Lipid reassembly is a key step in the transfer of cytosolic lipid precursors to the plastid.
Abstract
In plants, fatty acids are synthesized within the plastid and need to be distributed to the different sites of lipid biosynthesis within the cell. Free fatty acids released from the plastid need to be converted to their corresponding coenzyme A thioesters to become metabolically available. This activation is mediated by long-chain acyl-coenzyme A synthetases (LACSs), which are encoded by a family of nine genes in Arabidopsis (Arabidopsis thaliana). So far, it has remained unclear which of the individual LACS activities are involved in making plastid-derived fatty acids available to cytoplasmic glycerolipid biosynthesis. Because of its unique localization at the outer envelope of plastids, LACS9 was regarded as a candidate for linking plastidial fatty export and cytoplasmic use. However, data presented in this study show that LACS9 is involved in fatty acid import into the plastid. The analyses of mutant lines revealed strongly overlapping functions of LACS4 and LACS9 in lipid trafficking from the endoplasmic reticulum to the plastid. In vivo labeling experiments with lacs4 lacs9 double mutants suggest strongly reduced synthesis of endoplasmic reticulum-derived lipid precursors, which are required for the biosynthesis of glycolipids in the plastids. In conjunction with this defect, double-mutant plants accumulate significant amounts of linoleic acid in leaf tissue.
Two discrete but intimately connected pathways are involved in plant glycerolipid biosynthesis (Roughan et al., 1980). Both pathways follow exactly the same scheme of synthesis within the plastid and at the endoplasmic reticulum (ER) to assemble phosphatidic acid (PA) by two consecutive acylation reactions of glycerol-3-phosphate. Essential substrates for both pathways are fatty acids that are synthesized exclusively in plastids. De novo synthesized fatty acids can feed directly into the so-called prokaryotic lipid synthesis pathway localized within the plastid to produce phosphatidylglycerol (PG), the so-called C16:3 plants (e.g., Arabidopsis [Arabidopsis thaliana]), and also, other thylakoid lipids, like sulfoquinovosyldiacylglycerol, monogalactosyldiacylglycerol (MGDG), and digalactosyldiacylglycerol (DGDG; Heinz and Roughan, 1983). In addition, plastid-derived fatty acids are also substrates for eukaryotic lipid biosynthesis at the ER to produce important membrane lipid precursors, like PA and diacylglycerol (DAG). The main products of the lipid biosynthesis pathway in the ER are, however, phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol. Recent studies revealed an additional mechanism to incorporate plastid-derived fatty acids at the ER by acyl editing of PC (Bates et al., 2007). In the proposed model (also designated as the Lands cycle; Lands, 1958), PC is continuously converted to lyso-PC, which becomes reacylated by newly exported fatty acids to generate PC again. However, irrespective of the route taken to attach the fatty acids to the glycerol backbone, the interconnection between plastidial and cytoplasmic lipid metabolism is, in most plant species, further complicated by the fact that the eukaryotic pathway is not only generating lipids for all extraplastidial compartments but also, synthesizing lipid precursors, which are delivered back to the plastid to become thylakoid lipids. Consequently, plastidial membrane lipids represent a mixture of molecules partially synthesized within the plastid and partially assembled at the ER. The contribution of ER and plastidial lipid synthesis to the overall mixture of thylakoid lipids differs strongly between different plant species, but in Arabidopsis, both sites of synthesis are responsible for approximately equal amounts of chloroplast lipids (Browse et al., 1986). Subtle biochemical differences reveal the site of synthesis of a specific lipid molecule. Because of different substrate specificities of the acyltransferases located at the ER and in the plastid, the resulting lipid molecules can be distinguished based on the fatty acids attached to the sn-2 position of the glycerol backbone. Whereas the lysophosphatidyl acyltransferase at the ER is highly specific for 18-carbon fatty acids, its plastidial homolog incorporates exclusively 16-carbon fatty acids into the sn-2 position.
Another important difference between plastidial and cytoplasmic lipid metabolism is defined by the nature of the fatty acid substrate. In both cases, fatty acid thioesters are used; however, within the plastid, the fatty acids are provided as acyl-acyl carrier proteins (acyl-ACPs), whereas in the cytoplasm, acyl-CoAs are the established substrates. Acyl-ACP produced by plastidial fatty acid synthase can be used directly by enzymes of the plastidial lipid biosynthesis pathway, but fatty acids need to be exported and converted to acyl-CoA by long-chain acyl-CoA synthetases (LACS) to become substrate for the pathway operating at the ER. The precise mechanism of the fatty acid transport through the plastidial membrane is still unknown; however, the findings of acyl-ACP thioesterase activity in the stroma of plastids (Ohlrogge et al., 1978, 1979) and LACS activity at the outer envelope (Andrews and Keegstra, 1983; Block et al., 1983) suggested both enzymes to be involved in the export of fatty acids from plastids. This model was challenged by the identification of LACS9 as the major plastidial LACS isoform in Arabidopsis and the finding that its inactivation did not result in any substantial changes in lipid composition (Schnurr et al., 2002). Because LACS activity is encoded in Arabidopsis by a small gene family comprising nine genes (Shockey et al., 2002), there must be other LACS isoforms involved in providing acyl-CoA substrate to cytoplasmic lipid metabolism. Surprisingly, none of the lacs mutant lines analyzed so far, including single mutants of all members of the enzyme family, showed pronounced effects on glycerolipid metabolism. The data seem to suggest a network of overlapping LACS activities concealing the effects of individual members of the enzyme family. It may also indicate that mutual interactions between the different LACS enzymes are still poorly understood. To elucidate such interactions and identify those LACS activities contributing to glycerolipid metabolism, we established a comprehensive mutant collection comprising all possible double-mutant lines based on nine members of the LACS gene family. The individual mutants of this collection were screened for visual phenotypes potentially associated with modifications in lipid biosynthesis.
Here, we show overlapping functions of LACS4 and LACS9 in Arabidopsis. The combined inactivation of both proteins results in severe morphological phenotypes of the adult plant that are tightly linked to changes in the fatty acid metabolism. The results suggest that both LACS activities are involved in fatty acid channeling and lipid processing. But instead of contributing to fatty acid export from the plastid, both proteins were found to be involved in the process of retrograde lipid flux from the ER to the plastid.
RESULTS
Isolation of lacs4 lacs9 Double-Mutant Lines
This study focused on the identification of LACS activities involved in the biosynthesis of glycerolipids. A complete collection of lacs double mutants was generated by systematic crossing of transfer DNA (T-DNA) insertion lines identified in the T-DNA Express database (http://signal.salk.edu/cgi-bin/tdnaexpress) with insertion lines for all members of the LACS family. Any substantial impairment of glycerolipid biosynthesis was expected to impact plant development, and therefore, the mutant collection was screened for plants with obvious morphological defects. We identified lacs4 lacs9 double-mutant plants by their striking phenotypes (Fig. 1). The plants developed slowly compared with the wild type and were significantly reduced in size (Fig. 1D). The time of germination was less synchronized in the double mutant compared with the wild type, but on average, germination of the mutant was delayed by 24 h. Seedling establishment and further development were both delayed in lacs4 lacs9 double-mutant plants. During early stages of development, the limited plant growth was the only abnormality observed. However, after about 4 weeks, the morphology of the mutant plants became easily distinguishable from the wild type. The leaves became curly, and the petioles of new leaves were significantly reduced in length (Fig. 1, B and C). The leaf area of the whole rosette 6 weeks after germination was about 40% smaller than in the wild type (Supplemental Fig. S1). After transition to reproductive growth, developing stems remained thin, and the overall plant size was strongly reduced. Interestingly, these morphological abnormalities could be observed only under long-day conditions (16-h-light/8-h-dark cycle) but were almost absent when plants were grown under a short-day light regime (8-h-light/16-h-dark cycle; Fig. 1, A and B). Additional tests with varying growth conditions showed that the development of symptoms was directly correlated with day length rather than light intensity. The single-knockout plants of lacs4 and lacs9 were indistinguishable from the wild type no matter which light regime was applied. Therefore, we concluded that only the combined inactivation of LACS4 and LACS9 led to the severe growth phenotype under long-day conditions.
Figure 1.
Phenotype of the lacs4 lacs9 double mutants. A and B, Six-week-old wild type (WT) and mutant plants grown under either short- (A) or long-day (B) conditions. C, Close-up views of rosette leaves from 6-week-old plants of the wild type and both lacs4 lacs9 double-mutant lines grown under long-day conditions. Bars = 0.3 cm. D, Morphology of 8-week-old plants of the wild type and both lacs4 lacs9 double-mutant lines grown under long-day conditions. Bars = 2 cm.
