Abstract
Genotype imputation has been the pillar of the success of genome-wide association studies (GWAS) for identifying common variants associated with common diseases. However, most GWAS have been run using only 60 HapMap samples as reference for imputation, meaning less frequent and rare variants not being comprehensively scrutinized. Next-generation arrays ensuring sufficient coverage together with new reference panels, as the 1000 Genomes panel, are emerging to facilitate imputation of low frequent single-nucleotide polymorphisms (minor allele frequency (MAF) <5%). In this study, we present a two-step imputation approach improving the quality of the 1000 Genomes imputation by genotyping only a subset of samples to create a local reference population on a dense array with many low-frequency markers. In this approach, the study sample, genotyped with a first generation array, is imputed first to the local reference sample genotyped on a dense array and hereafter to the 1000 Genomes reference panel. We show that mean imputation quality, measured by the r2 using this approach, increases by 28% for variants with a MAF between 1 and 5% as compared with direct imputation to 1000 Genomes reference. Similarly, the concordance rate between calls of imputed and true genotypes was found to be significantly higher for heterozygotes (P<1e-15) and rare homozygote calls (P<1e-15) in this low frequency range. The two-step approach in our setting improves imputation quality compared with traditional direct imputation noteworthy in the low-frequency spectrum and is a cost-effective strategy in large epidemiological studies.
Introduction
The genome-wide association studies (GWAS) approach has been useful in identifying thousands of single-nucleotide polymorphisms (SNPs) associated with hundreds of complex traits and human diseases.1, 2, 3 This has been made possible by the increase in power achieved by the meta-analysis of different studies where imputation of missing genotypes is essential to harmonize and share data.4, 5, 6 Low-frequency variants have not been scrutinized by most GWAS based on HapMap content.7 Using newer reference panels for imputation will allow this (eg, the 1000 Genomes project based on resequenced data sets) with more variants with lower frequencies for the association analysis with traits8 hereby increasing resolution and improving power9, 10 within the so-called next-generation GWAS. Newer arrays have been designed including marker content in the low-frequency spectrum.11 These arrays are expensive (at the time of writing this paper, the approximate price for 5 M array was around 600$ per sample) and it is not clear so far if worth the investment in already GWAS'ed populations.
Here, we propose a two-step imputation approach seeking the optimization of imputations of low-frequency variants when using arrays based on HapMap content. This approach consists of first genotyping a local reference population with an array with dense marker content and a high coverage of markers in the low-frequency spectrum. Second, the whole study population on arrays with HapMap content is imputed to the array content of the local reference set, before imputing to the 1000 genomes references panel. This strategy results in improved imputation accuracy and quality in what constitutes a cost-effective strategy for genotyping very large populations.
Materials and methods
Design
This study was nested within the Rotterdam study, a prospective study of 14 926 participants over 45 years of age living in a suburb of Rotterdam. The study has been approved by the institutional review board (Medical Ethics Committee) of the Erasmus Medical Center and by the review board of The Netherlands Ministry of Health, Welfare and Sports. Over 11 000 samples have been genotyped either with Illumina (San Diego, CA, USA) Humanhap microarray beadchips 550 or 610 K.12
Strategy overview
A subset of individuals from the Rotterdam study was chosen as the study sample for this project. This subset consisted of trios genotyped on two different platforms: the Illumnia 550 K and the Omni2.5 (N=88). Another subset of individuals, consisting of women without major disease conditions during follow-up (N=397), was genotyped on the Illumnia Omni5 array and used as the local reference panel as described below. These individuals were also genotyped using the Illumina 550 K array. For baseline comparison of the Omni5 and 550 K arrays, these two data sets were imputed to 1000 Genomes (1000 G) in a traditional direct one-step approach (Supplementary Figure S1).
For the two-step approach, we first used our local reference panel and imputed our study sample using the 550 K data, while the Omni2.5 data for the same individuals were used as gold standard for later accuracy analyses. After this first imputation, the best-guess imputed genotypes were hereafter used as input for imputation using the 1000 G reference panel (Version 3,20101123, European panel) (Figures 1 and 2). For comparison, we also directly imputed our study sample to 1000 G in a traditional direct one-step approach.
