anti-rabbit IgG AlexaFluor 594 conjugated antibodies (all from Life Technologies).
Hoechst 33342 nuclear staining solution (Sigma, 10 mg/ml stock diluted 1:5000 in water).
2.3 Flow cytometry
FC fixation buffer: 1% (v/v) PFA in PBS.
Methanol (Fisher) diluted to 90% (v/v) solution with water.
FACS Buffer: PBS with 0.5% (w/v) bovine serum albumin (Sigma), 0.1% (v/v) Triton X-100 (Fisher), and 0.1% (w/v) sodium azide (Sigma) added. Store at 4°C for up to 2 weeks.
Primary antibodies: Same as those used for immunofluorescent staining. Mouse and rabbit IgG isotype controls are also recommended for gating negative populations during analysis.
Secondary antibodies: goat anti-mouse IgG1 AlexaFluor 488 conjugated antibody, goat anti-mouse IgG2 AlexaFluor 488 conjugated antibody, donkey anti-rabbit IgG AlexaFluor 633 conjugated antibodies (all from Life Technologies).
3. Methods
Differentiation of hPSCs to epithelial cells involves a stepwise process where simple epithelial cell populations are first derived from hPSC populations. Here, this first step of simple epithelial cell differentiation is initiated using a Src family kinase inhibitor, SU6656, which modulates β-catenin localization [9]. The second step in this process involves the maturation of simple epithelial cells to epidermal keratinocyte progenitors, which can then form terminally-differentiated epidermal keratinocytes, marked by the expression of several proteins, including cytokeratin 10 (K10).
Throughout this differentiation process, cells can be characterized by monitoring expression of protein markers for different cell populations at various stages of differentiation. Here, immunofluorescence and flow cytometry methods are described for how to analyze cell populations and confirm successful epithelial differentiation. Overall, the method described here provides an efficient method where epidermal keratinocyte progenitors can be produced from hPSCs via a simple epithelial progenitor that can be expanded indefinitely.
3.1 Cell growth and differentiation
hPSCs are expanded on a Matrigel-coated substrate in mTeSR1 medium and passaged every 4-5 days. To passage cells, incubate cells in Versene for 3 minutes to passage cells in colonies or 5 minutes to passage single cells. Aspirate Versene then shear off cells using fresh mTeSR1 medium. Split cells at desired ratio, 1:6 or 1:12. To passage singularized cells, add ROCK inhibitor to fresh mTeSR1 medium. Change medium every day.
Begin treatment with hPSC differentiation medium 1 when the cells reach 70-80% confluence (~100-150k cells/cm2) by adding 2.5 ml/well for 6-well plates. Change medium daily for 3 days. Some cell death will occur during these three days and a 20-40% loss of cells should be expected.
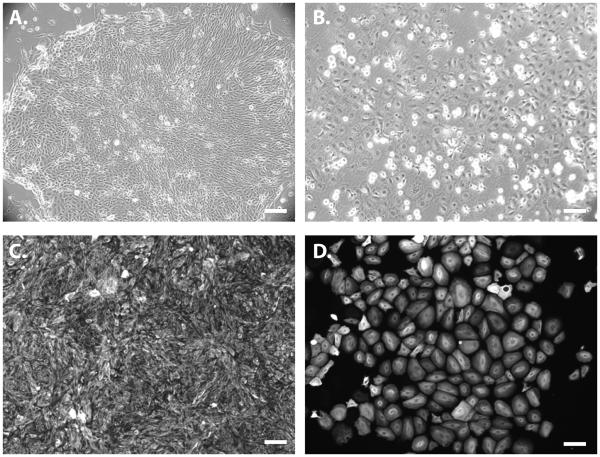
On the fourth day, add 2 ml/well fresh K-DSFM for a 6-well plate. Change the medium daily until cells reach 80-90% confluence (~80-120k cells/cm2). Cells should acquire a simple epithelial morphology (Fig. 1A). At this point, these cells have lost expression of pluripotency markers such as Oct4 and Nanog and have acquired expression of cytoplasmic K18 (Fig. 1C).
Passage simple epithelial cells by aspirating medium and incubating cells with 1 ml/well of Accutase at 37°C for 10 min. Quench Accutase using 2 ml/well Epithelial cell expansion medium. Pipet to shear off any remaining adhered cells. Split at a ratio of 1:3. Epithelial cells can be plated on either Matrigel or gelatin-coated substrates. When passaging cells to new substrate, resuspend cells in 2 ml/well Epithelial expansion medium with ROCK inhibitor.