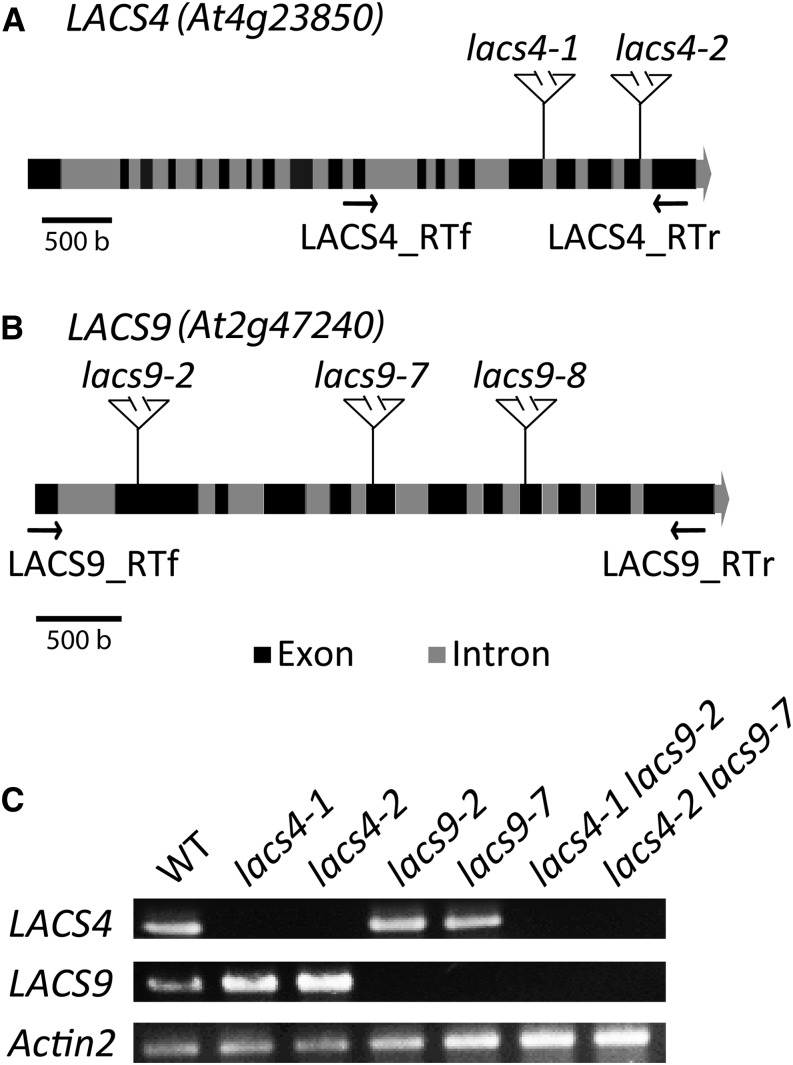
To address the problem of potentially unrecognized second-site mutations in the T-DNA lines used, two independent alleles of lacs4 and three alleles of lacs9 were used to produce altogether six lacs4 lacs9 double-mutant lines (Fig. 2, A and B). All double-mutant lines, designated as lacs4-1 lacs9-2, lacs4-1 lacs9-7, lacs4-1 lacs9-8, lacs4-2 lacs9-2, lacs4-2 lacs9-7, and lacs4-2 lacs9-8, showed similar phenotypes as described above that distinguished them clearly from the parental lines. For all of the following experiments, the two double-mutant lines lacs4-1 lacs9-7 and lacs4-2 lacs9-2 were used. To ensure that only null alleles were included in the analyses, total RNA was isolated from leaves of the different plant lines and used for reverse transcription (RT)-PCR analysis (Fig. 2C). Pairs of primers flanking the different T-DNA insertion sites were used, resulting in strong PCR products from wild-type RNA. In RNA of the single mutants of LACS4 and LACS9, the corresponding LACS transcripts were not detected, indicating that all lines investigated contained null alleles. In a previous report, lacs9-7 was shown to contain significant transcript levels (Zhao et al., 2010). Using the pair of primers stated in the study, we were able to obtain the corresponding PCR product from lacs9-7 RNA (Supplemental Fig. S2). Because this pair of primers is not flanking the T-DNA insertion site, the PCR product might represent a truncated transcript not encoding functional LACS activity.
Figure 2.
Identification of lacs4 and lacs9 mutant lines. A and B, Schematic gene structures of LACS4 (A) and LACS9 (B) depicting exons in dark gray and introns in light gray. For both genes, the independent T-DNA insertion sites are indicated. Arrows indicate positions of primers used for RT-PCR. C, RT-PCR analyses showing the absence of full-length transcripts of LACS4 in both lacs4 mutant alleles and LACS9 in both lacs9 mutant alleles. The two lacs4 lacs9 double-mutant lines are deficient in both transcripts. WT, Wild type.
Impaired Oil Accumulation in Seeds of lacs4 lacs9
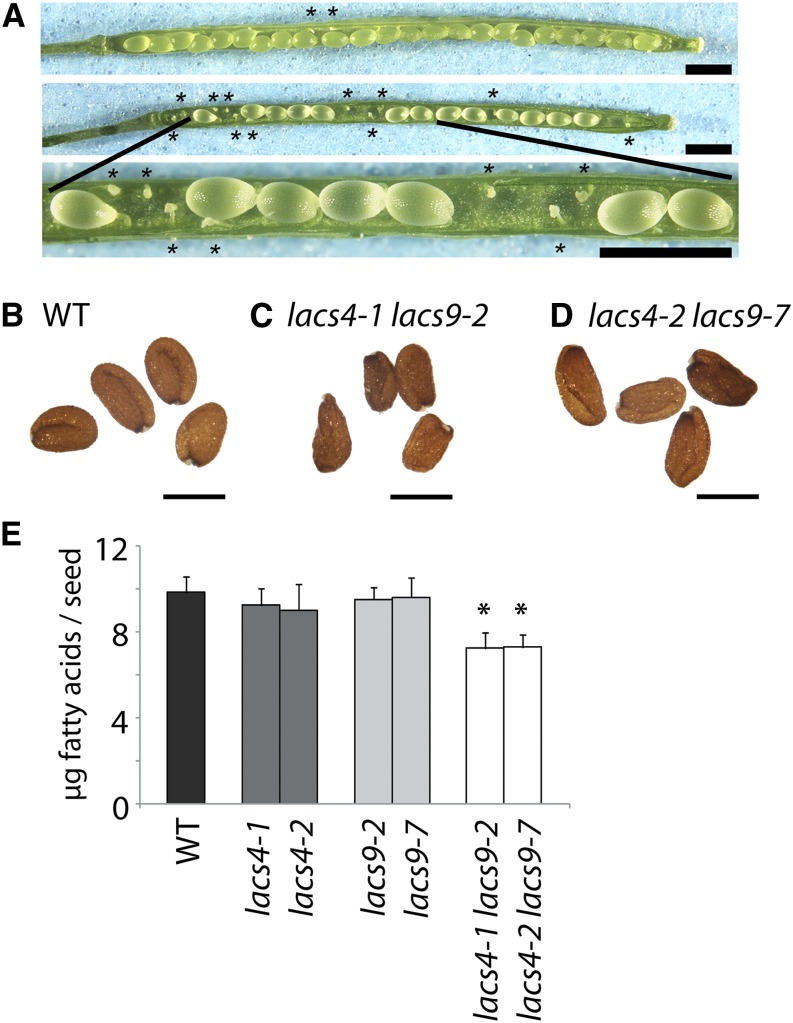
The abnormal vegetative growth phenotype of the adult plant was not the only anomaly in lacs4 lacs9. Closer inspection of the seeds revealed that gametogenesis as well as seed development were impaired. The siliques of the double mutants contained elevated levels of undeveloped ovules randomly dispersed inside the silique. Obviously, external factors influenced gametogenesis of lacs4 lacs9, because the numbers of undeveloped ovules in individual siliques were highly variable (Fig. 3A). On the same plant, the ratio of undeveloped ovules to developed seeds per carpel ranged from 0:30 to 22:9. On average, 6.7 undeveloped ovules and 21.7 developed seeds were observed per carpel (n = 662). The developed seeds of the double mutants showed predominantly aberrant appearance (Fig. 3, B–D). Also, in this case, external factors influenced the seed phenotype, because varying degrees of deformations were observed within a batch of seeds from a single plant. The wrinkled appearance was accompanied by a reduction of lipid content by about 26% (unless otherwise stated, the given values always refer to the average of both mutant alleles compared with the wild type; Fig. 3E). More detailed analysis revealed that seeds of lacs4 lacs9 contained reduced levels of 18:2 (−22%) but increased levels of 18:3 (+37%; Supplemental Fig. S3). These data are remarkable, because a reciprocal shift of the 18:2 to 18:3 ratio was observed between the wild type and lacs4 lacs9 when membrane lipids of leaves were analyzed (see below). All other fatty acids were not significantly different from the wild type, and as with the other phenotypes reported for lacs4 lacs9, the seed phenotype was not observed in either of the single-mutant parental lines.