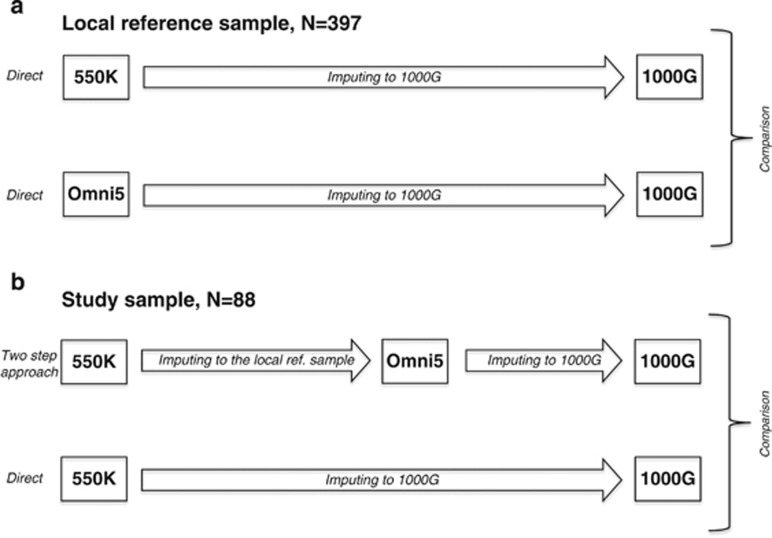
Figure 1.
Panel a illustrates the baseline comparison of the local reference sample genotyped on both the 550 K and Omni5 arrays imputed to the 1000 Genomes reference panel. Panel b illustrates imputation of the study sample applying the two-step approach and comparison with traditional direct one-step imputation approach.
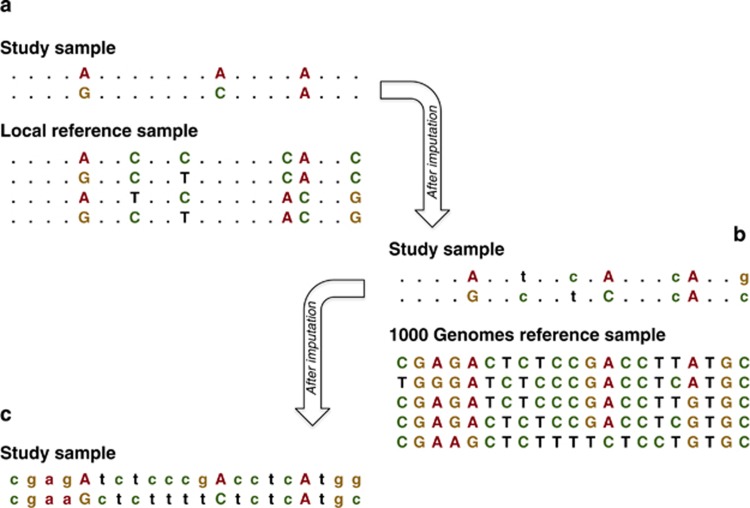
Figure 2.
Two-step imputation approach. Uppercase characters represent genotyped single-nucleotide polymorphisms (SNPs), while lowercase characters represent imputed SNPs. Panel a illustrates the first step: imputation of the study sample using the local reference panel. Panel b illustrates the second imputation step: imputing the output best-guess imputed genotypes from step 1 by using the 1000 Genome reference panel. The final output data set is illustrated in panel c. Illustration adapted from Li et al.6
Imputation software
We employed the MACH/Minimac software13 using a pre-phasing step before the actual imputation as described at the Minimac website (see Web resources) adjusted with an extra intermediate imputation step as described above. The local reference sample was phased with MACH and hereafter used as a reference panel.
