Abstract
HIV-associated sensory neuropathy (HIV-SN) is a common neurological complication of HIV infection. The TNF block is a region within the central MHC that contains many immunoregulatory genes. Polymorphisms and haplotypes of the TNF block have been associated with increased risk of HIV-SN in Asians and whites. Here we investigated genetic associations with HIV-SN in 342 black Southern Africans (190 cases and 152 neuropathy-free controls) using single nucleotide polymorphisms (SNPs) spanning the TNF block and a set of haplotypes defined by 31 SNPs in Asian and white populations (denoted FVa). We included population-appropriate tagSNPs derived from an African population (Yoruban, YRI, HapMap) and derived extended haplotypes comprising 61 SNPs (denoted FVa_ext b). We found no association between HIV-SN and carriage of two SNPs (TNF-1031/rs1799964*C and BAT1 (intron10)/rs9281523*C) associated with HIV-SN in whites and Asians. Additionally, a haplotype containing TNF-1031/rs1799964*C associated with increased risk of HIV-SN in Asians, but was not present in this African population. However, alleles of seven SNPs associated with reduced risk of HIV-SN (corrected for age, height and multiple comparisons). These were rs11796*A, rs3130059*G, rs2071594*C, NFKBIL1-62/rs2071592*A, rs2071591*A, LTA+252/rs909253*G, rs1041981*C. One haplotype (FV18_ext1), not containing these alleles, was associated with increased risk of HIV-SN after correction for age, height and multiple comparisons. Our results confirm the involvement of genes in the TNF block in altering risk for HIV-SN, but genotypes critical in this African population differed from those affecting HIV-SN in whites and Asians. These differences support the need for genetic association studies in diverse populations.
Introduction
The prevalence of HIV-associated sensory neuropathy (HIV-SN) is higher in black individuals than in whites and Asians on similar antiretroviral therapy (ART) regimens.1, 2 This debilitating and often painful condition is predicted to remain a significant long-term complication of HIV infection in Africa, irrespective of changes to treatment guidelines.3, 4, 5 Two recent studies in South Africa reported that about 60% of HIV-positive out-patients on ART had HIV-SN, and over two-thirds of these patients had a painful neuropathy.6, 7 While not fatal, HIV-SN has a significant negative impact on psychological, social and economic well-being4 and there are no proven treatments for the pain.8
The pathogenesis of HIV-SN has not been fully explained, but human9, 10, 11 and animal studies12 indicate that inflammation plays a central role. Genetic studies support an inflammatory etiology, providing evidence of associations with polymorphisms in TNFA, IL4 and IL10.13, 14, 15, 16
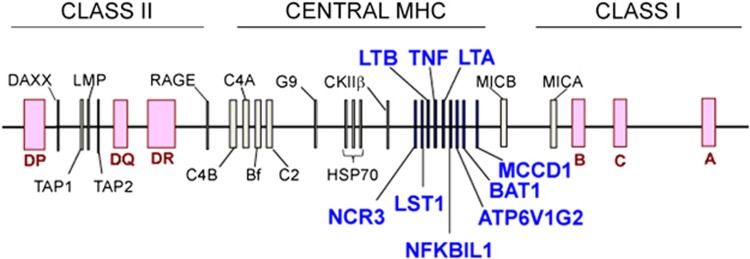
The TNF block (Figure 1) is a roughly 60 kbp region on chromosome six in the central MHC that contains many immunoregulatory genes. Alleles within the block have been associated with several inflammatory diseases including rheumatoid arthritis, type 1 diabetes and venous leg ulcers.17, 18, 19 Alleles of two single nucleotide polymorphisms (SNPs) within the block (TNF-1031/rs1799964*C and BAT1(intron10)/rs9281523*C) have been associated with increased risk of HIV-SN in whites and Malays. TNF-1031*C was also part of a 6-SNP haplotype associated with an increased risk of HIV-SN in these populations.15 BAT1(intron10)*C marks the 8.1 ancestral haplotype (HLA-A1, B8, DR3), a haplotype that has been associated with other immunopathological disorders.20
Figure 1.
The TNF block is a region of the major histocompatibility complex on chromosome six that contains many immune-related genes.
Building on this evidence that polymorphisms within the TNF block alter the risk for HIV-SN, we investigated an African population. We have defined a series of 37 haplotypes of 31 SNPs that explain >90% of variance in white, Chinese, Indian, Malay, Aboriginal (Australian and Malay) and Southern African populations.21 These have been defined with a consistent nomenclature (FV haplotypes).22 As African populations have greater genetic diversity compared with non-African populations,23 we included 30 additional population-appropriate tagSNPs. Each of the 61 SNPs individually, the 31-SNP FV haplotypes and novel 61-SNP haplotypes (FV ext haplotypes) were used to assess associations with HIV-SN in black Southern Africans.
