The transcription factor TaNFYA-B1 is up-regulated by low-nitrogen and low-phosphorus treatment in wheat seedlings, and overexpressing this gene increases the grain yield of wheat under differing nitrogen and phosphorus supply levels.
Abstract
Increasing fertilizer consumption has led to low fertilizer use efficiency and environmental problems. Identifying nutrient-efficient genes will facilitate the breeding of crops with improved fertilizer use efficiency. This research performed a genome-wide sequence analysis of the A (NFYA), B (NFYB), and C (NFYC) subunits of Nuclear Factor Y (NF-Y) in wheat (Triticum aestivum) and further investigated their responses to nitrogen and phosphorus availability in wheat seedlings. Sequence mining together with gene cloning identified 18 NFYAs, 34 NFYBs, and 28 NFYCs. The expression of most NFYAs positively responded to low nitrogen and phosphorus availability. In contrast, microRNA169 negatively responded to low nitrogen and phosphorus availability and degraded NFYAs. Overexpressing TaNFYA-B1, a low-nitrogen- and low-phosphorus-inducible NFYA transcript factor on chromosome 6B, significantly increased both nitrogen and phosphorus uptake and grain yield under differing nitrogen and phosphorus supply levels in a field experiment. The increased nitrogen and phosphorus uptake may have resulted from the fact that that overexpressing TaNFYA-B1 stimulated root development and up-regulated the expression of both nitrate and phosphate transporters in roots. Our results suggest that TaNFYA-B1 plays essential roles in root development and in nitrogen and phosphorus usage in wheat. Furthermore, our results provide new knowledge and valuable gene resources that should be useful in efforts to breed crops targeting high yield with less fertilizer input.
Nitrogen (N) and phosphorus (P) fertilizers have made a significant contribution to the improvement of crop yields (Godfray et al., 2010; Gregory and George, 2011). To meet the food demands for the estimated nine billion humans on Earth, the consumption of N and P fertilizers will likely more than double by 2050 (Tilman et al., 2001). However, the overuse of fertilizers has resulted in low fertilizer use efficiency, resource waste, and environmental problems (Tilman et al., 2001; Liu et al., 2013). As such, systematic approaches to efficiently utilize N and P fertilizers, such as the development of N- and P-efficient crop varieties, should be made. As N and P fertilizers are applied together in many agriculture systems, a promising approach would be to breed crops that can efficiently use both N and P; this strategy could lead to high yields with lower economic and environmental costs.
Conventional plant breeding has made remarkable improvements to both N and P use efficiency. For instance, modern wheat (Triticum aestivum) and barley (Hordeum vulgare) varieties significantly out-yield the old varieties at all N levels tested (Sylvester-Bradley and Kindred, 2009; Cormier et al., 2013). The International Maize and Wheat Improvement Centre Wheat Program has selected wheat varieties that can efficiently acquire P under low-P conditions but also respond to P application (Manske et al., 2000). In addition to conventional breeding, molecular breeding that employs marker-assisted selection and transgenic technologies has also improved N and P use efficiencies in crops. Marker-assisted selection for PHOSPHORUS UPTAKE1, a major rice (Oryza sativa) quantitative trait locus that confers tolerance to P deficiency in soils, significantly increases rice grain yields in P-deficient soils (Chin et al., 2010). In recent years, transgenic technology has been widely used to engineer crops with improved N use efficiency by manipulating amino acid biosynthesis, N translocation and remobilization, and N signaling (McAllister et al., 2012; Xu et al., 2012). Such methods have also been used to develop crops with efficient P use by enhancing P acquisition capacities (Tian et al., 2012; Zhang et al., 2014). However, to date, most of these transgenic studies have targeted the efficient use of a single nutrient, not the efficient use of both N and P.
Complicated gene networks control both N and P use in plants. A variety of transcription factors in N and P signaling networks have been identified recently; these are known to modulate the expression of genes involved in root development and genes involved in the uptake, assimilation, remobilization, and storage of N and P (Chiou and Lin, 2011; Alvarez et al., 2012; Gutiérrez, 2012; Zhang et al., 2014). Some of these transcription factors have been shown to be of value in the engineering of crops for improved N or P use efficiency. For example, the plant-specific transcription factor Dof zinc finger protein1 from maize (Zea mays) can increase the expression of phosphoenolpyruvate carboxylase and several genes involved in the tricarboxylic acid cycle, which results in the production of more carbon skeletons for the assimilation of N in Arabidopsis (Arabidopsis thaliana; Yanagisawa et al., 2004). This transcription factor has been used successfully to improve N use efficiency and enhance growth in rice under low-N conditions by increasing carbon flow toward N assimilation (Kurai et al., 2011). The MYB-CC-type transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1) plays a central role in the signaling network for sensing P availability (Rubio et al., 2001; Chiou and Lin, 2011). PHR1 and its homologs in Arabidopsis and rice regulate root development and a set of inorganic phosphate (Pi) starvation-inducible genes, including those involved in Pi uptake and redistribution (Rubio et al., 2001; Nilsson et al., 2007; Zhou et al., 2008; Bustos et al., 2010). Recently, a PHR1 homolog in wheat (TaPHR1) was shown to stimulate lateral branching, increase P uptake and grain yield, and up-regulate a subset of Pi starvation response genes in wheat (Wang et al., 2013). In addition to transcription factors, microRNAs also play vital roles in N and P signaling (Chiou et al., 2006; Pant et al., 2009; Fischer et al., 2013; Zeng et al., 2014), and some microRNAs are regulated by both N and P supply levels (Pant et al., 2009; Kant et al., 2011). The E3 ligase NITROGEN LIMITATION ADAPTATION (NLA) was initially shown to function as a positive regulator of the adaptability of Arabidopsis to N limitation (Peng et al., 2007). More recently, NLA was found to regulate Pi signaling/homeostasis through its interaction with the E2 conjugase PHOSPHATE2 (PHO2), a key negative regulator in the Pi response pathway (Park et al., 2014). NLA and PHO2 are, respectively, the targets of two Pi starvation-induced microRNAs, miR827 and miR399 (Bari et al., 2006; Lin et al., 2013). These two microRNAs were also recently found to be regulated by N starvation (Fischer et al., 2013). These results indicate that NLA, PHO2, miR827, and miR399 function in regulating the cross-talk between the N and P signaling pathways. Modulation of these factors may facilitate the development of crops with more efficient N and P use.
The microRNA169 (miR169) family is known to be regulated by both N and P starvation (Pant et al., 2009; Lundmark et al., 2010; Xu et al., 2011, 2013; Zhao et al., 2011; Liang et al., 2012). Different members of this microRNA family show distinct response patterns to N or P starvation (Pant et al., 2009; Liang et al., 2012; Xu et al., 2013). As such, miR169 and its targets may be valuable resources for engineering plants with altered N and/or P use efficiencies. For example, under N starvation treatment in Arabidopsis, miR169 is down-regulated while its potential targets NFYA2, NFYA3, NFYA5, and NFYA8 are up-regulated (Zhao et al., 2011). Overexpression of miR169a inhibited the expression of several of NFYAs and accumulated less N in transgenic Arabidopsis (Zhao et al., 2011).
CCAAT box-binding transcription factors are conserved among all eukaryotes; they are named Nuclear Factor Y (NF-Y) in plants, CCAAT-binding factor in mammals, and Heme Activator Protein (HAP) in yeast (Saccharomyces cerevisiae; Mantovani, 1999). NF-Y transcription factors are heterotrimers composed of three subunits: NF-YA, NF-YB, and NF-YC (hereafter, NF-YA, NF-YB, and NF-YC are referred as NFYA, NFYB, and NFYC, respectively). Several studies suggested that members of the NF-Y family play essential roles in the control of flowering (Kumimoto et al., 2008, 2010; Wei et al., 2010; Li et al., 2011; Yan et al., 2011), seed development (Yamamoto et al., 2009), photosynthesis (Stephenson et al., 2010, 2011), and nodule development (Combier et al., 2006, 2008; Zanetti et al., 2010). Other NF-Y family members are known to enhance tolerance to abiotic stresses such as drought (Nelson et al., 2007; Stephenson et al., 2007; Li et al., 2008; Ni et al., 2013), salinity (Zhao et al., 2009; Leyva-González et al., 2012), and cold (Leyva-González et al., 2012; Shi et al., 2014). A relationship between NF-Ys and N utilization was first reported in yeast: the HAP complex regulates the expression of Glucose dehydrogenase1 and an N-starvation-specific gene, vacuolar serine protease (isp6+; Dang et al., 1996; Nakashima et al., 2002; Riego et al., 2002). Although miR169 has been reported to potentially function in N and P signaling, the functions of its targets (NFYAs) in mediating N and P use are not well understood in crops such as wheat.