Samples QC
We used regular QC procedures before imputation steps as described in Supplementary Table S1, including removal of markers out of Hardy–Weinberg Equlibrium (P<1.0E-06) with very low MAF (<0.001) and low SNP call rate (<0.98). We further removed individuals from the study sample related to individuals in the local reference population based on IBD estimation (proportion IBD >0.2). The study sample consisted of 88 samples genotyped on both Illumina 550 K and Omni2.5. The concordance rate between these two arrays was higher than 99.9%. The local reference sample consisted of 397 individuals genotyped on the Omni5 platform.
Imputation quality assessment
The r2 statistic from Minimac was used as a quality measure with r2>0.3 considered as sufficient imputation quality4 and high imputation quality defined as r2>0.8. After the first imputation step in the two-step strategy, SNPs were removed using three different filters for comparison: (1) no filter; (2) sufficient quality (r2 filter on 0.3) and (3) high quality (r2 filter on 0.8). Accuracy was defined as the proportion of correct imputed genotypes over the total number of imputed genotypes and estimated by comparing the imputed best-guess data to the gold standard data (Omni2.5) using Calcmatch (see Web resources). This analysis was stratified per genotype class as the allelic concordance rate for homozygotes major allele (major–major), heterozygotes (major–minor) and for homozygotes minor allele (minor–minor). Mendelian errors were calculated per study sample family in Plink.14 Only imputed markers (ie, those markers not genotyped in the 550 K array) were taken into account while assessing and comparing the imputation quality. To evaluate the influence of number of individuals included in the local reference sample and its impact on imputation quality, we randomly selected two subsets of individuals with 100 and 200 individuals, respectively and compared these with the full local reference using all 397 samples.
The performance of our approach using different imputation parameters was assessed by analyzing chromosomes 2 and 20. Information on samples genotyped across different platforms and allele frequencies is presented in Table 1. After the evaluation of imputation parameters, we imputed all autosomal chromosomes using the two-step approach (r2 filter on first step 0.3) as well as using the direct approach. We then compared number of SNPs with sufficient imputation quality in the low-frequency spectrum by both approaches. Annotation was done using publicly available data (see web resources).
Table 1. Description of number of markers and samples per array.
| Local reference sample | Study sample | ||
|---|---|---|---|
| Array | Omni5 | 550 K | Omni2.5 |
| Samples | 397 | 88 | |
| SNPs, CHR2+20 | 309 320 | 51 050 | 157 924 |
| SNPs, MAF >0–1% | 61 815 | 413 | 7738 |
| SNPs, MAF 1–5% | 83 588 | 3130 | 25 912 |
| SNPs, MAF 5–10% | 33 718 | 5681 | 20 814 |
| SNPs, overlapa | — | 119 965 | |
Abbreviations: MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
For the study sample, the 550-K data was used for imputation.
Overlap: refers to the common SNPs between 550-K imputed data and Omni2.5 genotypes used in gold standard analysis.
When comparing quality between the two-step and the direct imputation strategy in means of r2 or concordance rates, we applied paired t-tests.
Results
Imputation quality (as measured by r2 statistic)
Description of number of markers per array is presented in Table 1. We compared the effect of the array's coverage in imputation quality by direct imputation of the local reference sample (N=397) using both the Omni5 and the 550 K array data (Supplementary Figure S1). This plot revealed improvement in imputation quality measured by r2 using the Omni5 array as compared with the 550 K with a mean r2=0.91 for the Omni5 and r2=0.71 for 550 K data in the minor allele frequency range between 1 and 5%.