Materials and methods
The study was approved by the Human Research Ethics Committee (Medical) of the University of the Witwatersrand, South Africa (protocol number: M080220 and M110754), and written informed consent was obtained from all participants. HIV-positive adults who had been on combination ART for at least 6 months were screened for neuropathy at the Virology Clinic of the Charlotte Maxeke Johannesburg Academic Hospital, South Africa between July 2008 and April 2009. At this time, stavudine (d4T) was routinely included in first-line therapy in South Africa. An interpreter fluent in English and local African languages facilitated consent and study procedures. The clinical and demographic associations with neuropathy status among the stavudine-exposed participants in this study have previously been published.6
Phenotyping procedure
Consecutive patients were invited to take part and those who consented to study involvement were screened using the AIDS Clinical Trials Group (ACTG) Brief Peripheral Neuropathy Screen, which is a validated tool for identifying symptomatic HIV-SN.24 Symptomatic HIV-SN was defined by at least one symptom experienced bilaterally (pain, aching or burning; numbness; pins-and-needles) and at least one clinical sign (reduced vibration sense or absent ankle reflexes). Vibration sense was assessed using a 128 Hz tuning fork placed on the interphalangeal joint of each great toe; <10 s was considered abnormal. Individuals free from neuropathy were assigned as controls. Individuals with other risks for neuropathy (eg alcoholism, vitamin B12 deficiency and exposure to chemotherapy) were excluded.
SNP selection
All 31 SNPs previously included in the TNF block (FV haplotypes)15 were included in the SNP list. These were supplemented with population-specific tagSNPs from the same chromosomal region genotyped in the Yoruba in Ibadan, Nigeria (YRI) population, available from the International HapMap dataset (HapMap Data Release 27, Phase II+III, February 2009, on the National Center for Biotechnology Information (NCBI) B36 assembly, dbSNP b126).25 A list of tagSNPs was selected using the software program Haploview (version4.2)26 using a pairwise approach at r2=1.0 and with minor allele frequency>0.01. The list was refined using the Assay Design Tool evaluation by Illumina (San Diego, CA, USA; Technical Note: DNA Analysis), which eliminated any SNPs which could not be genotyped. This tagSNP selection procedure produced 46 SNPs, 12 of which were already part of the original 31 SNPs that define the original FV haplotypes. Three additional SNPs (rs1128640, rs3093661 and rs909253) were included based on a review of the literature. Of these new SNPs, two were monomorphic in our Southern African samples (rs2239707 and rs3093982) and were excluded. Three SNPs (rs1129640, rs3093544 and rs3093661) that failed Hardy–Weinberg equilibrium (HWE) were excluded. This left 30 new SNPs in addition to the original 31 SNPs. Alleles 1 and 2 of the original 31 SNPs were designated based on the white population, with allele 1 being the major allele and allele 2 the minor allele. All alleles in the new SNPs are designated respective to the forward (+) strand of the genome assembly, and were obtained using BioMart (Ensembl release 67, May 2012).27, 28 Details of the SNPs are shown in Supplementary Table 1.
Genotyping
DNA extraction was performed on 5 ml venous whole-blood for the majority of samples included in this study. However, the first 39 individuals' DNA samples were extracted from saliva. This technique was subsequently replaced following concerns over DNA quantity. DNA was extracted from saliva samples using the QIAamp DNA mini kit (QIAGEN, Valencia, CA, USA) and from blood using the salting-out method.29 SNPs were genotyped using the Goldengate assay on the Illumina BeadXpress genotyping platform (Illumina). Initial genotyping of three of the SNPs (rs3179003, rs2516478 and rs2523502) failed; rs3179003 was re-genotyped by allelic discrimination using a Taqman SNP genotyping assay (Applied Biosystems, Foster City, CA, USA). DNA extraction and genotyping was carried out in the Division of Human Genetics, National Health Laboratory Services & University of the Witwatersrand (Johannesburg, South Africa). Genotype data have been submitted to GWAS central (www.gwascentral.org; expected release May 2014).
Quality control
Raw genotype data were examined using the genotyping module of BeadStudio (Framework version 3.1.3.0; module version 3.2.32). Data quality was assessed using Illumina-designed built-in assay controls and samples failing more than two such controls (out of five) were excluded from further analysis. To minimize the possibility of technical error influencing the association analysis, we excluded SNPs with HWE <1 × 10−4.30
Statistical analyses
Clinical and demographic associations with SN status were analyzed in Stats11 (StataCorp, College Station, TX, USA) as previously described.6 Statistical analyses of genetic data were carried out using PLINK.31, 32 χ2-analysis employing allelic, genotypic, dominant and recessive models were performed to test for association between the SNPs and the presence of HIV-SN. Haplotypes were also analyzed using χ2. SNPs and haplotypes achieving a P-value <0.05 on univariate analysis were included in multivariate analyses. Logistic regression was used to test for association, while correcting for factors previously associated with risk of HIV-SN in this cohort: age and height.6 We employed empirically calculated P-values (EMP) in all analyses, using 1000 permutations. We report two EMP values: EMP1 an empirical but uncorrected value and EMP2 which corrects for multiple comparisons.