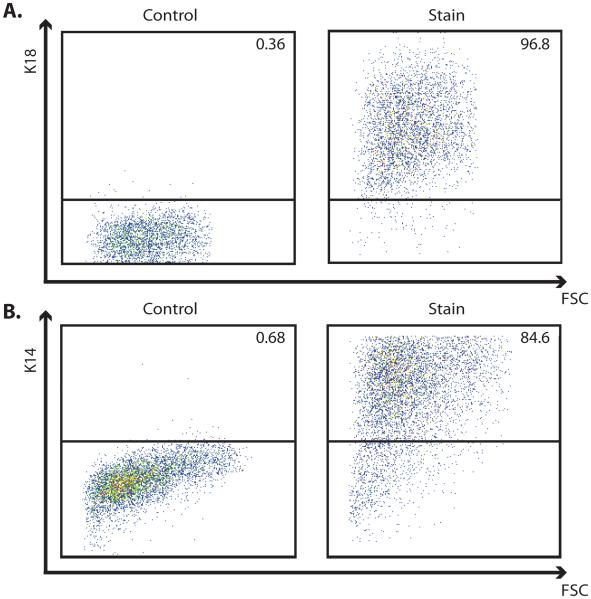
Change Epithelial cell expansion medium every other day. Simple epithelial cells can be passaged for > 10 passages in this state. Cells will maintain their expression of K18 and cell populations will remain highly enriched (>90%) in K18-expressing cells (Fig. 2A).
To further differentiate simple epithelial cells to epidermal keratinocyte progenitors, aspirate medium and add 2 ml/well of hPSC differentiation medium 2 when the cells reach at least 50% confluence (~40-50 cells/cm2). Change the medium daily for 4 days. On the fifth day, change the medium to 2 ml/well of Epithelial expansion medium.
Change the Epithelial expansion medium every other day and expand until the desired number of cells are obtained. These cells obtain a slightly different cobblestone morphology (Fig. 1B) and should express nuclear p63 and cytoplasmic K14 (Fig. 1D). Cell populations at this stage will remain highly enriched (>80%) in K14-expressing cells (Fig. 2B). Occasionally, <5% cells can be found in culture expressing corneal marker, K3.
These K14+ cells can be terminally-differentiated into epidermal keratinocytes using one of several previously-reported protocols or assays [6,5].
Figure 1.
Images of epithelial cell populations derived from the H9 human embryonic stem cell line using a Src family kinase inhibitor, Su6656. (A,B) Phase contrast images of (A) simple epithelial cell populations and (B) epidermal keratinocyte progenitor populations. (C,D) Immunofluorescence images demonstrating (C) homogeneous expression of K18 throughout the simple epithelial cell population and (D) K14 homogeneously expressed throughout the epidermal keratinocyte population. Scale bars in all panels are 100 μm.
Figure 2.
Representative flow cytometry plots of epithelial cell populations derived from the H9 human embryonic stem cell line using a Src family kinase inhibitor, Su6656. (A) Plots demonstrating highly enriched populations of K18-expressing simple epithelial cells and (B) highly enriched populations of K14-expressing epidermal keratinocyte progenitor cells. Panels show plots of fluorescence versus forward scatter for both isotype controls and K18 or K14 stains. Percentage of positive cells is indicated in the upper right corner of each panel.
3.2 Immunofluorescent staining
Aspirate cell culture medium and rinse once with 2 ml/well of PBS for a 6-well plate.
Fix cells in 2 ml/well of IF fixation buffer for 15-20 minutes at room temperature (RT) on a shaker.
Rinse cells 2X with 2 ml/well of PBS and incubate cells in 1 ml/well of blocking buffer for 2 hours at RT.
Incubate cells with primary antibodies in 300 μl of fresh blocking buffer at the appropriate dilution overnight on a shaker at 4°C.
Rinse cells 3-5X with 2 ml/well of PBS.
Incubate cells with secondary antibodies at a dilution of 1:1000 in 1 ml/well of blocking buffer for 1 hour at RT on shaker in the dark.
Rinse cells 2X with 2 ml/well of PBS.
Incubate cells with 1 ml/well nuclear staining solution for 5 minutes at RT.
Rinse cells 2X with PBS and view on epifluorescent microscope, using a mercury lamp and dichroic filters to excite AlexaFluor 488 and 594 dyes as well as the Hoechst nuclear stain.
3.3 Flow cytometry
Aspirate cell culture medium and rinse once with 2 ml/well of PBS for a 6-well plate.
Incubate cells with 1 ml/well of Trypsin-EDTA for 5-10 minutes at 37°C.
Mechanically dissociate larger cell aggregates by pipetting in the Trypsin-EDTA solution and inactivate trypsin with an equal volume of any medium containing at least 20% FBS.