Figure 3.
Morphology and lipid phenotype of seeds. A, Seed development in two randomly chosen siliques of lacs4-1 lacs9-2 showing highly variable numbers of abortive ovules (indicated by asterisks). Bars = 1 mm. B to D, Photographs of randomly chosen seeds of the wild type (B) and both lacs4 lacs9 double-mutant lines (C and D). Bars = 500 µm. E, Total fatty acid content of mature seeds of the wild type and all mutant lines under investigation. *, Significantly different values between the wild type (WT) and the respective mutant line (P ≤ 0.05).
LACS4 and LACS9 Are Both Expressed in Leaves but Differ in Their Subcellular Localization
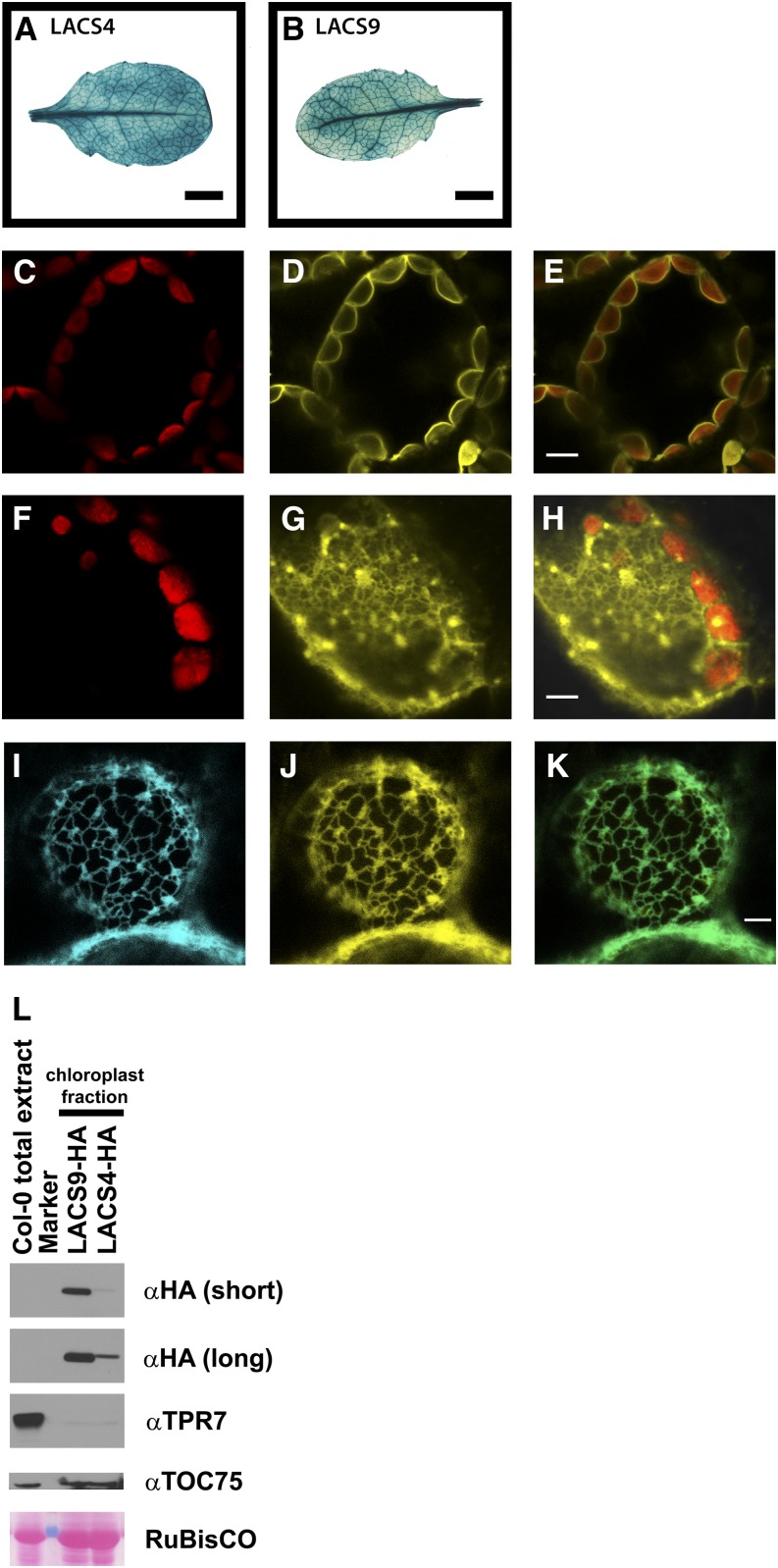
Phenotypic and genetic analyses suggested partially overlapping functions of LACS4 and LACS9. To investigate if this redundancy is reflected by overlapping expression patterns, promoter:GUS analyses were performed. For LACS4, a 2,268-bp genomic fragment containing the 5′ untranslated region and part of the coding region was fused to the β-glucuronidase reporter gene GUS. For LACS9, a genomic fragment of 1,108 bp located upstream of the start codon was used to establish a corresponding construct, and both plasmids were transformed into Arabidopsis wild type plants. Histochemical examination of different independent transgenic lines revealed strong leaf expression for LACS4 and LACS9 (Fig. 4, A and B), indicating that the encoded proteins might be able to complement each other. Interestingly, the results suggested that the expression in leaf veins is higher than in the surrounding tissue.
Figure 4.
LACS4 and LACS9 are expressed in leaf tissue but show different subcellular localization. A and B, Analysis of gene expression in leaves using promoter:GUS reporter constructs for LACS4 (A) and LACS9 (B). Bars = 0.5 cm. C to E, Subcellular localization of LACS9 in leaf mesophyll cells of stable transgenic Arabidopsis plants was investigated by confocal fluorescence microscopy. Images show chloroplast autofluorescence (C), the signal of LACS9 fused to enhanced yellow fluorescent protein (EYFP) under control of the endogenous promoter (D), and the merged image (E). Bar = 5 µm. F to H, Subcellular localization of LACS4 in leaf mesophyll cells of stable transgenic Arabidopsis plants. Confocal images show chloroplast autofluorescence (F), the signal of LACS4 fused to EYFP under control of the endogenous promoter (G), and the merged image (H). Bar = 5 µm. I to K, Colocalization of LACS4 with an ER marker in leaf mesophyll cells upon transient expression in N. benthamiana. Confocal images show the signal of an ER marker fused to enhanced cyan fluorescent protein (ECFP; I), the signal of LACS4 fused to EYFP under control of the 35S promoter (J), and the merged image (K). Bar = 5 µm. L, Immunoblot analysis of LACS proteins in chloroplast fractions of transgenic plants expressing human influenza hemagglutinin (HA)-tagged LACS4 or LACS9 under control of the respective native promoter. Upon short exposure, only LACS9-HA is detected in chloroplast preparations (αHA [short]). Small amounts of HA-tagged LACS4 in purified chloroplasts can only be detected after longer exposure time (αHA [long]). TOC75 (for translocon at the outer envelope membrane of chloroplasts, 75 kD) was used as the chloroplast outer envelope marker, and tetratricopeptide repeat protein7 (TPR7) was used as an ER marker. Total leaf extract of wild-type plants (Col-0 total extract) was used as the positive control for TPR7 and TOC75 immunoblots and the negative control for the HA immunoblot. Ponceau S staining of Rubisco was used as the loading control.
To extend the analyses of overlapping functions, the subcellular localization of both proteins was investigated. For LACS9, localization to the outer envelope of the plastid has been established (Schnurr et al., 2002), but the localization of LACS4 has not been elucidated. To visualize both proteins within the cell, fluorescent fusion proteins were constructed. For this approach, genomic fragments, including the respective promoter region and the complete intron/exon structures of LACS4 and LACS9, were fused in frame to the sequence encoding an EYFP, and the constructs were transformed into wild type plants. Transgenic lines were analyzed by confocal laser-scanning fluorescence microscopy. As shown in Fig. 4, C–E, the endogenous expression levels of LACS9 in Arabidopsis resulted in fluorescence signals decorating the surface of plastids. The data confirmed the localization of LACS9 at the plastidial envelope of leaf mesophyll cells. The same analysis of EYFP-tagged LACS4 resulted in reticular structures with some patch-like areas as often observed for ER-localized proteins (Fig. 4, F–H). To confirm the localization of LACS4 at the ER, the EYFP-LACS4 fusion protein was transiently coexpressed with an ECFP-labeled ER marker (Nelson et al., 2007) in leaves of Nicotiana benthamiana. Microscopic analysis revealed virtually complete colocalization of EYFP-LACS4 with the ER marker (Fig. 4, I and K). No obvious LACS4 fluorescence signal was observed in association with plastids. The results suggest discrete subcellular localization of LACS9 and LACS4 at the plastidial envelope and the ER, respectively.