PHASE analysis and reconstruction of FV haplotypes
Haplotypes were defined by carriage of allele 2 at each SNP, where allele 2 referred to the minor allele in whites. The definition of alleles 1 and 2 refer to whites and were held constant here to allow comparison between ethnicities.15, 21 Haplotype reconstruction and population frequency estimations were performed using PHASE v2.1,33, 34 which is recognized as the best tool for this purpose.35, 36 Default parameters were used (100 iterations, a burn-in value of 100 and a thinning interval of 1) along with performing a case-control permutation test with 1000 permutations. The algorithm was run five times, with different seeds for the random number generator in each run. All haplotypes with a frequency of <1% were excluded from further analyses. Haplotypes were aligned with the 31-SNP FV haplotypes and 61-SNP FV ext haplotypes were generated to include the additional population-specific tagSNPs. Linkage Disequilibrium (LD) between SNPs was visualized using Haploview, where the confidence interval method was implemented.26
Results
Three hundred and forty two black Southern African HIV-positive patients were recruited and successfully genotyped. Of these, 75% (257/342) were female with a mean age of 39 (SD 8) and median CD4 T-cell count of 388 cells/μl (range 27–1091) at the time of assessment. Ninety-eight percent (334/342) were treated with ART regimens that included stavudine (d4T). Antiretroviral regimens of the remaining eight patients were zidovudine based (n=1), tenofovir based (n=6) or nucleoside sparing (n=1, on ritonavir-boosted lopinavir with efavirenz). We note that the rate of SN among patients who had never used stavudine (3/8) was not different from the overall cohort (P=0.5, Fisher's exact test) and that neither the demographic or genetic associations with SN presented here were influenced by whether or not these individuals are included in the cohort. Fifty-six percent (56%, 190/342) had HIV-SN. Most were South African (93%, 318/342) with the remaining 7% from other Southern African countries in the Southern, Narrow-Bantu sub-group of the Niger-Kordofanian ethno-linguistic grouping (Zimbabwe n=12, Mozambique n=9, Malawi n=2, Zambia n=1). Increasing age and increasing height were the only demographic factors associated with SN, as previously described.6 None of sex, weight, body mass index, current or nadir CD4 T-cell count nor a history of TB were associated with SN status in this cohort.
Associations between HIV-SN and individual SNPs were assessed. No association was detected between HIV-SN and carriage of the C allele at TNFA-1031/rs1799964 or BAT1(intron10)/rs9281523 (alleles associated with HIV-SN in white and Asian populations).13, 14, 15 However, associations were found with 10 other SNPs on allelic and dominant models of univariate analysis, seven of which remained significant after correction for age and height (independent risk factors for HIV-SN in this cohort6), and multiple comparisons (Table 1).
Table 1. SNPs that were significantly associated with altered risk for HIV-SN.
| Univariate | Multivariatea | |||||
|---|---|---|---|---|---|---|
| Model | SNP | PEMP1 | OR (95% CI) | PEMP1 | PEMP2 | OR (95% CI) |
| Allelic | rs2075582b | 0.029 | 0.58 (0.35, 0.96) | 0.020 | 0.083 | 0.54 (0.32, 0.93) |
| rs3130059b | 0.016 | 0.67 (0.49, 0.93) | 0.032 | 0.090 | 0.68 (0.49, 0.96) | |
| rs2523504b | 0.033 | 0.63 (0.41, 0.98) | 0.020 | 0.059 | 0.57 (0.36, 0.91) | |
| rs2071594b | 0.008 | 0.66 (0.48, 0.91) | 0.020 | 0.072 | 0.66 (0.47, 0.94) | |
| rs2071592 | 0.009 | 0.64 (0.46, 0.88) | 0.015 | 0.040 | 0.64 (0.45, 0.91) | |
| rs2071591b | 0.015 | 0.68 (0.49, 0.93) | 0.026 | 0.085 | 0.68 (0.48, 0.96) | |
| rs909253b | 0.024 | 0.68 (0.50, 0.94) | 0.040 | 0.118 | 0.70 (0.49, 0.98) | |
| Dominant | rs11796 | 0.017 | 0.58 (0.37, 0.91) | 0.019 | 0.040 | 0.56 (0.35, 0.90) |
| rs3130059 | 0.007 | 0.53 (0.34, 0.83) | 0.004 | 0.012 | 0.50 (0.31, 0.79) | |
| rs2071594 | 0.002 | 0.51 (0.32, 0.80) | 0.005 | 0.009 | 0.48 (0.29, 0.77) | |
| rs2071592 | 0.005 | 0.49 (0.31, 0.77) | 0.004 | 0.008 | 0.46 (0.29, 0.75) | |
| rs2071591 | 0.006 | 0.54 (0.34, 0.85) | 0.006 | 0.016 | 0.51 (0.32, 0.82) | |
| rs4947324 | 0.024 | 0.56 (0.35, 0.92) | 0.069 | 0.179 | 0.63 (0.38, 1.04) | |
| rs909253 | 0.003 | 0.50 (0.31, 0.79) | 0.006 | 0.009 | 0.48 (0.29, 0.77) | |
| rs1041981 | 0.009 | 0.56 (0.35, 0.88) | 0.015 | 0.031 | 0.54 (0.34, 0.88) | |
The minor allele of each SNP is associated with a decreased risk for developing HIV-SN relative to the major allele following correction for covariates (age and height) and multiple comparisons.