Here, we show that both N and P starvation cause the down-regulation of miR169 but the up-regulation of the NFYAs in wheat. Overexpressing TaNFYA-B1 in wheat stimulated lateral branching, up-regulated the expression of nitrate and phosphate transporters, increased N and P uptake, and increased grain yields under control, low-N, and low-P conditions. Thus, NFYAs represent valuable genetic resources for efforts to breed crops that utilize both N and P more efficiently.
RESULTS
Cloning and Phylogenetic Analysis of NF-Y Transcription Factors
In a microarray experiment to identify N and P starvation-responsive genes in wheat, we found that the probes corresponding to NF-Y transcription factors were induced by both low-N and low-P treatments. In order to more comprehensively understand the responses of different NF-Y transcription factors to N and P supply levels, we first isolated NF-Y genes using both complementary DNA (cDNA) cloning and sequence mining from public databases, including the National Center for Biotechnology Information dbEST, the genome sequences of the wheat cv Chinese Spring (International Wheat Genome Sequencing Consortium, 2014), and the two grasses thought to be the A genome and D genome donors of wheat (Triticum urartu and Aegilops tauschii; Jia et al., 2013; Ling et al., 2013). We identified a total of 80 unique NF-Y genes in the wheat genome. These included 18 NFYAs, 34 NFYBs, and 28 NFYCs (Supplemental Table S1), which encode the A, B, and C subunits of NF-Y, respectively. The chromosomal locations of 76 NF-Y genes were determined according to the draft sequence of the 21 individual chromosomes of cv Chinese Spring (Supplemental Table S1), while the putative chromosomal locations of the remaining four genes (e.g. TaNFYB-B2, TaNFYB-D3, TaNFYB-A13, and TaNFYB-D13), which were isolated by cDNA cloning, were assigned according to their similarities with NFYBs from cv Chinese Spring, T. urartu, and A. tauschii. Of the 18 NFYAs, six each were from the A, B, and D genomes; of the 34 NFYBs, 11, 12, and 11 were from the A, B, and D genomes, respectively; of the 28 NFYCs, seven, nine, and 12 were from the A, B, and D genomes, respectively (Supplemental Table S1).
Phylogenetic trees for NFYA (Supplemental Fig. S1), NFYB (Supplemental Fig. S2), and NFYC (Supplemental Fig. S3) were created after alignment of the putative NF-Y protein sequences from wheat, T. urartu, A. tauschii, barley, maize, rice, Brachypodium distachyon, Arabidopsis, and yeast. The phylogenetic trees clearly showed that the NF-Y proteins of wheat were most closely related to the homologs from T. urartu and A. tauschii, followed by those of barley. The phylogenetic tree of NFYAs was divided into three clades, and each clade contained NFYAs from all the plant species that were used to create this phylogenetic tree (Supplemental Fig. S1). For the 18 NFYAs from wheat, TaNFYA5 (TaNFYA-A5, TaNFYA-B5, and TaNFYA-D5) fell into clade I; TaNFYA1 (TaNFYA-A1, TaNFYA-B1, and TaNFYA-D1), TaNFYA3 (TaNFYA-A3, TaNFYA-B3, and TaNFYA-D3), and TaNFYA4 (TaNFYA-A4, TaNFYA-B4, and TaNFYA-D4) were in clade II; and TaNFYA2 (TaNFYA-A2, TaNFYA-B2, and TaNFYA-D2) and TaNFYA6 (TaNFYA-A6, TaNFYA-B6, and TaNFYA-D6) were in clade III (Supplemental Fig. S1).
Expression of TaNFYAs Is Induced by N and P Starvation
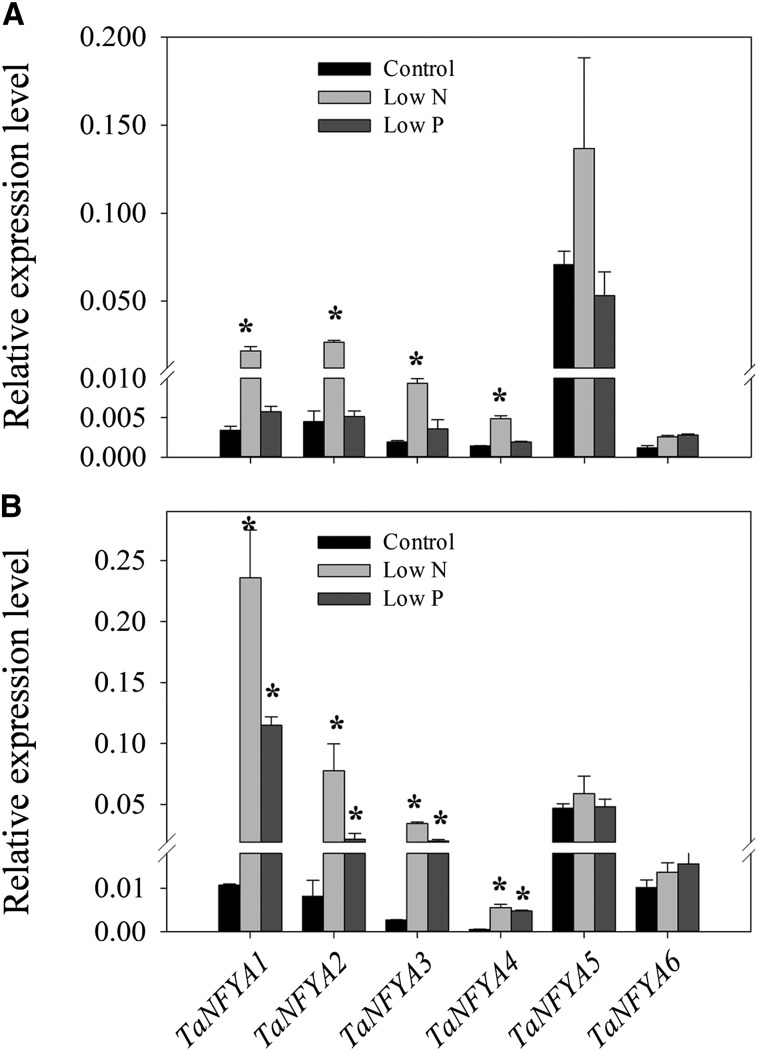
Quantitative real-time reverse transcription (RT)-PCR was used to evaluate the mRNA expression levels of the NF-Y genes in the roots and shoots of wheat seedlings grown under different N and P supply levels. Primers were designed to amplify the homologous alleles at a particular locus; for example, the relative expression level of TaNFYA1 might represent that of all three homologous alleles of TaNFYA1 (TaNFYA-A1, TaNFYA-B1, and TaNFYA-D1). Compared with the control treatment, low-N treatment significantly increased the expression levels of TaNFYA1 to TaNFYA4 in shoots (Fig. 1A), and both low-N and low-P treatments significantly increased the expression levels of TaNFYA1 to TaNFYA4 in roots (Fig. 1B). However, neither low-N nor low-P treatment significantly affected the expression of TaNFYA5 or TaNFYA6 in shoots or roots (Fig. 1). We also investigated the responses of the TaNFYB and TaNFYC genes to N and P supply levels. The expression of TaNFYB1 in shoots was up-regulated by low-P treatment. The expression of TaNFYB3 in shoots and the expression of TaNFYB2 to TaNFYB5 in roots were repressed by low-N and low-P treatments (Supplemental Fig. S4). Most of the tested TaNFYC genes did not respond significantly to alternation of N or P supply levels. The exceptions to this were that TaNFYC3 was up-regulated in shoots by low-N treatment and TaNFYC1 was down-regulated in roots by low-N treatment (Supplemental Fig. S5). These results indicated that TaNF-Y genes responded differentially to N and P availability and that low-N and low-P treatments mainly induced the expression of TaNFYA genes.