To investigate the localization of both LACS proteins by an alternative method, purified plastids were analyzed by protein immunoblotting. Independent transgenic Arabidopsis lines were generated expressing either C-terminally HA-tagged LACS4 or HA-tagged LACS9 proteins under control of the respective endogenous promoters. The purity of the chloroplast fractions was verified by detection of the chloroplast outer envelope marker TOC75 (Tranel et al., 1995) and a very faint immunoblot signal for the ER membrane marker TPR7 detectable only upon extended exposure times (Schweiger et al., 2012). In samples of isolated chloroplasts, HA-tagged LACS9 was clearly detected (Fig. 4L), whereas small amounts of HA-tagged LACS4 became detectable only after long exposure times. For LACS9, the result is in agreement with the findings obtained by fluorescence microscopy. For LACS4, the immunoblot might be able to trace small protein amounts associated with plastids that are not detectable by fluorescence microscopy. Alternatively, this might be attributed to low amounts of ER that copurify with chloroplasts, because a very faint signal for the ER marker protein TPR7 was also observed in chloroplast fractions of transgenic LACS4-HA and LACS9-HA plants.
Membrane Lipids of lacs4 lacs9 Exhibit Only Moderate Anomalies in Their Fatty Acid Profiles Compared with the Wild Type
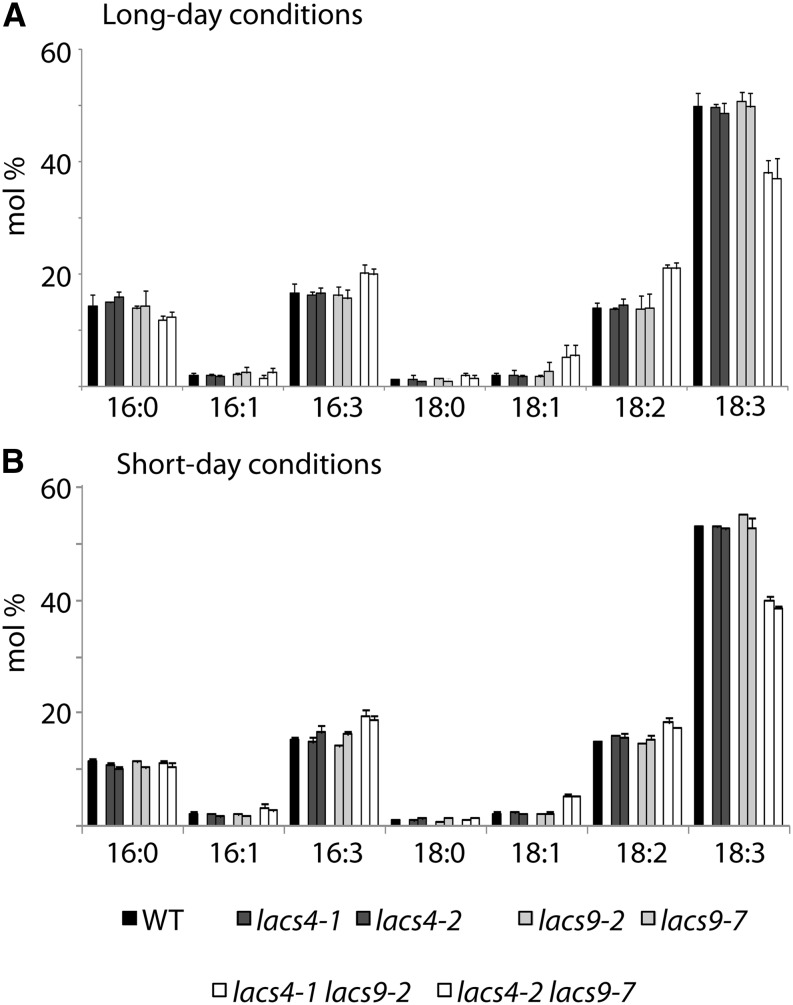
To elucidate the underlying biochemical aspects of the observed visual phenotypes, a detailed analysis of lipids was initiated. The lipid profiles of the double-mutant lines and the corresponding lacs4 and lacs9 single knockouts were analyzed. As shown in Supplemental Figure S4, no statistically significant differences between the lipid class compositions of the wild type and the mutant plants could be detected. The relative amounts of PC, PE, and PG as well as those of the galactolipids MGDG and DGDG were comparable with the wild type in all cases, no matter if the plants were grown under short- or long-day conditions (Supplemental Fig. S4, A and B). To investigate the impact of the combined inactivation of LACS4 and LACS9 on the lipid metabolism in more detail, the fatty acid profiles of the mutants were analyzed. Lipid extracts of mature leaves from 5-week-old plants were used. The single mutants of lacs4 and lacs9 showed little to no change compared with the wild type (Fig. 5). In contrast, the fatty acid profiles of both lacs4 lacs9 double mutants were considerably different from the wild type, showing alterations for 18:1 (+230%), 18:2 (+65%), and 18:3 (−20%) in plant tissue grown under long-day conditions (Fig. 5A). Similar variations have been observed in leaf material grown under a short-day light regime as well (Fig. 5B), suggesting that these differences are unlikely to cause the observed morphological anomalies. Surprisingly, changes in the fatty acid profiles closely resemble those reported for the mutant of the omega-3 fatty acid desaturase3 (fad3; Browse et al., 1993). There is no established direct metabolic link between the activities of LACS and desaturases. To test if compromised acyl-CoA pools in the lacs4 lacs9 double mutant might indirectly impact FAD3 or FAD7, real-time PCR gene expression analyses were performed. The results showed no significant differences between the transcript levels of FAD3 or FAD7 in the wild type and the lacs4 lacs9 double mutant (Supplemental Fig. S5).
Figure 5.
Alterations of leaf fatty acid composition in lacs4 lacs9 double mutants are not affected by light regime. The fatty acid profiles of leaf total lipids of the wild type (WT) and all mutants under investigation were analyzed from plants grown under long- (A) or short-day (B) conditions. Each value represents the mean of three independent replicates, with error bars indicating sd.