Centrifuge samples at 200 × g for 5 minutes and aspirate supernatant.
Resuspend cell pellet in 1 ml FC fixation buffer and incubate at 37°C for 10 minutes.
Centrifuge samples at 200 × g for 5 minutes and decant supernatant.
Resuspend pellet in 1 ml ice-cold methanol solution and hold on ice for 30 minutes or store at 4°C for up to 2 weeks.
Aliquot 1-2 × 105 cells per sample and add 2 ml FACS buffer to each sample.
Centrifuge cells and decant supernatant.
Add 2 ml FACS buffer to each sample and repeat centrifugation.
Decant supernatant leaving ~50 uL in each sample.
Pre-dilute primary antibodies (0.5 uL per sample) in 50 uL of FACS buffer and add primary antibody solution to each sample. Pipet to resuspend pellet. Incubate samples at 4°C overnight. Include control samples for each biological sample lacking primary antibodies in each combination for sample compensation.
Add 2 ml FACS buffer to each sample, centrifuge, and decant supernatant.
Pre-dilute secondary antibodies (0.1 uL per sample) in 50 uL of FACS buffer and add secondary antibody solution to each sample. Pipet to resuspend pellet. Incubate 30-60 minutes at RT in the dark.
Add 2 ml FACS buffer to each sample, centrifuge, and decant supernatant.
Resuspend each sample in 300 uL FACS buffer and hold on ice or at 4°C until analysis.
Analyze samples on a BD FACSCalibur, LSRII, or equivalent cytometer capable of excitation at 488 and 633 nm. Appropriate cell populations should be gated under forward scatter vs. side scatter plots to exclude debris from analysis. Isotype controls should be used to gate ‘negative’ populations. 488 and 633 fluorophores used for the same samples do not have significant spectral overlap, but fluorescence compensation should be applied for good practice.
Acknowledgements
The protocols and methods described here were developed under the support of National Science Foundation Grant CBET-1066311 (S.P.P.) and a National Institutes of Health training grant NIDCD T32 DC009401 (J.A.S.).
References
- 1.Thomson J, Itskovitz-Eldor J, Shapiro S, Waknitz M, Swiergiel J, Marshall V, Jones J. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik M, Smuga-Otto K, Antosiewicz-Bourget J, Frane J, Tian S, Nie J, Jonsdottir G, Ruotti V, Stewart R, Slukvin I, Thomson J. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. doi:DOI 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. doi:DOI 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Hewitt KJ, Shamis Y, Carlson MW, Aberdam E, Aberdam D, Garlick JA. Three-dimensional epithelial tissues generated from human embryonic stem cells. Tissue Eng Part A. 2009;15(11):3417–3426. doi: 10.1089/ten.tea.2009.0060. doi:10.1089/ten.TEA.2009.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selekman JA, Grundl NJ, Kolz JM, Palecek SP. Efficient generation of functional epithelial and epidermal cells from human pluripotent stem cells under defined conditions. Tissue Engineering Part C. 2013 doi: 10.1089/ten.tec.2013.0011. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metallo C, Ji L, de Pablo J, Palecek S. Directed differentiation of human embryonic stem cells to epidermal progenitors. Methods Mol Biol. 2010;585:83–92. doi: 10.1007/978-1-60761-380-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aberdam E, Barak E, Rouleau M, de LaForest S, Berrih-Aknin S, Suter D, Krause K, Amit M, Itskovitz-Eldor J, Aberdam D. A pure population of ectodermal cells derived from human embryonic stem cells. Stem Cells. 2008;26(2):440–444. doi: 10.1634/stemcells.2007-0588. [DOI] [PubMed] [Google Scholar]
- 8.Metallo C, Ji L, de Pablo J, Palecek S. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells. 2008;26(2):372–380. doi: 10.1634/stemcells.2007-0501. [DOI] [PubMed] [Google Scholar]
- 9.Lian X, Selekman J, Bao X, Hsiao C, Zhu K, Palecek SP. A small molecule inhibitor of SRC family kinases promotes simple epithelial differentiation of human pluripotent stem cells. PLoS One. 2013;8(3):e60016. doi: 10.1371/journal.pone.0060016. doi:10.1371/journal.pone.0060016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chickarmane V, Peterson C. A computational model for understanding stem cell, trophectoderm and endoderm lineage determination. PLoS One. 2008;3(10):e3478. doi: 10.1371/journal.pone.0003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128(3):445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koster M, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop D. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci U S A. 2007;104(9):3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koster M, Kim S, Mills A, DeMayo F, Roop D. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18(2):126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, Aberdam D. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3(10):e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]