Figure 1.
The responses of TaNFYA genes to N and P starvation. Wheat seedlings 7 d after germination were grown for 20 d in nutrient solutions that contained 2 mm N + 0.2 mm P (control), 0.2 mm N + 0.2 mm P (low N), and 2 mm N + 0.02 mm P (low P). The shoots (A) and roots (B) were then collected for gene expression analysis. cDNA was standardized by reference to an actin (TaACTIN) standard. Data represent means ± se of three independent biological replicates. Asterisks indicate that differences between the control and low N or low P were significant at the 0.05 level.
Expression of T. aestivum (tae)-miR169 Is Reduced by N and P Starvation
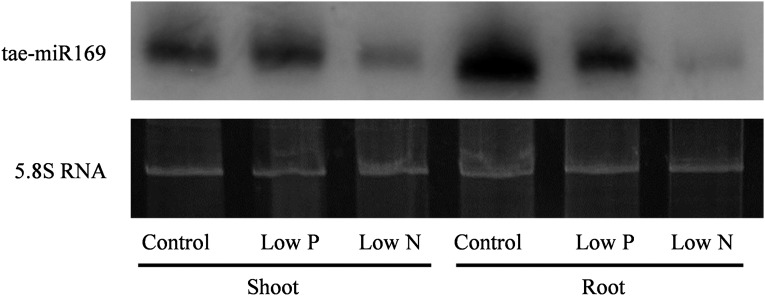
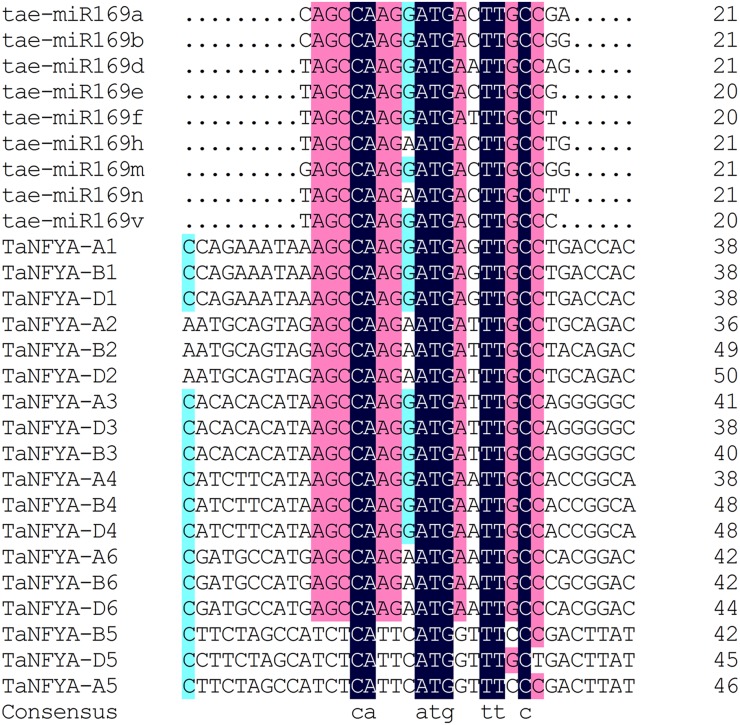
It has been reported that miR169 can regulate the expression of NFYAs by cleaving the mRNA of NFYAs (Li et al., 2008; Zhao et al., 2011). To determine whether tae-miR169 responds to N and P starvation, small RNAs were extracted from the roots and shoots of Xiaoyan 54 plants grown under control, low-N, and low-P conditions and sequenced. The clean sequence reads for individual small RNA samples varied between 6,024,563 and 12,328,574. Bioinformatics analysis of the sequence data identified nine tae-miR169 members and revealed that the expression of tae-miR169a, tae-miR169b, tae-miR169e, tae-miR169f, tae-miR169h, and tae-miR169n was inhibited in both shoots and roots by both low-N and low-P treatments. The other three members (tae-miR169d, tae-miR169m, and tae-miR169v) were present only in very low numbers of transcripts; thus, whether their expression was regulated by N and P supply levels will require further investigation (Table I). To confirm the results of the small RNA sequencing, microRNA northern blotting was used to detect the expression of tae-miR169 by using tae-miR169b as a probe. The results showed that the expression of tae-miR169 in shoots was repressed by low-N treatment and that in roots was repressed by both low-N and low-P treatments (Fig. 2). This repression was stronger in roots than in shoots and more dramatic with low-N treatment than with low-P treatment (Supplemental Fig. S6). Our subsequent analysis showed that all nine tae-miR169 members nearly perfectly matched with their putative target sites in the 3′ untranslated region of TaNFYA1 to TaNFYA4 and TaNFYA6 but not in TaNFYA5 (Fig. 3). TaNFYA-B1 was degraded by tae-miR169f (Supplemental Fig. S7).
Table I. Abundance of the miR169 family in wheat.
Xiaoyan 54 seedlings were grown for 7 d in nutrient solutions that contained 2 mm N + 0.2 mm P (control), 0 mm N + 0.2 mm P (low N), and 2 mm N + 0 mm P (low P). Total RNA was isolated from roots and shoots separately for small RNA sequencing. Transcript abundance is presented as transcripts per million.
| Seedling | Transcript Abundance |
|||||
|---|---|---|---|---|---|---|
| Shoot |
Root |
|||||
| Control | Low P | Low N | Control | Low P | Low N | |
| tae-miR169a | 406.92 | 133.62 | 133.23 | 74.54 | 45.59 | 36.26 |
| tae-miR169b | 1,399.30 | 573.98 | 488.27 | 180.80 | 69.00 | 42.36 |
| tae-miR169d | 0.00 | 0.00 | 0.00 | 0.32 | 0.16 | 0.12 |
| tae-miR169e | 39.86 | 22.74 | 13.45 | 49.97 | 20.78 | 4.46 |
| tae-miR169f | 28.28 | 9.30 | 6.58 | 13.71 | 0.99 | 0.82 |
| tae-miR169h | 163.06 | 51.46 | 51.52 | 52.16 | 10.60 | 2.70 |
| tae-miR169m | 0.40 | 0.50 | 0.14 | 0.00 | 0.00 | 0.00 |
| tae-miR169n | 14.00 | 4.65 | 3.86 | 1.30 | 0.25 | 0.12 |
| tae-miR169v | 0.13 | 0.00 | 0.14 | 0.16 | 0.08 | 0.00 |
Figure 2.
Northern blot of tae-miR169b from the shoots and roots of wheat plants grown under different N and P supply levels. Wheat seedlings were grown for 20 d in the control, low-P, and low-N nutrient solutions, and then total RNA from the shoots and roots was separated by PAGE; RNA blots were probed with [γ-32P]ATP-labeled oligonucleotides. The 5.8S RNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls.
Figure 3.
Putative tae-miR169 binding sites in the 3′ untranslated region of TaNFYA genes. The target sites of TaNFYA genes were aligned with miR169 mature sequences with DNAMAN6.0. The mature sequences of tae-miR169 were obtained through sequencing small RNA libraries of wheat seedlings with Illumina Genome Analyzer IIx.
The Molecular Cloning and Characterization of TaNFYA-B1
To further determine the roles of TaNFYA genes in mediating N and P use in wheat, TaNFYA1 was chosen as the representative subunit for further study. TaNFYA1 was selected because it had the strongest responses to N and P supply levels among the investigated TaNFYA genes (Fig. 1). The genome sequences of TaNFYA-A1 and TaNFYA-D1 both have five exons and four introns in the same arrangement, but the second intron is absent in TaNFYA-B1 (Supplemental Fig. S8). By using the length polymorphisms of the first intron, we successfully located TaNFYA-A1, TaNFYA-B1, and TaNFYA-D1 on the long arms of chromosomes 6A, 6B, and 6D, respectively (Supplemental Fig. S9), confirming the results of the cv Chinese Spring draft genome sequence (Supplemental Table S1). After sequencing the cDNAs corresponding to the homologous TaNFYA1 alleles, we found that TaNFYA-B1 accounted for about 60% of the cDNA sequences (data not shown), suggesting that TaNFYA-B1 had higher expression levels than did TaNFYA-A1 or TaNFYA-D1. By overexpressing a 35S::TaNFYA-B1::GFP fusion in Arabidopsis, we observed that TaNFYA-B1 was localized to the nucleus (Supplemental Fig. S10A). A transcriptional activity assay in yeast cells showed that TaNFYA-B1 had transcriptional activation activity (Supplemental Fig. S10B).