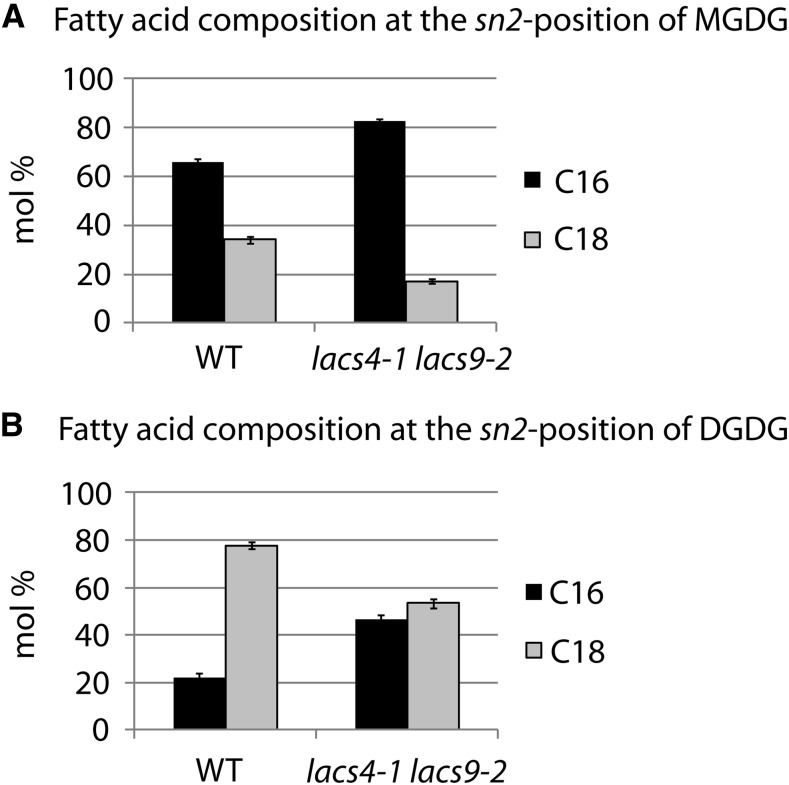
To elucidate the origin of the changes in the total fatty acid composition of the lacs4 lacs9 double mutants, individual lipid classes were analyzed (Supplemental Fig. S6). The phospholipids PC and PE showed a reduction in 18:3 (−28% and −21%, respectively) and a strong increase in 18:2 (+40% and +40%, respectively; Supplemental Fig. S6, A and B). All other fatty acids were not significantly different from the wild type. In PG, a reduction of 18:3 (−26%) was accompanied by elevated levels of 18:0 (+138%; Supplemental Fig. S6C). All of these changes showed a similar tendency as reported for the fad3 mutant (Browse et al., 1993), although the changes are much more moderate in lacs4 lacs9. In galactolipids, however, such similarities to fad3 were not detected. MGDG and DGDG of the double mutants both showed reduced levels of 18:3 (−10% and −18%, respectively), which were compensated for in MGDG by elevated levels of 16:3 (+17%) and in DGDG by higher concentrations of 16:0 and 18:2 (+27% and +51%, respectively; Supplemental Fig. S6, D and E). Most of the changes described were also observed to a lesser degree in plants grown under short-day conditions (Supplemental Fig. S7, A–E) and are, therefore, unlikely to cause the morphological phenotypes of the lacs4 lacs9 double mutants directly. However, the evaluation of the complete set of changes in the fatty acids profiles of phospholipids and glycolipids revealed some similarities with the fatty acid profiles of trigalactosyldiacylglycerol1-1 (tgd1-1) and tgd2-1 (Xu et al., 2003; Li et al., 2012), two mutant lines with defects in the transport of lipids from the ER to plastids. The tgd mutants were identified by the accumulation of trigalactosyldiacylglcyerol (TGDG) and named accordingly. In the lacs4 lacs9 double mutants, no TGDG was detected, and also, no accumulation of triacylglycerol (TAG) in leaves was observed as in the tgd mutant lines, but similar to these mutants, the 16-carbon to 18-carbon fatty acid ratio of MGDG and DGDG is significantly increased in lacs4 lacs9. Such modification is compatible with possible changes in the balance between the plastidial and the ER pathway of glycerolipid biosynthesis. 16-carbon fatty acids in the sn-2 position of galactoglycerolipids are indicative for the plastidial pathway of lipid biosynthesis, whereas molecular species carrying an 18-carbon fatty acid at this position were shown to derive from the ER pathway (Heinz and Roughan, 1983). To determine the contribution of both pathways to the biosynthesis of the major galactoglycerolipids in lacs4 lacs9, we analyzed the amounts of 16-carbon fatty acids in the sn-2 position of the glycerol backbone by using a position-specific lipase from Rhizopus sp. The results show that, in MGDG as well as in DGDG, the concentration of 16-carbon fatty acids in the sn-2 position is significantly higher in lacs4 lacs9 compared with the wild type (Fig. 6). The detailed fatty acid profiles are given in Supplemental Table S1. The data are compatible with a compromised ER pathway of galactoglycerolipids biosynthesis in the lacs4 lacs9 double mutant as it was described for the tgd mutant lines (Xu et al., 2008). The results prompted us to have a closer inspection of the flux of fatty acids between the ER and chloroplasts in the lacs4 lacs9 double-mutant lines (see below).
Figure 6.
Fatty acid composition at the sn-2 position of MGDG (A) and DGDG (B). The lipid classes were purified by TLC and digested by Rhizopus sp. lipase. The fatty acid compositions of the resulting lysoderivatives were determined by gas chromatography analysis. Black bars represent the sum of 16-carbon fatty acids, and gray bars represent the sum of 18-carbon fatty acids. The values represent the mean of three independent replicates, with error bars indicating sd. WT, Wild type.
Leaf Tissue of lacs4 lacs9 Contains Elevated Levels of Free Fatty Acids
The enzymatic product of both LACS proteins is acyl-CoA. To investigate if the combined inactivation of LACS4 and LACS9 influenced the cellular acyl-CoA pool, extracts from 5-week-old leaves were analyzed. No statistically significant differences were found for either the total acyl-CoA pool or individual acyl-CoAs between the wild type and the different single-mutant plants. In contrast, the acyl-CoA pool concentrations of both lacs4 lacs9 double mutants were about 20% lower than in the wild type under long-day conditions and 28% lower under short-day conditions (Supplemental Fig. S8, A and B). A more detailed analysis of the individual metabolites revealed an almost uniform reduction of acyl-CoAs in lacs4 lacs9 throughout the measured profile (Supplemental Fig. S8, C and D). Consequently, the relative profile of acyl-CoAs remained almost unchanged (Supplemental Fig. S8, E and F). Altogether, it can be concluded that none of the observed differences in the acyl-CoA pools were correlated specifically with the growth phenotypes observed under long-day conditions. Therefore, it is unlikely that the reported morphological symptoms of the double-mutant plants were caused by limitations of the total cellular acyl-CoA pool.
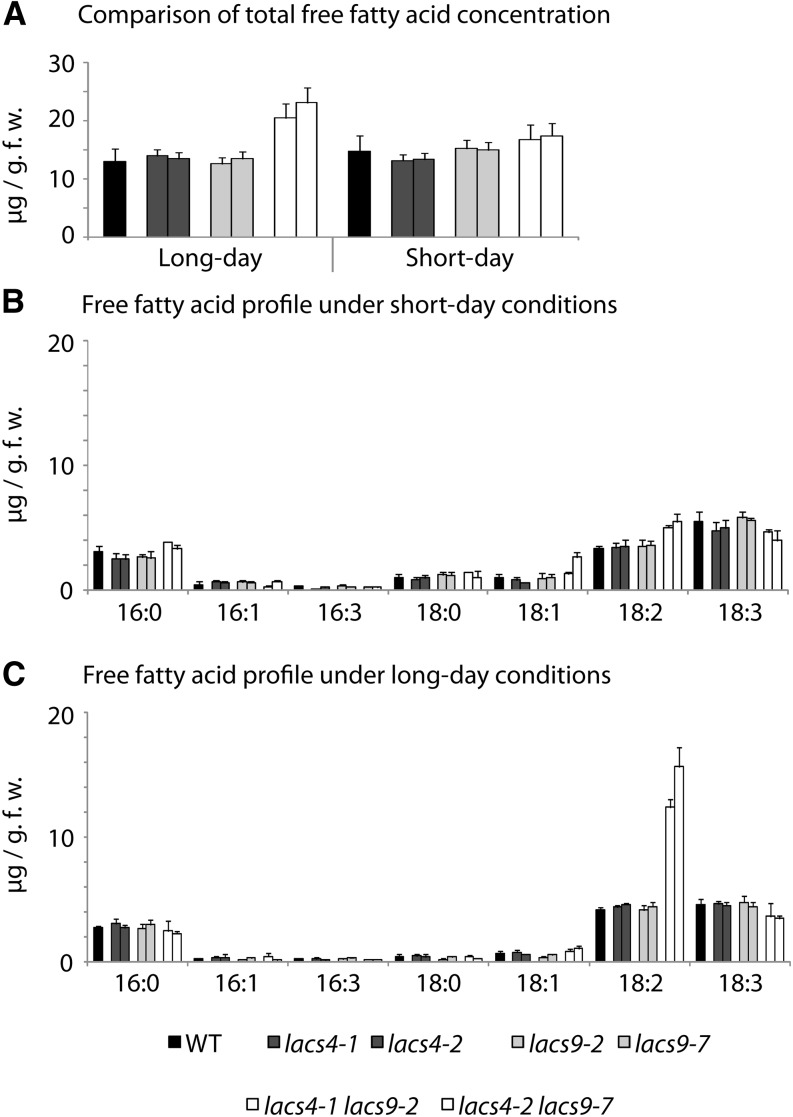
Another class of compounds relevant to LACS mutant lines is the pool of free fatty acids, because they are the direct educts of the LACS enzymes. Free fatty acids are able to act as detergents, and therefore, their cellular concentration is normally kept at very low levels. In Saccharomyces cerevisiae, it has been shown that compromised LACS activities resulted in elevated levels of cellular free fatty acids, which in turn, induced severe damages to intracellular membranes (Scharnewski et al., 2008). Lipid extracts of 5-week-old leaf material were analyzed to evaluate the effect of the combined inactivation of LACS4 and LACS9 on the level of free fatty acids. All single-mutant lines investigated showed free fatty acid concentrations comparable with the wild type (Fig. 7A). In both lacs4 lacs9 double-mutant lines, however, the concentrations of free fatty acids were about 67% higher than in the wild type. Especially interesting was the observation that such values were detected only in plants grown under long-day conditions, whereas plants cultivated under short-day conditions showed only a very modest increase in the concentration of free fatty acids. Therefore, the elevated concentration of free fatty acids was the first biochemical parameter identified that directly correlated with the long day-specific morphological phenotypes observed for lacs4 lacs9 double-mutant plants. Closer inspection of the complete profile of free fatty acids revealed another striking result. In contrast to the uniform changes in the acyl-CoA profiles, the alteration of the fatty acid profile is highly specific. The change in total free fatty acids is accounted for specifically by a strong increase in 18:2 in lacs4 lacs9 double mutants (Fig. 7, B and C). Under long-day conditions, the 18:2 content in lacs4 lacs9 double-mutant lines is increased by 300% and 380%, respectively, suggesting involvement of both LACS proteins in a highly specific process of lipid metabolism involving linoleic acid.