SNPs that are significant following correction for covariates (age and height) only.
SNPs that are significant following correction for covariates (age and height) and multiple comparisons are indicated in bold.
To illustrate the unique patterns of LD in this South African population, an LD plot of the TNF block was constructed using Haploview (Supplementary Figure 1). Nine LD blocks within the region were identified, six of which contained SNPs associated with HIV-SN in this cohort.
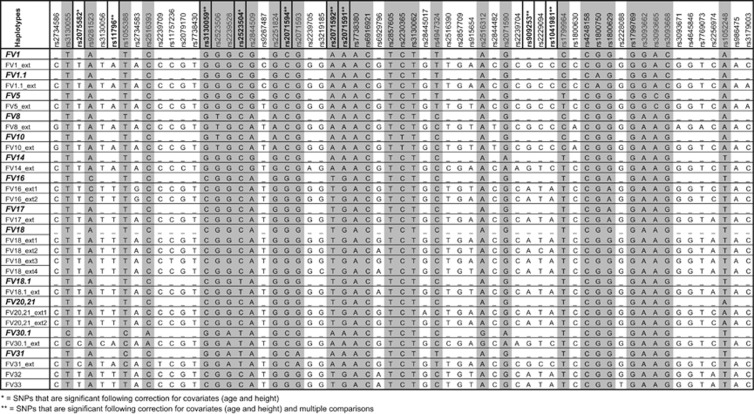
PHASE analysis reconstructed 62 31-SNP FV haplotypes and 126 61-SNP FV ext haplotypes, of which 13 and 20, respectively, had a frequency >1%. The 13 31-SNP FV haplotypes accounted for 87% of the cohort and the 20 61-SNP FV haplotypes accounted for 74% of the cohort. Figure 2 shows 61- and 31-SNP FV haplotypes aligned according to shared SNPs. Univariate analysis identified one 61-SNP haplotype (FV18_ext1) that was associated with HIV-SN (EMP1 <0.05) (Supplementary Table 2). The association with increased risk of HIV-SN remained after correction for age, height and multiple comparisons (EMP2=0.009; OR (95% CI)=2.09 (1.26–3.50)).
Figure 2.
Original FV haplotypes15, 22 and FV sub-haplotypes (generated with population-specific tagSNPs) in this Southern African population. SNPs found to be associated with HIV-SN are shown in bold. SNPs included in the original FV haplotypes are shown shaded.
Discussion
This is the largest study to date of the TNF block and risk of HIV-SN, and the first in an African population. We found no association with SNPs previously associated with HIV-SN or with 31-SNP FV haplotypes described in this population.21 However we found strong novel associations between HIV-SN and seven SNPs from across the TNF block and a 61-SNP haplotype that included Yoruban tagSNPs (FV18_ext1). As expected, we also observed a unique LD pattern across the TNF block in this Southern African population, as compared with other non-African groups.22 While it would be premature to assign a particular biological phenotype to carriage of a particular haplotype, the associations with HIV-SN demonstrate an inflammatory etiology and in time will help to elucidate pathogenic pathways. Importantly, improving our understanding of SN pathogenesis may lead to effective preventative strategies, or even pathogenesis-based treatments for a condition that currently lacks effective analgesic options.8
Of the individual SNPs associated with neuropathy, allele 2 was associated with reduced risk in each case (Table 1). The FV18_ext1 haplotype was associated with increased risk of HIV-SN. Consistent with the results for the SNP analyses, for each associated SNP, the allele not associated with protection was contained in FV18_ext1. The parent 31-SNP haplotype (FV18) was not associated with HIV-SN but this was subdivided into four 61-SNP extension haplotypes by the Yoruban tagSNPs. FV18_ext1 was defined by alleles carried at rs11796, rs909253 and rs1041981, which were associated with HIV-SN. The 31-SNP FV18 haplotype associates with increased TNFα and LTα production by blood leukocytes in whites.37 These differences may have been clearer if the donors had been genotyped at the Yoruban tagSNPs.