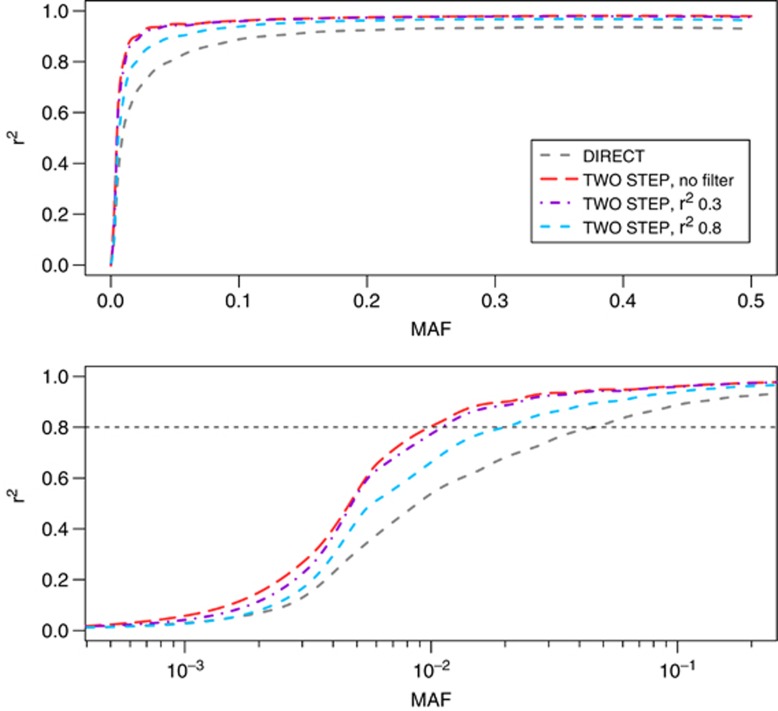
To assess the effect of the two-step approach, we compared the imputation quality, as measured by the mean r2 across all imputed SNPs (chromosome 2 and 20), over the whole-allele frequency spectrum (Figure 3). First, our results indicate a gain in mean r2 while using the two-step approach compared with the direct imputation, especially in the low-frequency ranges. Second, evaluating the results when applying different imputation quality (r2) cutoffs on the first imputation showed that using no filter and using a filter on r2>0.3 increased the quality as compared with applying the stricter filter on r2>0.8.
Figure 3.
Imputation quality metric, r2 per minor allele frequency (MAF) for the two different imputation strategies, chromosome 2 and 20 for the study sample. The two-step strategy further stratified by first imputation step filter. A gray horizontal dashed line at r2=0.8 is included to indicate high imputation quality. Lines were created by meaning r2 per 100 single-nucleotide polymorphisms (SNPs) and applying local polynomial regression fitting (loess function in R-project).
The comparison of the statistical differences in r2 values revealed highly significant improvement in imputation quality for the two-step approach using paired t-test (all P<1e-15) (Table 2). In the 1–5% MAF range, the two-step approach using a r2 filter on 0.3 (mean r2=0.87) had a 28% higher mean r2 compared with the direct imputation (mean r2=0.68). For this analysis, filtering out SNPs genotyped in the local reference panel resulted in a slightly lower r2 increase between approaches on 21%.
Table 2. Imputation quality metric (r 2) for the two-step approach compared with direct imputation.
| Two stepa | Direct | |||
|---|---|---|---|---|
| Minor allele frequency | N | r2, mean (SD) | N | r2, mean (SD) |
| >0–1%b | 684 334 | 0.16 (0.28) | 839 141 | 0.10 (0.20) |
| 1–5%b | 309 759 | 0.87 (0.23) | 303 608 | 0.68 (0.31) |
| 5–10%b | 144 718 | 0.94 (0.17) | 143 065 | 0.85 (0.24) |
| 10–50%b | 538 142 | 0.97 (0.11) | 539 322 | 0.92 (0.17) |
First imputation filter on r2 0.3.
Comparing two step and direct imputation in each minor allele frequency bin revealed P-values <10−15.
Once we set a fixed threshold of r2>0.3 (sufficient quality) to remove badly imputed markers in the first imputation step, we imputed all autosomal chromosomes using both the two-step and direct imputation approaches. This analysis revealed 2 528 598 SNPs (intergenic: 78.49%, intronic: 20.47%, exonic: 1.04%) with MAF 1–5% and r2 above 0.3 using the two-step approach (r2>0.3 on first step) compared with 2 147 168 (intergenic: 78.42%, intronic: 20.58%, exonic: 1.00%) when imputing directly to the 1000 G. This represents an increment of 18% in the number of low-frequency SNPs using the two-step approach (synonymous: 21.10%, non-synonymous: 25.89% and UTR 20.39% more variants). The r2 distribution of the extra imputed SNPs for the two-step approach in this low-frequency spectrum was highly right-skewed toward an r2 of 1 depicted in the supplement (Supplementary Figure S2).