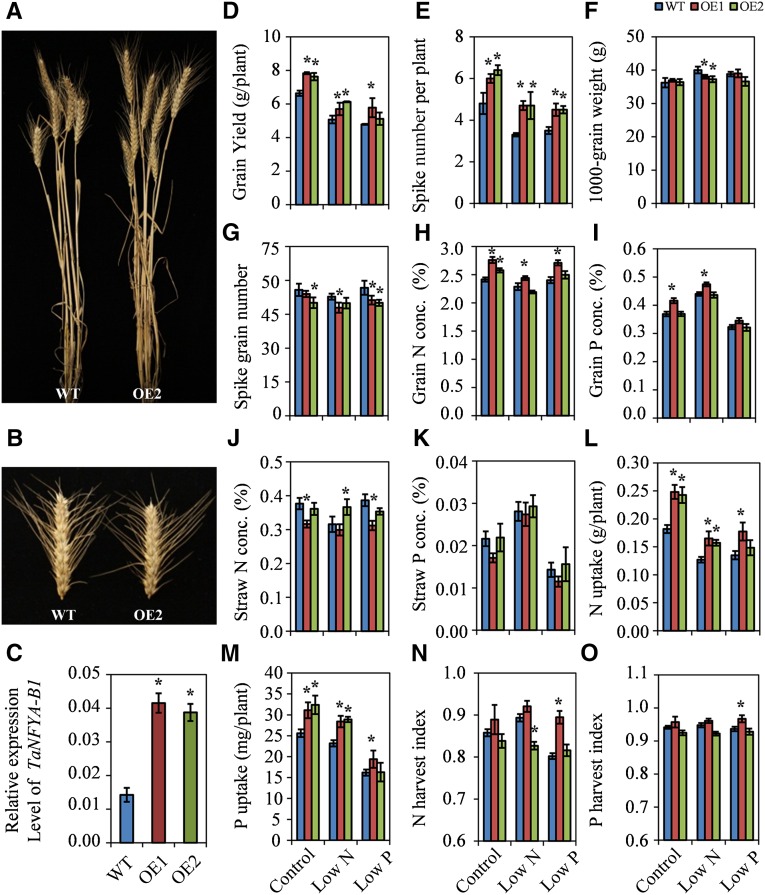
Overexpressing TaNFYA-B1 in Wheat Increases Grain Yield under Different N and P Supply Levels in a Field Experiment
In order to investigate the functions of TaNFYA genes, we developed TaNFYA-B1 transgenic wheat plants using a maize ubiquitin promoter-controlled construct. Two homozygous transgenic lines (OE1 and OE2) were obtained. TaNFYA-B1 expression in these lines was severalfold higher than in the wild-type Longchun 23 plants (Fig. 4C). The performance of the wild-type and transgenic lines were evaluated under control, low-N, and low-P conditions in a field experiment. Both of the transgenic lines had significantly higher grain yield and spike numbers than the wild type in all three of the treatments (Fig. 4, A, D, and E). However, the transgenic lines had similar or lower 1,000-grain weights and spike grain numbers than the wild type (Fig. 4, B, F, and G). These results indicated that overexpression of TaNFYA-B1 increased grain yield by increasing spike number.
Figure 4.
Agronomic traits and N and P use of the wild type and TaNFYA-B1 transgenic lines in the field experiment. The wild type (WT) and the transgenic lines (OE1 and OE2) were grown in soils that were treated with 18 g N m−2 + 13.5 g P m−2 (control), 0 N + 13.5 g P m−2 (low-N treatment), and 18 g N m−2 + 0 P (low-P treatment). A, Representative plants of the wild type and OE2 under control conditions. B, Primary spikes of the plants in A. C, Expression levels of TaNFYA-B1 in the seedlings grown under control conditions. D to O, Grain yield per plant (D), spike number per plant (E), 1,000-grain weight (F), grain number per spike (G), grain N concentration (H), grain P concentration (I), straw N concentration (J), straw P concentration (K), N uptake (L), P uptake (M), N harvest index (N), and P harvest index (O). Data represent means ± se of three replicates. Asterisks indicate that differences between the wild type and the transgenic lines were significant at the 0.05 level.
We also evaluated N and P use in the transgenic lines. The two transgenic lines had higher grain N concentration than the wild type under control conditions, and OE1 had higher grain N concentration than the wild type under low-P conditions (Fig. 4H). OE1 also had higher grain P concentration than the wild type in all three treatments (Fig. 4I). Overexpressing TaNFYA-B1 did not consistently affect straw N concentration or straw P concentration (Fig. 4, J and K). The two transgenic lines had higher N and P uptake than the wild type under all three treatments (with the exception of P uptake of OE2 under low-P conditions; Fig. 4, L and M). For all three treatments, the transgenic lines and the wild type had similar N and P utilization efficiencies (Supplemental Fig. S11); the transgenic lines and the wild type also had similar N and P harvest indexes (Fig. 4, N and O). These results suggested that overexpression of TaNFYA-B1 increased N and P uptake under different N and P supply levels but did not apparently affect N and P utilization efficiency.
Overexpressing TaNFYA-B1 Promotes Lateral Root Growth
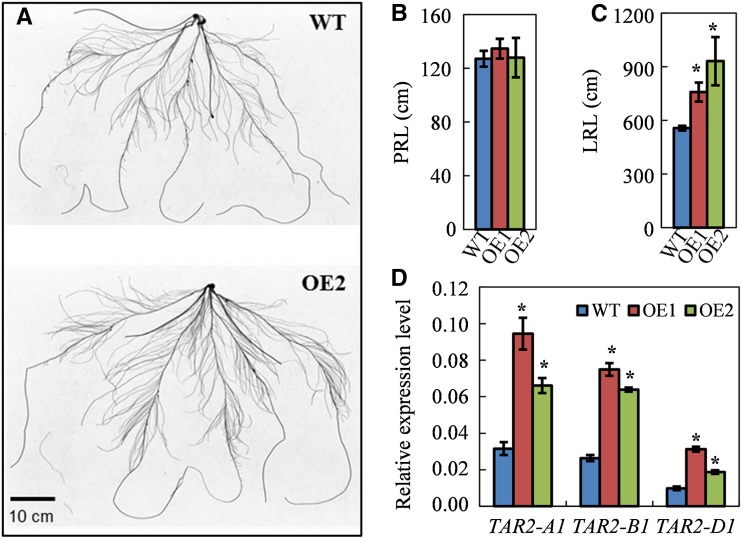
To understand the mechanisms by which TaNFYA-B1 increases N and P uptake, we first investigated how TaNFYA-B1 affected root growth. In a hydroponic culture system with plants at the seedling stage, we observed that the two transgenic lines had higher lateral root length than the wild type (Fig. 5, A and C). The transgenic plants had similar primary root length to the wild type (Fig. 5B). In a soil-pot experiment, we also observed that overexpressing TaNFYA-B1 increased N uptake and P uptake under both low-N and high-N conditions and root dry weight under low-N conditions (Supplemental Fig. S12). Therefore, both the hydroponic culture and soil-pot experiments demonstrated that overexpressing TaNFYA-B1 enhanced root development, possibly by increasing lateral root growth.
Figure 5.
Root morphological parameters of the wild type (WT) and the transgenic lines (OE1 and OE2) hydroponically grown under control nutrient conditions. Wheat seedlings were grown for 20 d in the control nutrient solution, and then the roots were scanned with an STD1600 scanner and analyzed for the root morphological parameters with WinRHIZO software (Regent Instruments). A, Root images of the wild type and OE2. B, Primary root length (PRL). C, Total lateral root length (LRL). D, Expression levels of TaTAR2 in roots. Data represent means ± se of at least five replicates. Asterisks indicate that differences between the wild type and the transgenic lines were significant at the 0.05 level.