Figure 7.
The combined inactivation of LACS4 and LACS9 results in elevated levels of free fatty acids in plants grown under long-day conditions. A, Concentration of total free fatty acids in leaf tissue of the wild type (WT) and all mutant lines under investigation. Values are given in micrograms per gram of fresh weight (g.f.w.). B and C, Profiles of free fatty acids in the wild type and mutant lines grown under short- (B) or long-day (C) conditions. Each value represents the mean of five independent replicates, with error bars indicating sd.
LACS4 and LACS9 Are Involved in Lipid Trafficking
The elevated levels of 16-carbon fatty acids in the sn-2 position of MGDG and DGDG and the accumulation of 18:2 in the pool of free fatty acids in lacs4 lacs9 could indicate a correlation between both LACS activities and the transfer of fatty acids from the ER to plastids. However, LACS9 was considered to be involved in the export of fatty acids from the plastid. The data prompted us to analyze possible participation of both LACS activities in fatty acid transport in general. The impact of the combined inactivation of LACS4 and LACS9 on the export of fatty acids from the plastid was analyzed. The wild type and lacs4 lacs9 were compared with respect to the incorporation of radiolabeled acetate into the lipid metabolism. The exogenously applied acetate is channeled into plastids, where it is used by the fatty acid synthase to synthesize fatty acids de novo (James, 1963). The generated fatty acids supply both plastidial lipid biosynthesis and (after export from the plastid) eukaryotic lipid biosynthesis at the ER. Impairment of fatty acid export is expected to delay the incorporation of labeled fatty acids into ER-derived phospholipids PC and PE. Leaves of lacs4 lacs9 and the wild type were incubated in buffer containing radiolabeled acetate. Material was harvested after 3, 6, and 9 min, and lipid extracts were prepared. The analysis of labeled lipids showed only small differences between samples from the wild type and both double mutants (Supplemental Fig. S9, A–C). We considered differences to be relevant only if they were observed in at least two consecutive samples. Application of this criterion established a moderately elevated level of label in free fatty acids in lacs4 lacs9 compared with the wild type. In addition, a very small delay in the incorporation of label into PC could be observed at very early times, but after only 9 min of incubation, the latter difference was no longer detectable. Because the observed differences were rather small, we concluded that the export of fatty acids from the plastid is, at most, marginally compromised in the lacs4 lacs9 double mutant. For unknown reasons, the incorporation of acetate into PC was (in our experiments) consistently lower than in a previous report (Xu et al., 2008).
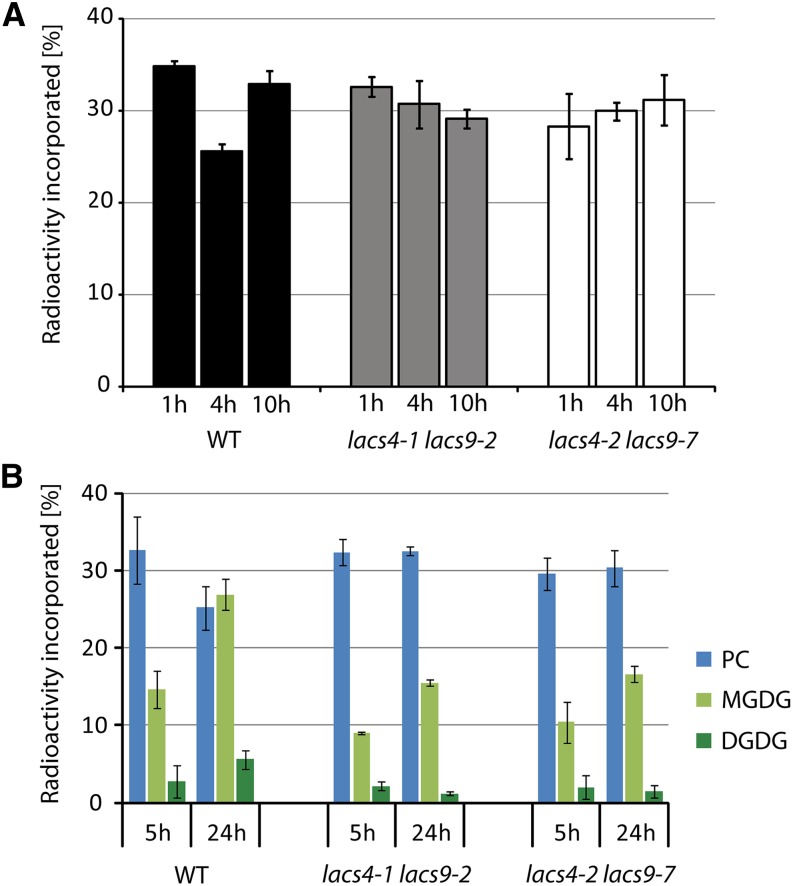
In another experiment, we analyzed whether retrograde lipid transport from ER to plastids is constrained. The question was addressed by applying an in vivo pulse-chase labeling approach using radiolabeled acetate. The assay conditions were refined over previous decades by many groups (Browse et al., 1986; Xu et al., 2003), and it was shown that the accumulation of label in MGDG over time is a qualified measure for assessing the integrity of the lipid transport from ER to the plastid (Xu et al., 2003). In the assay, de novo synthesized fatty acids are incorporated shortly after the pulse into MGDG by the prokaryotic lipid biosynthesis pathway. The concentration of label is diluted most rapidly in MGDG with time during chase conditions, resulting in reduced levels of label in MGDG after a few hours. After a short delay, the trend is temporarily reversed when labeled fatty acids, which detoured through the eukaryotic lipid biosynthesis pathway, reenter the plastid. The mechanism results in a second peak of label in MGDG about 10 h after the initial pulse. Leaves of the wild type and both double mutants were floated on buffer containing radiolabeled acetate for 60 min before being transferred to buffer without acetate. Samples were harvested after 1, 4, and 10 h, and the lipid extracts were analyzed by two-dimensional thin-layer chromatography (TLC). As shown in Figure 8A, extracts from the wild type showed the expected time-dependent incorporation of label into MGDG, resulting in a decrease of label between 1 and 4 h after exposure to acetate followed by a subsequent increase of label after 10 h. In contrast, in both double mutants, the concentration of label in MGDG remained nearly constant throughout the experiment, indicating that lipid transfer from ER to the plastid is compromised in lacs4 lacs9 double-mutant lines.
Figure 8.
Compromised lipid transfer from the ER to plastids in both lacs4 lacs9 double mutants. A, In vivo pulse-chase acetate labeling of MGDG. Detached leaves of the wild type (WT) and both lacs4 lacs9 double-mutant lines were floated on buffer containing [14C]acetate for 1 h before they were chased for the times indicated. Leaf material was collected, and lipid extracts were prepared and separated by two-dimensional TLC. The developed TLC plate was exposed to imaging plates, and the radiolabeled MGDG was quantified using a phosphorimager. B, In vivo pulse-chase labeling of lipids using [14C]oleic acid. The experimental setup was similar to that described for the labeling by acetate. The incorporation of radiolabeled oleic acid into the lipid classes PC, MGDG, and DGDG is shown for the wild type compared with both lacs4 lacs9 double-mutant lines. Each value represents the mean of three independent replicates. Error bars indicate sd.