Accuracy
Overall accuracy estimates are presented in Supplementary Table S2. The accuracy is also presented per genotype class in Table 3 and in Supplementary Figure S3. Supplementary Figure S3 shows higher accuracy for the heterozygotes and rare homozygotes when using the two-step approach in low frequencies (MAF<10%). Again, using a filter for the first step on r2>0.3 performed best. The mean accuracy was statistically significantly higher in the lower MAF (1–5% and 5–10%) bins when using the two-step approach (first-step filter r2>0.3) compared with the direct imputation. Comparing the accuracy of the heterozygotes-imputed genotypes from the two-step approach using the r2 filter on 0.3 with the approach without filter also revealed a significant difference (P<1e-15) and better accuracy of applying a filter on 0.3.
Table 3. Imputation accuracy for the two-step approach compared with direct imputation stratified per genotype class.
| Minor allele frequency | N | Two stepa accuracy, mean (SD) | Direct accuracy, mean (SD) | P-valueb |
|---|---|---|---|---|
| Common homozygotes (major–major) | ||||
| >0–1% | 25 087 | 0.998 (0.005) | 0.999 (0.004) | — |
| 1–5% | 25 087 | 0.998 (0.006) | 0.998 (0.006) | — |
| 5–10% | 18 613 | 0.996 (0.009) | 0.996 (0.01) | — |
| Heterozygotes (minor–major) | ||||
| >0–1% | 25 087 | 0.830 (0.237) | 0.806 (0.244) | 3.4e-6 |
| 1–5% | 25 071 | 0.915 (0.145) | 0.886 (0.172) | <1e-15 |
| 5–10% | 18 613 | 0.955 (0.083) | 0.942 (0.106) | <1e-15 |
| Rare homozygotes (minor–minor) | ||||
| >0–1% | 0 | — | — | — |
| 1–5% | 1248 | 0.895 (0.241) | 0.826 (0.294) | <1e-15 |
| 5–10% | 6746 | 0.925 (0.204) | 0.888 (0.245) | <1e-15 |
First imputation filter on r2>0.3.
Comparing groups, heterozygotes and rare homozygotes in each minor allele frequency bin.
Mendelian consistency
There were no differences between direct and two-step imputation in the number of mendelian errors per family in the imputed SNPs (Supplementary Table S3).
Evaluating the size of the local reference panel
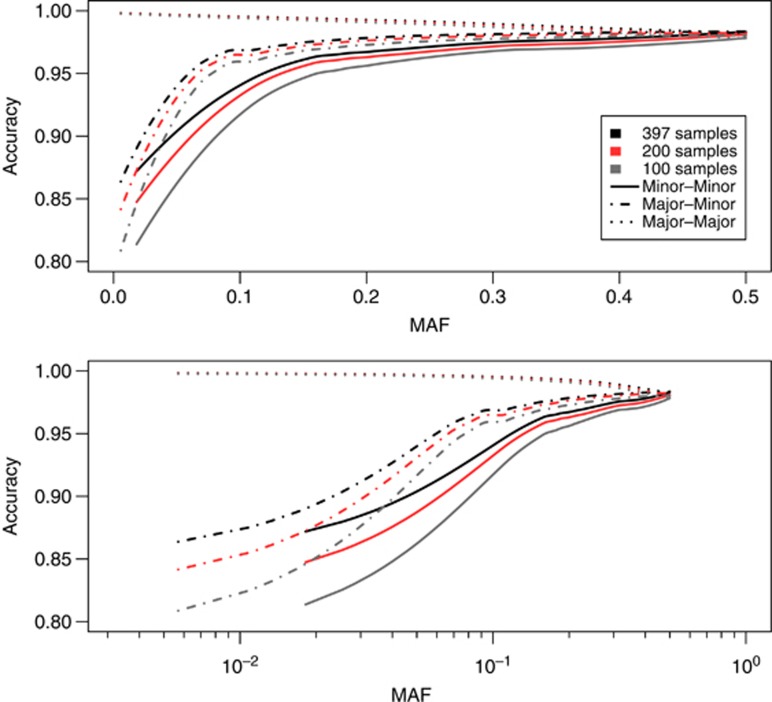
Accuracy estimates are presented in Figure 4, Supplementary Figures S4 and S5 for different sizes of the local reference panel. As expected, the larger the local reference panel, the better the quality of the imputation. In the low-frequency spectrum with MAF from 1 to 5%, accuracy measures (two step, r2>0.3 at first step) dropped in the heterozygotes when lowering the number from all 397 samples (accuracy=0.915) to 200 (accuracy=0.901) and 100 (accuracy=0.879) individuals in the local reference panel. Similar drops were seen in the rare homozygotes in the same MAF bin from all 397 samples (accuracy=0.895) to 200 (accuracy=0.868) and 100 (accuracy=0.838) individuals used in the local reference panel.