To analyze the mechanism by which overexpression of TaNFYA-B1 enhanced root development, we compared the expression of auxin biosynthesis genes in the wild-type and transgenic lines. It has been reported that overexpressing TRYPTOPHAN AMINOTRANSFERASE RELATED2 (AtTAR2), an auxin biosynthetic gene, promoted lateral growth in Arabidopsis (Ma et al., 2014). We analyzed the effect of overexpressing TaNFYA-B1 on the expression of TAR2 homologs in wheat. We determined the expression of the three homologous alleles of TaTAR2 located on chromosomes 5A, 5B, and 5D and found that overexpressing TaNFYA-B1 up-regulated the expression of TaTAR2-A1, TaTAR2-B1, and TaTAR2-D1 in roots (Fig. 5D).
Overexpressing TaNFYA-B1 Increases the Expression of Nitrate and Phosphate Transporter Genes
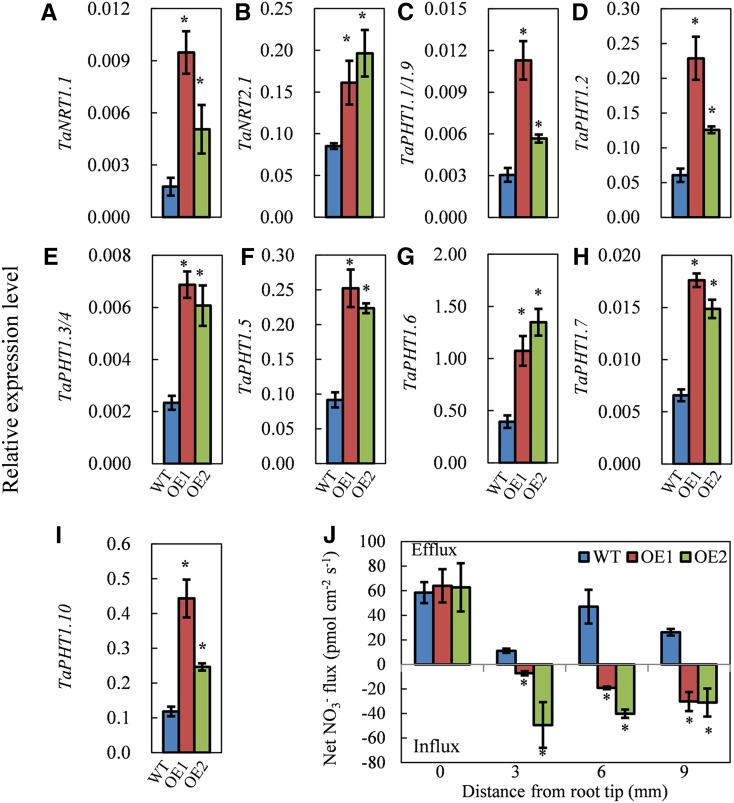
Nitrate transporters of the NRT1 and NRT2 families and phosphate transporters of the PHT1 family are the key membrane proteins that function in nitrate and phosphate uptake from soil, respectively (Schachtman et al., 1998; Xu et al., 2012). As OE1 and OE2 plants absorbed more N and P than did the wild-type plants, and there is at least one putative CCAAT box in the promoters of NRT and PHT genes (Supplemental Fig. S13), we evaluated the expression of these genes in the roots of plants that were grown under control conditions. The results showed that both of the transgenic lines had higher mRNA expression levels of NRT1.1, NRT2.1, PHT1.1/1.9, PHT1.2, PHT1.3/1.4, PHT1.5, PHT1.6, PHT1.7, and PHT1.10 than did the wild type (Fig. 6, A–I).
Figure 6.
Expression levels of nitrate and phosphate transporter genes in the wild type (WT) and the transgenic lines (OE1 and OE2). Wheat seedlings were grown for 20 d in the control nutrient solution, and then total RNA was isolated from the roots and used for gene expression analysis. A to I, Expressions levels of TaNRT1.1 (A), TaNRT2.1 (B), TaPHT1.1/1.9 (C), TaPHT1.2 (D), TaPHT1.3/4 (E), TaPHT1.5 (F), TaPHT1.6 (G), TaPHT1.7 (H), and TaPHT1.10 (I). J, Net NO3− flux rate. NO3− fluxes were measured by SIET with 0.2 mm NO3− in the measuring solution. Positive and negative flux values indicate efflux and influx, respectively. Data represent means ± se of at least three replicates. Asterisks indicate that differences between the wild type and the transgenic lines were significant at the 0.05 level.
Overexpressing TaNFYA-B1 Increases Root Nitrate Influx
As overexpression of TaNFYA-B1 increased the expression of nitrate transporters in roots, we measured the net nitrate flux rates using the scanning ion-selective electrode technique (SIET; Luo et al., 2013; Zheng et al., 2013). The roots of wild-type and transgenic lines (OE1 and OE2) grown for 20 d in the control nutrient solution (2 mm nitrate) were used to measure net nitrate flux rates in measuring solution containing 0.2 mm nitrate. At the root tip (0 mm from the root tip), wild-type and transgenic line roots all exhibited net nitrate efflux; at sites 3, 6, and 9 mm from the root tip, the roots of the wild type exhibited net nitrate efflux, while the roots of the transgenic lines exhibited net nitrate influx (Fig. 6J). These results indicated that overexpressing TaNFYA-B1 enhanced root nitrate influx.
DISCUSSION
The NF-Y Family in Wheat
The CCAAT box is one of the most ubiquitous elements existing upstream of eukaryotic promoters. The three subunits of NF-Y transcript factors form a heterotrimer that binds to these CCAAT boxes and thus act as a transcriptional activator or repressor (Laloum et al., 2013). Unlike the situation in animals, each NF-Y subunit is encoded by a single gene; a large expansion in the members of the NF-Y family has occurred in plants (Laloum et al., 2013). For example, there are nine NFYAs, 10 NFYBs, and 10 NFYCs in Arabidopsis (Gusmaroli et al., 2001, 2002). In a previous study in wheat, 10 NFYAs, 11 NFYBs, and 14 NFYCs were identified (Stephenson et al., 2007). In this study, a total of 80 wheat NF-Y genes were identified (Supplemental Table S1). By analyzing the draft genome of the wheat A genome progenitor T. urartu (Ling et al., 2013), we identified six NFYAs, 12 NFYBs, and eight NFYCs (Supplemental Table S1). In a previous study of A. tauschii, the D genome donor of common wheat, six NFYAs, 12 NFYBs, and 11 NFYCs were identified (Jia et al., 2013). After comparing the copy numbers of NF-Y genes in wheat, T. urartu, and A. tauschii, we concluded that there are six NFYAs, and theoretically as many as 12 NFYBs and 15 NFYCs, in each of the three genomes of wheat. In the sequenced cereals rice, maize, and B. distachyon, the copy numbers of the NF-Y genes vary between seven and 13 for NFYA genes, 11 and 17 for NFYB genes, and seven and 12 for NFYC genes (Supplemental Table S1). Thus, the copy numbers of NF-Y genes in each wheat genome fell into the range of that known for other cereals.
The members of the NF-Y family in plants have undergone functional diversification (Laloum et al., 2013). It has been reported that NF-Ys play roles in regulating photosynthesis, flowering, seed development, nodule development, and adaptions to abiotic stresses such as drought, salinity, and nutrient deficiency (for references, see the introduction). However, the functions of NF-Ys in wheat are still largely unknown, mainly due to the complex and large genome of wheat. The information regarding the chromosomal locations of the NF-Y genes from wheat (Supplemental Table S1) and phylogenetic trees of the families of the three NF-Y subunits (Supplemental Figs. S1–S3) will help in analyzing the collinear relationships for the NF-Y genes between wheat and model cereals such as rice. The information about NF-Y functions obtained in studies of model cereals promises to facilitate the study in wheat.