To obtain additional support for the idea that LACS4 and LACS9 might be involved in retrograde lipid transport from ER to the plastid, another experiment was performed to assess the fate of exogenously supplied fatty acids. For this approach, detached leaves of the wild type and both double mutants were incubated for a pulse-chase experiment in buffer containing radiolabeled oleic acid. It had been shown previously that, under these conditions, the fatty acids enter lipid metabolism almost exclusively through the eukaryotic pathway at the ER (Roughan et al., 1987). Starting from PC as the major site of incorporation, all further channeling of the labeled fatty acid throughout the branches of lipid metabolism can be followed. The analysis revealed only small differences between the wild type and both double mutants 5 h postlabeling (Fig. 8B). Importantly, the label of PC is almost identical between the wild type and the double mutant, indicating that the combined inactivation of LACS4 and LACS9 is not affecting the initial incorporation of oleic acid into cytoplasmic lipid metabolism. Only the channeling of labeled oleic acid into MGDG of lacs4 lacs9 was significantly impaired at this time point compared with the wild type (−34%). However, 24 h postlabeling, the differences between both double mutants and the wild type became more conspicuous. In the wild type, the label in PC decreased between 5 and 24 h by about 23%, whereas the label in MGDG and DGDG increased (+83% and +99%, respectively). The data reflect the efficient transfer of PC-derived lipid precursors into plastids, where they serve as substrates for glycolipid biosynthesis. In lacs4 lacs9, however, the concentration of labeled oleic acid in PC remained almost constant between 5 and 24 h, and the transfer toward plastidial lipids was severely compromised. In MGDG, the concentration of labeled oleic acid increased by only 65%, whereas in DGDG, no increase was detected at all. Because DGDG is mainly synthesized by the eukaryotic pathway (Browse et al., 1986), the poor increase of label associated with DGDG is strongly supporting the assumption of inefficient transfer of lipid precursors between ER and plastids in the lacs4 lacs9 double mutant.
LACS8 Shows Overlapping Functions with LACS4 and LACS9
In summary, all of the different labeling experiments showed conclusively that the combined inactivation of LACS4 and LACS9 is significantly inhibiting the transfer of lipid precursors from the ER to the plastid. However, strong interference with ER to plastid lipid transfer was shown to cause more severe symptoms than observed in the lacs4 lacs9 double-mutant lines. A complete disruption of the process even resulted in embryo lethality (Xu et al., 2005). One possible explanation for this discrepancy could be extended functional redundancy between additional members of the LACS enzyme family in Arabidopsis. To test this hypothesis, we generated a complete set of all five lacs triple-mutant lines based on the lacs4 lacs9 double mutant, excluding only the two peroxisomal isoforms LACS6 and LACS7. The combinations of lacs4 lacs9 with mutant alleles of LACS1 to LACS5 yielded homozygous triple mutants that were able to set seeds. However, the combination of lacs4 lacs8 lacs9 could not be obtained as a triple homozygous mutant line, and additional analysis suggested that this was because of gametophyte lethality. To confirm the initial results, two independent lacs4 lacs8 lacs9 triple-mutant lines were generated by crossing the double homozygous mutant lines lacs4-1 lacs8-1 to lacs4-1 lacs9-7 and lacs4-2 lacs8-5 to lacs4-2 lacs9-2. PCR-based analysis of individual F1 plants confirmed the expected genotypes specified by homozygosity for the respective lacs4 allele and heterozygosity for lacs8 and lacs9. Progeny from self-pollinated F1 plants were grown on soil, and 384 F2 plants were genotyped. The experiment identified plants homozygous for two of the mutant alleles and heterozygous for the third allele, but not a single triple homozygous mutant line could be detected. The siliques of plants homozygous for lacs4 and lacs9 and heterozygous for lacs8 remained even smaller than siliques of lacs4 lacs9 (Fig. 9A), and analysis of seed development revealed that the siliques contained almost exclusively unfertilized ovules; only very rarely, siliques with one or two seeds were observed (Fig. 9B). Genotyping of 42 of those rare seeds identified again not a single homozygous triple-mutant line. Together, these results strongly suggest that lacs4 lacs8 lacs9 triple mutants are, indeed, nonviable and show overlapping functions of LACS4, LACS8, and LACS9. It was shown by analyses of tgd mutant alleles of different strengths that the pathway of lipid trafficking between ER and plastid is absolutely essential in Arabidopsis, and its inactivation results in lethality (Xu et al., 2005). Our data show LACS4 and LACS9 to be involved in this lipid trafficking pathway as well, and additional limitations of LACS activity in lacs4 lacs9 by the additional inactivation of LACS8 are also resulting in lethality. One possible explanation for this observation is the involvement of all three LACS proteins in ER to plastid lipid trafficking, but other pleiotropic effects of the triple mutant cannot be ruled out at present.
Figure 9.
The reproductive development of the lacs4 lacs8 lacs9 triple mutant. A, Siliques (from left to right) of the wild type, lacs4-1 lacs9-2, and lacs4-1 LACS8/lacs8-1 lacs9-2, respectively. Bar = 5 mm. B, Seed development in siliques of lacs4-1 LACS8/lacs8-1 lacs9-2. The majority of the siliques contained only abortive ovules and only very rarely set seed (indicated by an arrow). Bar = 2 mm.
DISCUSSION
LACS4 and LACS9 Are Involved in Lipid Transfer between ER and Plastids
Analyses of a complete collection of all Arabidopsis lacs double-mutant lines identified LACS4 and LACS9 as strongly overlapping activities, and the corresponding double mutants showed severe morphological phenotypes. In contrast to our expectations, the limitations in LACS activity had only modest effects on the composition of extraplastidial membrane lipids. Instead, we found strong indications for interaction of both proteins with plastidial lipid biosynthesis. Analyses of lacs4 lacs9 double-mutant plants provided several lines of evidence, suggesting that both LACS activities are involved in a process resulting in the transfer of fatty acid moieties from the cytoplasm to the plastid. Initial support for this idea was obtained from inspections of the fatty acid profiles of the galactolipids. In the lacs4 lacs9 double-mutant line, the 16- to 18-carbon fatty acid ratio of MGDG and DGDG was significantly increased compared with the wild type. More thorough compositional analysis revealed higher concentrations of 16-carbon fatty acids in the sn-2 positions of both galactolipids, indicating a shift from ER- toward plastid-derived molecular species in the double mutant (Fig. 6). Similarly, more severe alterations were reported earlier for several mutant lines defective in ER to plastid lipid trafficking (Xu et al., 2003, 2008; Awai et al., 2006; Lu et al., 2007). Over the last decade, a comprehensive set of data established four TGD proteins as essential components of a lipid transport machinery mediating the transfer of ER-derived lipid precursors to plastidial galactolipid biosynthesis. In respective mutant plants, the insufficient supply of plastids by ER-derived lipid precursors is partially compensated by elevated prokaryotic lipid synthesis within the plastids.
The changes in the fatty acid profiles of lacs4 lacs9 were just a hint on a contribution of both LACS proteins to ER to plastid lipid trafficking. Strong support for such involvement was obtained by comparing the dynamics of lipid flux in the wild type with those in the double-mutant lines. In vivo pulse-chase labeling experiments with acetate and oleic acid both showed very clearly that fatty acids are not efficiently transferred from the ER to plastids in the lacs4 lacs9 double mutants (Fig. 8). Interestingly, the combined inactivation of LACS4 and LACS9 was not affecting the incorporation of fatty acids into membrane lipids in general but was specifically hindering their transfer from cytoplasmic PC to plastidial glycolipids. Similar results were reported previously for tgd mutant lines defective in components of the established lipid transport machinery (Xu et al., 2003, 2008). The analyses of the tgd mutant lines revealed that the ER to plastid lipid trafficking is an essential pathway in Arabidopsis. It was shown for TGD1 that only plants containing less severe alleles were viable and amenable to biochemical analyses, but total inactivation of TGD1 resulted in lethality (Xu et al., 2005). In the lacs4 lacs9 double mutant, the observed symptoms for compromised ER to plastid lipid trafficking, like the enrichment of plastid-derived molecular species of galactolipids or the decreased transfer of labeled fatty acids to the plastid, were clearly less severe than in tgd1. The analyses of lacs triple-mutant lines revealed that this is most likely caused by the partially overlapping activity of LACS8, which might still enable the plant to supply the plastid with ER-derived lipid precursors. Consequently, the additional inactivation of LACS8 in the lacs4 lacs9 background resulted in lethality, suggesting that LACS activity is, indeed, essential for ER to plastid lipid trafficking (Fig. 9).