Figure 4.
Imputation accuracy for different local reference sizes. Imputation accuracy stratified per genotype for the different sizes of the local reference panel. This is presented using a first step r2 filter on 0.3.
Discussion
Principal findings
In this study, we have compared the traditional direct one-step imputation procedure with a proposed two-step imputation approach to improve the quality of the imputed data obtained after the use of dense reference panels, as the ones emerging nowadays. Our most important finding was that the two-step approach increases both the quality and the accuracy of imputation, even increasing the number of high-quality markers in 18% for the lower frequency spectrum (MAF 1–5%) as compared with the traditional direct imputation approach. Accuracy improvement was primarily seen for the low-frequency variants and only in the heterozygotes and rare homozygotes genotype classes.
Limitations and strengths
We evaluated the two-step imputation approach by means of different measures of quality, specifically r2 and imputation accuracy. Comparison to a gold standard data (in our case: 88 samples genotyped with two different arrays) must be seen as one of the primary strengths of this study, enabling the evaluation of the approaches in low-frequency markers. Nonetheless, one limitation is the relative low number of samples in our study sample.
To evaluate the gain of power and improvement in means of novel findings in the GWAS setting, a future effort should focus on applying the two-step approach to entire data sets, as earlier applied when analyzing the difference between HapMap and 1000 G imputation.9 This comparison would surely require large sample sizes and the combination of several studies before an assessment of its value at meta-analysis level can be done.
Further estimation of minimum marker content of the input for the study sample would also be necessary to evaluate and analyze by testing if this two-step approach would surpass the quality of direct imputation in studies that are already genotyped in arrays with lower content than the 550 K array.
Meaning of the study
We found that the proposed two-step imputation approach excels the imputation quality (r2) and accuracy (concordance) as compared with the direct imputation to 1000 G. One of the main reasons for this could be that we used a dense array with many low-frequency markers at the intermediate imputation step. As these low-frequency markers can create population-specific haplotypes,8 we would expect that the markers imputed in our study sample using the local reference panel provide more precise haplotypes of both common and rare variants over which imputation to a more comprehensive panel such as the 1000 G can be improved. The main reasoning to this improvement being that a local reference panel must be closer to the study sample in means of ancestry and that the overall number of haplotypes for some markers are doubled (794 haplotypes in the local reference panel in step 1+758 from the 1000 G panel in step 2). However, we did not apply the two-step approach imputing other European populations and using this Dutch local reference panel. The reasoning on closeness in ancestry between the study sample and local reference panel is therefore speculations.
It is possible that the increasing quality of future panels of the 1000 G (or other sequencing projects used as reference) may decrease the boost provided by our proposed two-step approach and thus be as efficient or even surpass the accuracy reached by the two-step approach suggested in this manuscript. Nevertheless, the rationale behind our approach is that by first imputing from a local reference panel can result in more accurate imputation of variants in the lower minor allele frequency spectrum, independent of the quality of the reference panel.