NF-Y Genes in Wheat Respond Variously to N and P Starvation
In plants, a subset of miR169 members are repressed by N and P starvation (Pant et al., 2009; Liang et al., 2012; Xu et al., 2013), and several miR169 NFYA targets are induced in such conditions (Pant et al., 2009; Zhao et al., 2011). We observed similar results in this study in wheat: low-N and low-P treatments decreased tae-miR169 abundance (Fig. 2; Table I) and increased the expression of TaNFYA1 to TaNFYA4 (Fig. 1). In correlation with the converse responses of tae-miR169 and TaNFYA1 to TaNFYA4 to low-N and low-P treatments, tae-miR169f was able to degrade its target TaNFYA-B1 (Supplemental Fig. S7). The lack of a miR169 target in TaNFYA5 was a possible reason for the lack of any response to low-N or low-P treatments for TaNFYA5 (Fig. 3). Although there was a putative miR169 target site in the three TaNFYA6 genes (Fig. 3), we did not detect any response to low-N and low-P treatment for these genes (Fig. 1). In contrast with the up-regulation of TaNFYA genes in low-N and low-P treatments, the majority of TaNFYB and TaNFYC genes did not positively respond to these same treatments (Supplemental Figs. S4 and S5). Previously, various TaNF-Y genes have been shown to differentially respond to drought stress in wheat (Stephenson et al., 2007). Therefore, TaNF-Y genes could play different roles in the adaptation to abiotic stresses.
Overexpression of TaNFYA-B1 Promotes Wheat Growth and Grain Yield
NF-Y transcription factors have been shown to improve drought tolerance in Arabidopsis (Li et al., 2008), maize (Nelson et al., 2007), and soybean (Glycine max; Ni et al., 2013) and to reduce N starvation-induced senescence in Arabidopsis (Leyva-González et al., 2012). Here, we found that overexpressing TaNFYA-B1 improved N uptake, P uptake, and grain yield in wheat under different N and P supply conditions in a field experiment (Fig. 4, D, L, and M). The increased N and P uptake in the transgenic lines appears to favor an increase in the number of tillers (Supplemental Fig. S12) and spikes (Fig. 4E) and ultimately in grain yield. In a recent 10-year period (2004–2013), the average wheat grain yield in China varied from 4,251.9 to 5,050.6 kg ha−1 (http://faostat.fao.org/). Under control conditions in the field experiment, the theoretical grain yield per square meter (the product of grain yield per plant and plant density per square meter) of the wild-type and transgenic lines fell into the range of the average wheat grain yield in China in that recent 10-year period (Supplemental Fig. S14A). As such, TaNFYA-B1 showed potential in engineering wheat with improved yield and fertilizer use efficiency under the growth conditions of average yield. The negative effect of overexpressing TaNFYA-B1 on spike grain number may limit the use of this gene in breeding high-yield wheat, as spike grain number greatly contributes to grain yield under the growth conditions of high yield. Although overexpressing AtNFYA2, AtNFYA3, AtNFYA7, and AtNFYA10 delayed N starvation-induced senescence, such overexpression of these genes impaired cell elongation and caused a dwarf phenotype in Arabidopsis (Leyva-González et al., 2012). Moreover, the expression of these four genes is induced by N and/or P limitation (Zhao et al., 2011; Leyva-González et al., 2012). Thus, it seems that the NFYA genes are positively responsive to N and P starvation but do not essentially promote plant growth. As such, more research is needed to evaluate the functions of the different NFYA genes in engineering wheat varieties with improved grain yield under different N and P availabilities.
Overexpression of TaNFYA-B1 Increases N and P Uptake, Possibly by Enhancing Root Growth and Up-Regulating the Expression of Nitrate and Phosphate Transporters
Breeding wheat with a large root system is an important strategy to improve N and P use efficiency (Manske et al., 2000; Liao et al., 2004; An et al., 2006; Ehdaie et al., 2010). Here, we showed that overexpressing TaNFYA-B1 enhanced root development by stimulating lateral root growth (Fig. 5, A–C; Supplemental Fig. S12D) and that TaNFYA-B1 overexpression also increased N and P uptake. It has been demonstrated that miR169/NFYA modules are important regulators of primary and lateral root development. For instance, expressing a miR169-resistant version of AtNFYA2 in Arabidopsis indirectly increased lateral root initiation and lateral root density but did not affect primary root growth (Sorin et al., 2014). However, the molecular mechanisms through which miR169/NFYA modules regulate root development are still not understood well. Our study shows that TaNFYA-B1 possibly acts on auxin biosynthesis to promote lateral root development. TAR2 functions in the indole-3-pyruvic acid biosynthetic pathway, the main route for the de novo synthesis of auxin in plants (Mashiguchi et al., 2011). In Arabidopsis, overexpressing AtTAR2 increases auxin accumulation in both shoots and roots and increases lateral root growth under both high-N and low-N conditions (Ma et al., 2014). The more abundant TaTAR2 transcripts in the roots of TaNFYA-B1-overexpressing plants (Fig. 5D) might favor auxin biosynthesis and, hence, lateral root development.
The up-regulated expression of nitrate and phosphate transporters in the TaNFYA-B1-overexpressing plants also might contribute to improved N and P uptake. Transporters of the NRT1 and NRT2 families mediate the uptake of nitrate (Xu et al., 2012), which is the main N source taken up by wheat. Thus, the elevated expression of TaNRT1.1 and TaNRT2.1 (Fig. 6, A and B) and the increased nitrate influx rate in roots (Fig. 6J) should facilitate N acquisition by the TaNFYA-B1-overexpressing plants. When the wheat plants prepared in 2 mm nitrate solution were used to measure nitrate flux in 0.2 mm nitrate solution by the SIET method, the wild type exhibited net nitrate efflux while the transgenic lines showed net nitrate influx at sites 3, 6, and 9 mm from the root tip (Fig. 6J). It has been reported that net nitrate efflux can occur when a plant grown in a high-nitrate solution is transferred to a low-nitrate solution (Zheng et al., 2013). The larger net nitrate influx of the transgenic lines was possibly due to the increased expression of TaNRT1.1 and TaNRT2.1 in the TaNFYA-B1 lines (Fig. 6, A and B). In line with the positive regulation of nitrate transporters by TaNFYA-B1, overexpression of miR169a down-regulated the expression of AtNRT1.1 and AtNRT2.1 (Zhao et al., 2011). To cope with P deficiency, plants increase the expression of PHT1 transporters in roots and thus increase the ability of their root systems to utilize P from soils (Zhang et al., 2014). Increasing the expression of PHT1 transporters through transgenic approaches has been shown to be successful in engineering crops with improved P uptake (Tian et al., 2012). As such, the increased mRNA levels of a number of PHT1 transporters (Fig. 6, C–I) in roots should account for the improved P uptake by the plants overexpressing TaNFYA-B1. It is already known that it is the NFYA subunit of the NF-Y complexes that binds to the CCAAT box (Laloum et al., 2013). Since the promoters of the investigated NRT and PHT1 transporters contain putative CCAAT boxes (Supplemental Fig. S13), it is worthy to check if TaNFYA-B1 directly binds to the promoters and thus activates the expression of these transporters.
In summary, we performed a genome-wide identification of miR169 and NF-Y transcription factors and evaluated their responses to N and P availability. TaNFYA-B1 was shown to be a valuable genetic resource/target for the engineering of wheat varieties with more efficient use of N and P and higher grain yields under different N and P availability conditions. The up-regulation of auxin biosynthetic genes and nutrient transporters by overexpressing TaNFYA-B1 suggested that TaNFYA-B1 may control a complex gene network. Further dissecting the gene network controlled by TaNFYA-B1 and other NF-Y members will facilitate our understanding of the roles of NF-Y transcription factors in mediating nutrient transport from soils to grains and in regulating root and aerial development and yield formation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The wheat (Triticum aestivum) varieties Xiaoyan 54 and Longchun 23 were used in this study. Xiaoyan 54 was used to isolate the sequences and evaluate the expression of the TaNF-Y genes, and Longchun 23 was used to generate the transgenic lines.