In contrast to Asians and whites,13, 14, 15 we found no association between TNFA-1031/rs1799964*C and HIV-SN. A mini-haplotype FVa6,7,8 comprising six SNPs and including TNFA-1031/rs1799964*C associated with increased risk of HIV-SN in Chinese and Malay individuals,15 but did not occur in the Southern Africans. This result is most likely owing to the unique LD pattern observed in this population (Supplementary Figure 1). It is possible that previous associations of TNFA SNPs with HIV-SN could be owing to indirect association. Genetic association studies in African populations have the distinct advantage that LD generally exists over a shorter genomic distance, resulting in an increased probability of identifying the true causal variants.38The suggested use of an algorithm combining patient age, height and carriage of the TNFA-1031/rs1799964*C genotype to predict likelihood of developing HIV-SN on stavudine13 would therefore be inappropriate in this African population.
BAT1(intron10)/rs9281523*C associated with increased risk of HIV-SN in whites14 but not in Southern Africans. In addition, we found no association with the well-known TNF-308/rs1800869*A, which has been associated with high production of TNFα39 and clinically with rheumatoid arthritis in Egyptians and Brazilians,40, 41 lupus nephritis in Portuguese individuals42 and type 1 diabetes in Asians.43 Both BAT1(intron10)*C and TNF-308*A mark FV16 and the 8.1 ancestral haplotype (HLA-A1, B8, DR3, DQ2).44 Although FV16 did not influence TNFα or LTα production,37 the 8.1 ancestral haplotype has been associated with higher TNFα production in vitro39, 45, 46 and with multiple autoimmune diseases.20, 47 Clearly BAT1(intron10)*C and TNF-308*A do not tag the causative SNPs in Southern Africans.
The cytokine LTα is closely related to TNFα and the SNP LTA+252/rs909253 has frequently been assessed in conjunction with TNF-308/rs1800869. LTA+252/rs909253*G has been associated with inflammatory and autoimmune conditions48, 49, 50 in non-African populations. Conversely, we found LTA+252/rs909253*G associated with reduced risk of HIV-SN. This might allude to the complexity of HIV-SN12 or suggest the different tagging of SNPs by rs909253 in Southern Africans. Unfortunately this SNP was not included in our earlier studies of TNF haplotypes.13, 14, 15 The C allele of another LTA SNP, rs1041981, also associated here with reduced risk of HIV-SN. Studies reporting associations with this SNP and risk of diabetes and myocardial infarction in various ethnicities have yielded conflicting results,51, 52, 53, 54, 55 consistent with differing LD patterns in different populations.
Of the other SNPs associated with HIV-SN, only NFKBIL1-62/rs2071592 has been studied previously. The paucity of functional data related to this gene hampers interpretation of any associations. The A allele of NFKBIL1-62/rs2071592, which associated with reduced risk of HIV-SN here, associated with protection against systemic lupus erythematosus in Colombians.56 In another study, the T allele was associated with increased risk of rheumatoid arthritis (suggesting relative protection with the A allele) in Taiwanese and Japanese individuals.57, 58 It has been suggested that the A allele may disrupt transcription of NFKBIL1.58, 59 rs2071591 also from NFKBIL1 and rs2071594 from ATP6V1G2 have also been associated with development of rheumatoid arthritis.58 However, there is no evidence of independent associations with disease or protein production and these alleles are in tight LD with NFKBIL1-62/rs2071592*A, even in this Southern African cohort (Supplementary Figure 1).
We consider the possibility that associations with TNF block haplotypes may be mediated by variations in genes outside the TNF block, most likely elsewhere in the MHC. While the TNF block has tight internal LD, it is less strongly linked with HLA genes or with alleles in the centromeric region of the central MHC (HSP, complement and so on). TNF blocks have been aligned with conserved MHC haplotypes found in Asians and whites.44 However Southern African MHC haplotypes have not been defined so we would need a cohort large enough to detect associations between 30+ TNF haplotypes and 20–50 MHC Class l and ll alleles. This is outside a study of HIV-SN.