The size of the local reference panel influenced the imputation quality of the two-step approach. Nonetheless, for heterozygous genotypes of variants in the MAF 1–5% range, the increase in quality seems to level-off with increasing sample size, as observed from doubling the local reference panel from 100 to 200 samples. This increment showed a bigger leap in imputation quality, than the one after doubling the number from 200 to 397 samples (Supplementary Figure S5). Probably, further increment in the local reference panel size (beyond the 397 samples) could still add to the imputation accuracy but in a less pronounced manner. Moreover, the current imputation quality metrics are within reasonable values expected across studies, raising the question if adding more samples will be a cost-efficient approach.
One other study by Sampson et al15 has assessed a two-platform approach, genotyping a subset of samples on a denser array (Omni2.5). These samples were combined with the 1000 G reference set before imputation. Increasing the number of samples to combine with the 1000 G improved r2.15 However, by using standard imputation tools, the combination of reference panels is only implemented in IMPUTE2,5 software not used in the current study. We were therefore not able to compare the two-step approach to a direct one-step approach with the combined use of the 1000 G reference panel and our local reference panel. Nonetheless, in a recent study we showed that combining 1000 G with a Dutch sequenced panel (GoNL) only increased imputation r2 by 1.4% compared with the GoNL alone, for low-frequency markers imputing a Dutch GWA data set (Deelen et al, accepted EJHG). Thus, we would not expect that combining our local reference panel with the 1000 G panel would surpass the increase showed in our previous study.
Our study highlights a possible cost-effective strategy for large studies undertaking genome-wide genotyping or a possible upgrade of existing genotyped data sets. We expect that in large studies (with thousands of individuals), a substantial reduction in costs would be achieved by genotyping only a subset of the samples on these relatively expensive arrays with many low-frequency markers (eg, Omni5) and subsequently use these samples as a local reference set in a two-step imputation approach. This extra load will represent an increase in imputation quality and hence, the uncertainty of imputation calls will decrease, yielding more precise dosages what will be reflected in the power to detect associations in future GWAS on complex diseases. Nonetheless, discovering low-frequency variants associated with complex traits in GWAS does not depend only on imputation quality, but also on sufficiently powered settings (large sample sizes). Thus, the proposed two-step approach needs to be applied by all participating studies of a GWAS meta-analysis in order to observe an increase in the power needed to detect rare variants.
Conclusion
Imputation is a useful approach to improve both coverage and power in genetic association studies. The two-step approach in our setting increased imputation quality compared with direct imputation especially in the low-frequency spectrum. Further, this imputation methodology is a cost-effective strategy for improving imputation quality in large samples.
Acknowledgments
The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam, The Netherlands Organization for Scientific Research (NWO), The Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), The Netherlands Genomics Initiative, the Ministry of Education, Culture and Science, the Ministry of Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The contribution of inhabitants, general practitioners and pharmacists of the Ommoord district to the Rotterdam Study is gratefully acknowledged. We further acknowledge the support of the European Respiratory Society, Fellowship STRTF 429-2011 and from the NWO Rubicon fellowship 825.12.023.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Hindorff L, Junkins H, Hall P, Metha J, Manolio T.A Catalog of Published Genome-Wide Association Studies. Available at www.genome.gov/gwastudies/ .
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genom Hum G. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Wang L, Rankinen T, Bouchard C, Rao DC. Performance of genotype imputations using data from the 1000 Genomes Project. Hum Hered. 2012;73:18–25. doi: 10.1159/000334084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Genomics: the search for association. Nature. 2010;467:1135–1138. doi: 10.1038/4671135a. [DOI] [PubMed] [Google Scholar]
- Hofman A, van Duijn CM, Franco OH, et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol. 2011;26:657–686. doi: 10.1007/s10654-011-9610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JN, Jacobs K, Wang Z, Yeager M, Chanock S, Chatterjee N. A two-platform design for next generation genome-wide association studies. Genet Epidemiol. 2012;36:400–408. doi: 10.1002/gepi.21634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.