Hydroponic Culture
The Xiaoyan 54 plants used for the TaNF-Y gene expression analysis and the transgenic lines that were phenotyped at the seedling stage were grown hydroponically. Seven days after germination, wheat seedlings were grown for 20 d in plastic pots containing 1 L of nutrient solution; this solution was refreshed every 2 d. The hydroponic culture was carried out in a growth chamber with the following conditions: 20°C ± 1°C, 50% to 70% relative humidity, a photon fluence rate of 300 µmol photons m−2 s−1, and a 16-h-day/8-h-night cycle. To analyze the expression of the TaNF-Y genes in Xiaoyan 54 plants, three treatments (control, low N, and low P) were used. The nutrient solution of the control treatment was described previously (Ren et al., 2012). The N and P supply levels of the control, low-N, and low-P treatments were set as 2 mm N [Ca(NO3)2] + 0.2 mm P (KH2PO4), 0.2 mm N + 0.2 mm P, and 2 mm N + 0.02 mm P, respectively. CaCl2 and KCl were used to balance the calcium and potassium concentrations of the different treatments. The roots and shoots of three plants were collected separately for gene expression analysis. To analyze the phenotypes of the transgenic lines overexpressing TaNFYA-B1, seedlings of the wild-type Longchun 23 and T4 transgenic lines OE1 and OE2 were grown in the control nutrient solution. The roots were used to measure root morphology, the expression of nitrate and phosphate transporter genes, and net nitrate flux rates. The root morphological parameters were measured as described by Ren et al. (2012). Nitrate influx rate was measured as described by Zheng et al. (2013).
Field Experiment
The field experiment was conducted in the experimental station of the Institute of Genetics and Developmental Biology in Beijing. The experiment contained three treatments, which each had three replications. The control treatment had 18 g m−2 N in the form of urea with 12 g m−2 applied prior to sowing and 6 g m−2 applied at the stem elongation stage, and 13.5 g m−2 P (calcium superphosphate) applied prior to sowing. The low-N treatment had no N application but 13.5 g m−2 P. The low-P treatment had no P application but 18 g m−2 N. For each genotype in each replicate, 90 seeds were sown in three 1.5-m-long rows, and the rows were spaced 23 cm apart (87 seeds m−2). The plant density was thinned to 22 plants per row (63.8 plants m−2). The grain yield per plant, spike number per plant, and grain number of the primary spike were recorded on 30 representative plants for each sample group. The 1,000-grain weight was determined according to the dry weight of 500 dried grains. Total N concentrations in straw and grain were measured using a semiautomated Kjeldahl method (Tecator Kjeltec Auto 1030 Analyzer; Tecator). To measure total P concentration, the straw and grain samples were digested with concentrated H2SO4 and H2O2 and then the P concentrations of these digestions were determined using a Molybdate Blue colorimetric method (Murphy and Riley, 1962).
Gene Cloning and Phylogenetic Analysis
The sequences of the NF-Y genes from wheat, Aegilops tauschii, rice (Oryza sativa), Brachypodium distachyon, and Arabidopsis (Arabidopsis thaliana; Stephenson et al., 2007; Petroni et al., 2012; Jia et al., 2013) were used as queries to search the EMBL-European Bioinformatics Institute GenBank to identify the NF-Y genes of wheat, Triticum urartu, barley (Hordeum vulgare), maize (Zea mays), and B. distachyon (http://www.ebi.ac.uk). In addition to sequence mining from public databases, cDNA cloning was also employed to isolate NF-Y genes by using the primers listed in Supplemental Table S2. After removing redundancies, the NFYA, NFYB, and NFYC subunits from all of the plants investigated were aligned using ClustalX 2.0 (Thompson et al., 2002). We used the neighbor-joining method to generate a phylogenetic tree, and the phylogenetic tree was drawn using MEGA 5.0 (Tamura et al., 2011).
To identify microRNA169 members, Xiaoyan 54 seedlings were grown for 7 d in nutrient solutions that contained 2 mm N + 0.2 mm P (control), 0 mm N + 0.2 mm P (low N), and 2 mm N + 0 mm P (low P). Total RNA was isolated from roots and shoots separately using TRIzol reagent (Thermo). The total samples were sent to the Beijing Genomics Institute for small RNA processing and sequencing. The small RNAs with lengths between 18 and 30 nucleotides were enriched and then reverse transcribed to cDNA as described previously (Liang et al., 2014). Total cDNA was sequenced by Illumina Genome Analyzer IIx. Raw reads were filtered to get clean reads. During this process, the following reads were removed: all low-quality reads including 3′ adapter reads and 5′ adapter contaminants, reads with poly(A), and reads larger than 30 nucleotides and smaller than 18 nucleotides.
Quantitative Real-Time PCR and MicroRNA Northern Blot
Total RNA from plant tissues was extracted with TRIzol reagent (Thermo). The first-strand cDNA was synthesized from 2 μg of DNase I-treated total RNA using murine leukemia virus reverse transcriptase (Promega). Quantitative real-time RT-PCR analysis was performed with a LightCycler 480 engine (Roche) using the LightCycler480 SYBR Green І Master Mix (Roche). ACTIN2 mRNA was used as an internal control. The primers for real-time RT-PCR are detailed in Supplemental Table S2.
For microRNA gel-blot analysis, 20 μg of RNA was separated by denaturing (7 m urea containing) 15% PAGE and then blotted onto a nylon membrane (Hybond NX; GE Healthcare); the RNA blot was cross-linked by exposing to UV light for 1 min. miR169b was used as the probe and was prepared by end labeling with [γ-32P]ATP using T4 polynucleotide kinase (Thermo Scientific). RNA gel blots were hybridized with the miR169b probe. Nonsaturated signals were quantified on a Molecular Dynamics Storm 840 PhosphorImager (Molecular Dynamics).
Vector Construction and Transformation
In order to generate the vector for wheat transformation, the maize ubiquitin promoter (pUbi) was subcloned into the PstI site of the pAHC25 vector (Christensen et al., 1992), resulting in a vector named pUbi-pAHC25. Then, the TaNFYA-B1 cDNA was inserted into the BamHI/KpnI site of pUbi-pAHC25, resulting in the construct pUbi::TaNFYA-B1-pAHC25. The construct was transformed into immature embryos of wheat variety Longchun 23 using the method described by Wang et al. (2013).
Measurement of Net Nitrate Flux Using a Noninvasive Microtest Technique
Wild-type and TaNFYA-B1 transgenic line plants grown in the control solution for 20 d were used to measure net nitrate fluxes. For measuring net nitrate fluxes, the roots were transferred to measuring solution (0.2 mm KNO3, 0.1 mm CaCl2, and 0.3 mm MES, pH 6) and allowed to balance for 10 min, and then the net nitrate fluxes were measured by using a SIET (systemBIO-003A; Younger USA Science and Technology) at Xuyue Science and Technology in Beijing, as described (Luo et al., 2013; Zheng et al., 2013).
Statistical Analysis
One-way ANOVA was performed with SPSS11.5 for Windows (SPSS).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TaNRT1.1, AY587265; TaNRT2.1, AF288688; TaPHT1.1-A1, KJ170110; TaPHT1.1-D1, KJ170111; TaPHT1.2-A1, KJ170112; TaPHT1.2-B1, KJ170113; TaPHT1.2-D1, KJ170114; TaPHT1.3-D1, KJ170115; TaPHT1.4-D1, KJ170116; TaPHT1.4-D2, KJ170117; TaPHT1.5-A1, KJ170118; TaPHT1.5-B1, KJ170119; TaPHT1.5-D1, KJ170120; TaPHT1.6-B1, KJ170121; TaPHT1.6-D1, KJ170122; TaPHT1.7-D1, KJ170123; TaPHT1.8-B1, KJ170124; TaPHT1.8-D1, KJ170125; TaPHT1.9-D1, KJ170126; TaPHT1.10-D1, KJ170127; TaPHT1.10-D2, KJ170128; TaTAR2-A1, KM078759; TaTAR2-B1, KM078760; and TaTAR2-D1, KM078761. Sequence data of the NF-Y genes in this study can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of NFYAs.
Supplemental Figure S2. Phylogenetic analysis of NFYBs.
Supplemental Figure S3. Phylogenetic analysis of NFYCs.
Supplemental Figure S4. The responses of TaNFYB genes to N and P starvation.
Supplemental Figure S5. The responses of TaNFYC genes to N and P starvation.
Supplemental Figure S6. The relative expression level of tae-miR169 under different N and P supply levels.
Supplemental Figure S7. The regulation of tae-miR169f on TaNFYA-B1 and the cleavage site mapping.
Supplemental Figure S8. The gene structures of TaNFYA1 genes.
Supplemental Figure S9. Chromosome localizations of TaNFYA1.
Supplemental Figure S10. Subcellular localization and transcription activation of TaNFYA-B1.
Supplemental Figure S11. The N and P utilization efficiency of the wild type and transgenic lines grown in the field experiment.