Considering the TNF block in isolation, this study of an African cohort allows us to narrow down the list of SNPs likely to be critical to the phenotype. We selected tagSNPs based on data from the Yoruban in Nigeria, accepting that they are not genetically identical to Southern Africans. However a 61-SNP haplotype augmented with these tagSNPs associated with HIV-SN risk, where no association with the 31-SNP FV haplotype was found. It would have been ideal to verify whether HIV-negative Southern Africans carry FV18_ext1, but the high prevalence of HIV in Southern Africa means that all donors would need to be tested. This would have ethical implications. It is worth noting that there were no differences in the TNF haplotypes with and without HIV in white and Asian haplotypes.15 The prevalence of HIV in Africa is greater than any individual risk allele, so there is no reason to suppose that Southern Africans differ from other ethnicities in this regard. Ethnic heterogeneity in our cohort may have influenced results, but was minimized by only including local black Southern Africans who declared no white ancestry and whose local ethnic affiliation belonged to the Southern, Narrow-Bantu sub-group of the larger Niger-Kordofanian language grouping. This grouping shows lower heterogeneity than other African population groups60 and local studies have revealed no significant substructure in the population.61, 62
The TNF block is known to be highly conserved but TNF block SNPs and haplotypes associated with HIV-SN are fundamentally different in this African population. The associations with individual SNPs and FV18_ext1 withstood correction for age, height and multiple comparisons. This is the strongest evidence to date that genes in the TNF block influence development of HIV-SN.
Acknowledgments
We wish to thank the staff and the patients of the Virology Clinic in the Charlotte Maxeke Johannesburg Academic Hospital and Florence Mtsweni for acting as the interpreter for the study. In addition we would like to thank Punita Pitamber for her assistance with genotyping. We gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute (CLC) and the International Association for the Study of Pain for a Developed-Developing Countries Collaborative Research Grant (CLC, PRK). Finally we wish to thank the Belgian Technical Cooperation (LMH) and the Hillel Friedland Trust (ALW) for Fellowships. This work was directly supported by the University of the Witwatersrand, South Africa (Faculty of Health Sciences and the University Research Council) and the National Research Foundation, South Africa (Rated Researchers Program).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Anziska Y, Helzner EP, Crystal H, et al. The relationship between race and HIV-distal sensory polyneuropathy in a large cohort of US women. J Neurol Sci. 2011;315:129–132. doi: 10.1016/j.jns.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry C, Affandi JS, Brew B, et al. Ethnicity is an independent predictor of neuropathy risk in HIV patients. Proceedings of the 18th Conference on Retroviruses and Opportunistic Infections; 27 February–2 March 2011.
- Cherry C, Kamerman PR, Bennett D, Rice A. HIV-associated sensory neuropathy: still a problem in the post-stavudine era. Future Virol. 2012;7:849–854. [Google Scholar]
- Ellis RJ, Rosario D, Clifford DB, et al. Continued high prevalence and adverse clinical impact of human immunodeficiency virus-associated sensory neuropathy in the era of combination antiretroviral therapy: the CHARTER Study. Arch Neurol. 2010;67:552–558. doi: 10.1001/archneurol.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerman PR, Wadley AL, Cherry CL. HIV-associated sensory neuropathy: risk factors and genetics. Curr Pain Headache Rep. 2012;16:226–236. doi: 10.1007/s11916-012-0257-z. [DOI] [PubMed] [Google Scholar]
- Wadley AL, Cherry CL, Price P, Kamerman PR. HIV neuropathy risk factors and symptom characterization in stavudine-exposed South Africans. J Pain Symptom Manage. 2011;41:700–706. doi: 10.1016/j.jpainsymman.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Maritz J, Benatar M, Dave JA, et al. HIV neuropathy in South Africans: frequency, characteristics, and risk factors. Muscle Nerve. 2010;41:599–606. doi: 10.1002/mus.21535. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Cherry CL, Cox S, Marshall SJ, Rice AS. Pharmacological treatment of painful HIV-associated sensory neuropathy: a systematic review and meta-analysis of randomised controlled trials. PLoS One. 2010;5:e14433. doi: 10.1371/journal.pone.0014433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn K, Robinson B, Anderson C, et al. Differential effects of HIV infected macrophages on dorsal root ganglia neurons and axons. Exp Neurol. 2008;210:30–40. doi: 10.1016/j.expneurol.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Zhu Y, Silva C, et al. Peripheral nerve-derived HIV-1 is predominantly CCR5-dependent and causes neuronal degeneration and neuroinflammation. Virology. 2005;334:178–193. doi: 10.1016/j.virol.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Nagano I, Shapshak P, Yoshioka M, Xin K, Nakamura S, Bradley WG. Increased NADPH-diaphorase reactivity and cytokine expression in dorsal root ganglia in acquired immunodeficiency syndrome. J Neurol Sci. 1996;136:117–128. doi: 10.1016/0022-510x(95)00317-u. [DOI] [PubMed] [Google Scholar]