Supplemental Figure S12. The performance of the wild-type and transgenic lines grown in a pot experiment.
Supplemental Figure S13. The putative CCAAT-box in the promoter of nitrate and phosphate transporters.
Supplemental Figure S14. Theoretical grain yield, spike number, and the N and P uptake per square meter of the wild-type and transgenic lines grown in the field experiment.
Supplemental Table S1. The identified NF-Y genes in wheat.
Supplemental Table S2. The primers used in this study.
Supplementary Material
Acknowledgments
We thank the members of Caixia Gao’s laboratory (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for developing the wheat transgenic lines.
Glossary
- N
nitrogen
- P
phosphorus
- Pi
inorganic phosphate
- cDNA
complementary DNA
- RT
reverse transcription
- SIET
scanning ion-selective electrode technique
Footnotes
This work was supported by the Ministry of Science and Technology of China (grant nos. 2011CB100304 and 2014CB138100), the Chinese Ministry of Agriculture (grant no. 2014ZX08002–005), and the European Community under the Seventh Framework Programme for Research, Technological Development, and Demonstration Activities Integrated Project (grant no. NUE–CROPS FP7–CP–IP 222645).
References
- Alvarez JM, Vidal EA, Gutiérrez RA (2012) Integration of local and systemic signaling pathways for plant N responses. Curr Opin Plant Biol 15: 185–191 [DOI] [PubMed] [Google Scholar]
- An DG, Su JY, Liu QY, Zhu YG, Tong YP, Li JM, Jing RL, Li B, Li ZS (2006) Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant Soil 284: 73–84 [Google Scholar]
- Bari R, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JH, Lu X, Haefele SM, Gamuyao R, Ismail A, Wissuwa M, Heuer S (2010) Development and application of gene-based markers for the major rice QTL Phosphorus uptake 1. Theor Appl Genet 120: 1073–1086 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 [DOI] [PubMed] [Google Scholar]
- Combier JP, de Billy F, Gamas P, Niebel A, Rivas S (2008) Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev 22: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier F, Faure S, Dubreuil P, Heumez E, Beauchêne K, Lafarge S, Praud S, Le Gouis J (2013) A multi-environmental study of recent breeding progress on nitrogen use efficiency in wheat (Triticum aestivum L.). Theor Appl Genet 126: 3035–3048 [DOI] [PubMed] [Google Scholar]
- Dang VD, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B (1996) The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J Bacteriol 178: 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehdaie B, Merhaut DJ, Ahmadian S, Hoops AC, Khuong T, Layne AP, Waines JG (2010) Root system size influences water-nutrient uptake and nitrate leaching potential in wheat. J Agron Crop Sci 196: 455–466 [Google Scholar]
- Fischer JJ, Beatty PH, Good AG, Muench DG (2013) Manipulation of microRNA expression to improve nitrogen use efficiency. Plant Sci 210: 70–81 [DOI] [PubMed] [Google Scholar]
- Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327: 812–818 [DOI] [PubMed] [Google Scholar]
- Gregory PJ, George TS (2011) Feeding nine billion: the challenge to sustainable crop production. J Exp Bot 62: 5233–5239 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R (2001) Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 264: 173–185 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R (2002) Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283: 41–48 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA. (2012) Systems biology for enhanced plant nitrogen nutrition. Science 336: 1673–1675 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ (2008) The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723 [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt BF III (2010) NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 63: 379–391 [DOI] [PubMed] [Google Scholar]
- Kurai T, Wakayama M, Abiko T, Yanagisawa S, Aoki N, Ohsugi R (2011) Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol J 9: 826–837 [DOI] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18: 157–166 [DOI] [PubMed] [Google Scholar]
- Leyva-González MA, Ibarra-Laclette E, Cruz-Ramírez A, Herrera-Estrella L (2012) Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7: e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Distelfeld A, Comis A, Dubcovsky J (2011) Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J 67: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Liu X, Sun Y, Yiu SM, Lim BL (2014) Global small RNA analysis in fast-growing Arabidopsis thaliana with elevated concentrations of ATP and sugars. BMC Genomics 15: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Yu D (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 7: e48951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Fillery IR, Palta JA (2004) Early vigorous growth is a major factor influencing nitrogen uptake in wheat. Funct Plant Biol 31: 121–129 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, et al. (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Y, Han W, Tang A, Shen J, Cui Z, Vitousek P, Erisman JW, Goulding K, Christie P, et al. (2013) Enhanced nitrogen deposition over China. Nature 494: 459–462 [DOI] [PubMed] [Google Scholar]
- Lundmark M, Kørner CJ, Nielsen TH (2010) Global analysis of microRNA in Arabidopsis in response to phosphate starvation as studied by locked nucleic acid-based microarrays. Physiol Plant 140: 57–68 [DOI] [PubMed] [Google Scholar]
- Luo J, Qin J, He F, Li H, Liu T, Polle A, Peng C, Luo ZB (2013) Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis. Planta 237: 919–931 [DOI] [PubMed] [Google Scholar]
- Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y (2014) Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J 78: 70–79 [DOI] [PubMed] [Google Scholar]
- Manske GGB, Ortiz-Monasterio JI, Van Ginkel M, Gonzalez RM, Rajaram S, Molina E, Vlek PLG (2000) Traits associated with improved P-uptake efficiency in CIMMYT’s semidwarf spring bread wheat grown on an acid Andisol in Mexico. Plant Soil 221: 189–204 [Google Scholar]
- Mantovani R. (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CH, Beatty PH, Good AG (2012) Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol J 10: 1011–1025 [DOI] [PubMed] [Google Scholar]
- Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27: 31–36 [Google Scholar]
- Nakashima A, Ueno M, Ushimaru T, Uritani M (2002) Involvement of a CCAAT-binding complex in the expression of a nitrogen-starvation-specific gene, isp6+, in Schizosaccharomyces pombe. Biosci Biotechnol Biochem 66: 2224–2227 [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol 82: 113–129 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF III, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YZ, He X, Liu DC, Li JJ, Zhao XQ, Li B, Tong YP, Zhang AM, Li ZS (2012) Major quantitative trait loci for seminal root morphology of wheat seedlings. Mol Breed 30: 139–148 [Google Scholar]
- Riego L, Avendaño A, DeLuna A, Rodríguez E, González A (2002) GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem Biophys Res Commun 293: 79–85 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Zhong B, Liu X, Jin R, Chan Z (2014) AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol 203: 554–567 [DOI] [PubMed] [Google Scholar]
- Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Brière C, Njo MF, Beeckman T, Crespi M, Hartmann C (2014) A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol 202: 1197–1211 [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP (2007) Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol 65: 77–92 [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP (2010) TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Funct Integr Genomics 10: 265–276 [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP (2011) TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct Integr Genomics 11: 327–340 [DOI] [PubMed] [Google Scholar]
- Sylvester-Bradley R, Kindred DR (2009) Analysing nitrogen responses of cereals to prioritize routes to the improvement of nitrogen use efficiency. J Exp Bot 60: 1939–1951 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Tian J, Wang X, Tong Y, Chen X, Liao H (2012) Bioengineering and management for efficient phosphorus utilization in crops and pastures. Curr Opin Biotechnol 23: 866–871 [DOI] [PubMed] [Google Scholar]
- Tilman D, Fargione J, Wolff B, D’Antonio C, Dobson A, Howarth R, Schindler D, Schlesinger WH, Simberloff D, Swackhamer D (2001) Forecasting agriculturally driven global environmental change. Science 292: 281–284 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, et al. (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot (Lond) 111: 1139–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Liu Q, Chen L, Kuang J, Walk T, Wang J, Liao H (2013) Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhong S, Li X, Li W, Rothstein SJ, Zhang S, Bi Y, Xie C (2011) Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 6: e28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, et al. (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4: 319–330 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM (2010) A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22: 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng HQ, Wang GP, Hu XY, Wang HZ, Du LQ, Zhu YY (2014) Role of microRNAs in plant responses to nutrient stress. Plant Soil 374: 1005–1021 [Google Scholar]
- Zhang Z, Liao H, Lucas WJ (2014) Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol 56: 192–220 [DOI] [PubMed] [Google Scholar]
- Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu JK, Zhang F, Li WX (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Han X, An Y, Guo H, Xia X, Yin W (2013) The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ 36: 1328–1337 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.