- Kamerman PR, Moss PJ, Weber J, Wallace VCJ, Rice ASC, Huang W. Pathogenesis of HIV-associated sensory neuropathy: evidence from in vivo and in vitro experimental models. J Peripher Nerv Syst. 2012;17:19–31. doi: 10.1111/j.1529-8027.2012.00373.x. [DOI] [PubMed] [Google Scholar]
- Affandi JS, Price P, Imran D, Yunihastuti E, Djauzi S, Cherry CL. Can we predict neuropathy risk before stavudine prescription in a resource-limited setting. AIDS Res Hum Retroviruses. 2008;24:1281–1284. doi: 10.1089/aid.2008.0045. [DOI] [PubMed] [Google Scholar]
- Cherry CL, Rosenow A, Affandi JS, McArthur JC, Wesselingh SL, Price P. Cytokine genotype suggests a role for inflammation in nucleoside analog-associated sensory neuropathy (NRTI-SN) and predicts an individual's NRTI-SN risk. AIDS Res Hum Retroviruses. 2008;24:117–123. doi: 10.1089/aid.2007.0168. [DOI] [PubMed] [Google Scholar]
- Chew CSN, Cherry CL, Imran D, et al. Tumour necrosis factor haplotypes associated with sensory neuropathy in Asian and Caucasian human immunodeficiency virus patients. Tissue Antigens. 2010;77:126–130. doi: 10.1111/j.1399-0039.2010.01570.x. [DOI] [PubMed] [Google Scholar]
- Wadley AL, Kamerman PR, Chew CS, Lombard Z, Cherry CL, Price P. A polymorphism in IL4 may associate with sensory neuropathy in African HIV patients. Mol Immunol. 2013;55:197–199. doi: 10.1016/j.molimm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Kumar N, Sharma G, Kaur G, Tandon N, Bhatnagar S, Mehra N. Major histocompatibility complex class I chain related gene-A microsatellite polymorphism shows secondary association with type 1 diabetes and celiac disease in North Indians. Tissue Antigens. 2012;80:356–362. doi: 10.1111/j.1399-0039.2012.01931.x. [DOI] [PubMed] [Google Scholar]
- Martinez A, Fernandez-Arquero M, Pascual-Salcedo D, et al. Primary association of tumor necrosis factor-region genetic markers with susceptibility to rheumatoid arthritis. Arthritis Rheum. 2000;43:1366–1370. doi: 10.1002/1529-0131(200006)43:6<1366::AID-ANR21>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Tan JH, Price P, Gut I, Stacey MC, Warrington NM, Wallace HJ. Characterization of tumor necrosis factor-alpha block haplotypes associated with susceptibility to chronic venous leg ulcers in Caucasian patients. Hum Immunol. 2010;71:1214–1219. doi: 10.1016/j.humimm.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Price P, Witt C, Allcock R, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- Chew CSN, Wadley AL, Lombard Z, Kamerman PR, Price P. TNF haplotypes in a Southern African population resemble those seen in Caucasians and Asians. Genes Immun. 2013;14:268–270. doi: 10.1038/gene.2013.8. [DOI] [PubMed] [Google Scholar]
- Valente FP, Tan CR, Temple SE, et al. The evolution and diversity of TNF block haplotypes in European, Asian and Australian Aboriginal populations. Genes Immun. 2009;10:607–615. doi: 10.1038/gene.2009.45. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Cherry CL, Wesselingh SL, Lal L, McArthur JC. Evaluation of a clinical screening tool for HIV-associated sensory neuropathies. Neurology. 2005;65:1778–1781. doi: 10.1212/01.wnl.0000187119.33075.41. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S.PLINK v1.07 http://pngu.mgh.harvard.edu/purcell/plink/ , 2009
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Cutler D, Patterson N, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–450. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JH, Temple SE, Kee C, et al. Characterisation of TNF block haplotypes affecting the production of TNF and LTA. Tissue Antigens. 2011;77:100–106. doi: 10.1111/j.1399-0039.2010.01582.x. [DOI] [PubMed] [Google Scholar]
- Teo YY, Small KS, Kwiatkowski DP. Methodological challenges of genome-wide association analysis in Africa. Nat Rev. 2010;11:149–160. doi: 10.1038/nrg2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pociot F, Briant L, Jongeneel CV, et al. Association of tumor necrosis factor (TNF) and class II major histocompatibility complex alleles with the secretion of TNF-alpha and TNF-beta by human mononuclear cells: a possible link to insulin-dependent diabetes mellitus. Eur J Immunol. 1993;23:224–231. doi: 10.1002/eji.1830230135. [DOI] [PubMed] [Google Scholar]
- Boechat AL, Boechat Nde O, Ogusku MM, et al. The influence of a TNF gene polymorphism on the severity of rheumatoid arthritis in the Brazilian Amazon. Cytokine. 2013;61:406–412. doi: 10.1016/j.cyto.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Hussein YM, Mohamed RH, Pasha HF, El-Shahawy EE, Alzahrani SS. Association of tumor necrosis factor alpha and its receptor polymorphisms with rheumatoid arthritis in female patients. Cell Immunol. 2011;271:192–196. doi: 10.1016/j.cellimm.2011.06.023. [DOI] [PubMed] [Google Scholar]
- Santos MJ, Carmona-Fernandes D, Caetano-Lopes J, et al. TNF promoter -308G>A and LTA 252A>G polymorphisms in Portuguese patients with systemic lupus erythematosus. Rheumatol Int. 2012;32:2239–2244. doi: 10.1007/s00296-011-1950-7. [DOI] [PubMed] [Google Scholar]
- Feng RN, Li Y, Sun CH. TNF 308 G/A polymorphism and type 1 diabetes: a meta-analysis. Diabetes Res Clin Pract. 2009;85:e4–e7. doi: 10.1016/j.diabres.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Valente FP, Tan C, Phipps M, et al. TNF block haplotypes associated with conserved MHC haplotypes in European, Asian and Australian Aboriginal donors. Tissue Antigens. 2009;74:57–61. doi: 10.1111/j.1399-0039.2009.01258.x. [DOI] [PubMed] [Google Scholar]
- Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562–566. doi: 10.1002/jlb.66.4.562. [DOI] [PubMed] [Google Scholar]
- Jacob CO, Fronek Z, Lewis GD, Koo M, Hansen JA, McDevitt HO. Heritable major histocompatibility complex class II-associated differences in production of tumor necrosis factor alpha: relevance to genetic predisposition to systemic lupus erythematosus. Proc Natl Acad Sci USA. 1990;87:1233–1237. doi: 10.1073/pnas.87.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candore G, Lio D, Colonna Romano G, Caruso C. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev. 2002;1:29–35. doi: 10.1016/s1568-9972(01)00004-0. [DOI] [PubMed] [Google Scholar]
- Bolstad AI, Le Hellard S, Kristjansdottir G, et al. Association between genetic variants in the tumour necrosis factor/lymphotoxin alpha/lymphotoxin beta locus and primary Sjogren's syndrome in Scandinavian samples. Ann Rheum Dis. 2012;71:981–988. doi: 10.1136/annrheumdis-2011-200446. [DOI] [PubMed] [Google Scholar]
- Vasconcelos DF, da Silva MA, Marques MR, de Brito Junior RB, Vasconcelos AC, Barros SP. Lymphotoxin-alpha gene polymorphism +252A/G (rs909253, A/G) is associated with susceptibility to chronic periodontitis: a pilot study. ISRN Dent. 2012;2012:617245. doi: 10.5402/2012/617245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Fu X, Zhang Y, et al. The +252A/G polymorphism in the lymphotoxin-alpha gene increases the risk of asthma: a meta-analysis. Respirology. 2012;17:1229–1236. doi: 10.1111/j.1440-1843.2012.02243.x. [DOI] [PubMed] [Google Scholar]
- Boraska V, Rayner NW, Groves CJ, et al. Large-scale association analysis of TNF/LTA gene region polymorphisms in type 2 diabetes. BMC Med Genet. 2010;11:69. doi: 10.1186/1471-2350-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Xu P, Bennett D, et al. Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet. 2006;2:e107. doi: 10.1371/journal.pgen.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid YH, Urhammer SA, Glumer C, et al. The common T60N polymorphism of the lymphotoxin-alpha gene is associated with type 2 diabetes and other phenotypes of the metabolic syndrome. Diabetologia. 2005;48:445–451. doi: 10.1007/s00125-004-1659-1. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, et al. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Bonnycastle LL, Conneely KN, et al. Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes. 2007;56:256–264. doi: 10.2337/db06-0461. [DOI] [PubMed] [Google Scholar]
- Castiblanco J, Anaya JM. The IkappaBL gene polymorphism influences risk of acquiring systemic lupus erythematosus and Sjogren's syndrome. Hum Immunol. 2008;69:45–51. doi: 10.1016/j.humimm.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Lin CH, Cho CL, Tsai WC, et al. Inhibitors of kB-like gene polymorphisms in rheumatoid arthritis. Immunol Lett. 2006;105:193–197. doi: 10.1016/j.imlet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Makino S, Yoshikawa Y, et al. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am J Hum Genet. 2003;72:303–312. doi: 10.1086/346067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boodhoo A, Wong AM, Williamson D, et al. A promoter polymorphism in the central MHC gene, IKBL, influences the binding of transcription factors USF1 and E47 on disease-associated haplotypes. Gene Expr. 2004;12:1–11. doi: 10.3727/000000004783992206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard Z, Crowther NJ, van der Merwe L, Pitamber P, Norris SA, Ramsay M. Appetite regulation genes are associated with body mass index in black South African adolescents: a genetic association study. BMJ Open. 2012;2:1–10. doi: 10.1136/bmjopen-2012-000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AB, Soodyall H, Arndt S, et al. Genetic substructure in South African Bantu-speakers: evidence from autosomal DNA and Y-chromosome studies. Am J Phys Anthropol. 2002;119:175–185. doi: 10.1002/ajpa.10097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.