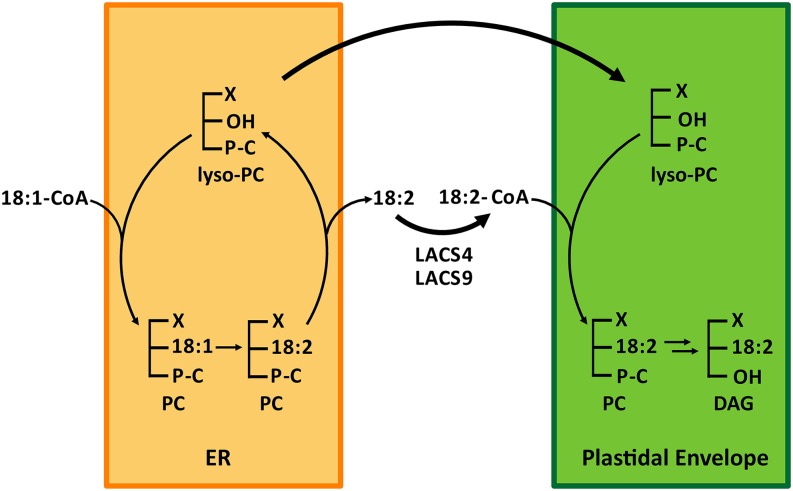
Because our data suggested that LACS4 and LACS9 are involved in ER to plastid lipid trafficking, it was necessary to clarify their precise role in this process. The correct functional assignment is complicated by the fact that many aspects of lipid transport remain hypothetical. Nevertheless, based on considerable progress in recent years, a reasonable model can be proposed. Four important features of the lipid transport process were established already more than 30 years ago by kinetic labeling experiments (Slack et al., 1977; Roughan et al., 1980): (1) PC is the major cytoplasmic lipid precursor of galactolipid biosynthesis, (2) label in the glycerol moiety of PC as well as label in the fatty acids are both transferred to glycolipids, (3) linoleic acid is the preferentially transferred fatty acid, and (4) DAG is the immediate substrate for plastidial glycolipid biosynthesis. From these data, it was concluded that mostly dilineoyl-PC from the ER is converted into dilineoyl-DAG, which finally serves as a direct substrate for galactolipid biosynthesis in the plastidial envelope. It remained unclear, however, which enzymes are involved in the conversion of PC to DAG, and also, the nature of the molecule that is actually transported has not yet been determined. It was not even possible to decide unambiguously if PC is broken down to provide substrate for the resynthesis of DAG or if glycerol and fatty acids are transferred in one entity. Advanced labeling studies provided several lines of evidence suggesting that the DAG moiety of PC is transferred not in one entity and that instead lyso-PC could be the transport form of lipids delivered to plastids (Bessoule et al., 1995). In vitro assays indicated that acyl-CoA-lyso-PC acyltransferase activity associated with plastids enabled the conversion of transferred lyso-PC to PC (Bertrams and Heinz, 1981; Bessoule et al., 1995). Additional support for lyso-PC as the precursor of plastidial lipids was obtained by comparing stereospecific labeling patterns of glycolipids with those of extraplastidial and plastidial PC (Mongrand et al., 2000). In conclusion, the data suggested that the glycerol backbone of PC is transferred with only one fatty acid esterified to its sn-1 position, whereas the sn-2 position is subjected to a process of deacylation/reacylation concomitant to the transfer. The transport process could greatly benefit by such a process, because the mobility of the lysolipid is strongly enhanced compared with regular phospholipids, and it has been shown that lyso-PC is able to partition rapidly between membranes and the aqueous phase (McLean and Phillips, 1984; Cassagne et al., 1991) and especially, from plant ER membranes through the aqueous phase to the plastidial envelope to become acylated in the presence of acyl-CoAs (Testet et al., 1999). However, this finding seems to be counterintuitive, because the fatty acid at the sn-2 position is considered as indicative for the site of lipid synthesis and should, in principle, stay tightly connected to the glycerol backbone throughout the whole transfer from ER to the plastid. The solution to this apparent contradiction could be a tightly coupled transport of lysolipids and fatty acid as first discussed (but considered unlikely) by Slack et al. (1977). Results of this study, however, support such coupled transport of fragmented lipid molecules and seem to match the proposed model perfectly. As discussed above, the analyses of the lacs4 lacs9 double mutant revealed that LACS activity is intimately connected to the lipid trafficking between ER and the plastid. If LACS activity is essential to this process, it is fair to assume that it is, in fact, the generated acyl-CoA, which is required for the lipid transfer. Obviously, this requirement fits nicely to the model postulating lyso-PC as transport molecule, which needs to be converted to PC upon arrival at the plastidial envelope. The conversion to PC is absolutely dependent on acyl-CoA, and based on our data, the used acyl-CoA is a very special pool generated by LACS4 and LACS9. Additional support for such a mechanism is provided by the most striking biochemical phenotype of lacs4 lacs9 plants. In leaves of the double mutant, the concentrations of all major free fatty acids were unchanged compared with the wild type, except for linoleic acid, which was dramatically increased (Fig. 7C). This highly specific alteration was surprising, because the data suggested the existence of a previously unknown transient pool of free 18:2 in the wild type that is efficiently converted to 18:2-CoA by overlapping activities of LACS4 and LACS9. Upon combined inactivation of both LACS proteins, the transfer of lipid precursors to the plastid is severely compromised, and instead, 18:2 shows up in a pool of free fatty acids. As described above, 18:2 is of special importance for ER to plastid lipid transport, because it has been shown in several reports to account for the bulk amount of the transferred fatty acids (Slack et al., 1977; Browse et al., 1986; Bates et al., 2007). Significant limitations for ER to plastid lipid transport were observed also in the fad2 mutant line devoid of microsomal 18:1 desaturase activity (Miquel and Browse, 1992). As possible explanation, Miquel and Browse (1992) proposed that 18:1-containing lipids might be a poor substrate for the transfer between the ER and the chloroplast, a concept that is in agreement with our findings in this study. In addition, it was shown that 18:2 is also the optimal substrate for plastidial acyl-CoA-lyso-PC acyltransferase responsible for the synthesis of PC in the envelope (Bessoule et al., 1995). In combination, the data suggest that 18:2 released from PC in the ER membrane is converted by LACS4 and LACS9 to 18:2-CoA, which is immediately used as substrate to resynthesize PC in the plastidial envelope. To integrate the data currently available, we propose the following model for ER to plastid lipid transport (Fig. 10). The transfer is initiated by action of a phospholipase A2 at the ER or in ER-plastid contact sites. The enzyme favors PC with 18:2 in the sn-2 position as substrate, resulting in the preferential release of linoleic acid. The generated lyso-PC may migrate to the envelope of plastids as proposed earlier (Bessoule et al., 1995), and the free fatty acid is activated by LACS4 or LACS9 to establish a local 18:2-CoA pool. This acyl-CoA pool must be protected from mixing with other fatty acids and is used specifically by plastidial acyl-CoA-lyso-PC acyltransferase to resynthesize PC molecules in the plastidial envelope. The conversion of diffusible lyso-PC into PC immediately anchors the molecule in the envelope membrane, thereby rendering the transport unidirectional. Subsequently, PC will be partially converted via PA to DAG to eventually become substrate for glycolipid biosynthesis.
Figure 10.
Working model describing the LACS-mediated transfer of fatty acids from the ER to the chloroplast envelope. By the Lands cycle at the ER, mainly de novo synthesized oleic acid is incorporated into the sn-2 position of PC. The oleic acid is converted by the desaturase FAD2 to linoleic acid (18:2), and PC molecules with this signature are the preferred substrate for lipid transfer toward the plastid. We propose that 18:2 of remodeled PC is released through deacylation by phospholipase A2 to establish a pool of lyso-PC and a pool of free linoleic acid, both destined for lipid transfer to the plastid. As shown earlier (Bessoule et al., 1995), the lyso-PC is well suited to migrate between membranes and is able to move from the ER to the plastid. In parallel, LACS4 and LACS9 are both involved in converting the free linoleic acid into a tightly coupled pool of linoleoyl-CoA, which is used by plastidial acyl-CoA-lyso-PC acyltransferase to resynthesize PC in the plastidial envelope. Parts of the resynthesized PC stay in the envelope, whereas another portion is converted via PA into DAG, which is the immediate substrate for glycolipid biosynthesis. X, C16, or C18 fatty acid.
The Functions of LACS4 and LACS9 Overlap, Despite Different Subcellular Localizations
Our genetic and biochemical data clearly indicate functional overlap between LACS4 and LACS9. Assays based on the analysis of both proteins fused to EYFP revealed different subcellular localizations to the envelope of chloroplasts and the ER. For LACS9, localization to the plastidial envelope was already well established (Schnurr et al., 2002), and our experiments confirmed this assignment. LACS4 was localized exclusively to the ER network when analyzed by confocal fluorescence microscopy, but small amounts of LACS4 were found to copurify with plastids when analyzed by cell fractionation combined with immunoblotting (Fig. 4). The latter finding is in agreement with data showing residual enzymatic LACS activity associated with purified plastids in lacs9 mutant plants (Schnurr et al., 2002), although we cannot fully exclude the possibility that low amounts of ER copurify with chloroplasts during cell fractionation. Therefore, it is difficult to decide to what extent elimination of LACS4 activity at the plastid might also be contributing to the observed phenotypes of lacs4 lacs9 double mutants. Based on the predominant cellular association of LACS4 with the ER, we believe that inactivation of ER-located LACS4 is, indeed, crucial for the phenotype of lacs4 lacs9. In this case, there are multiple explanations that could reconcile the phenotypic data with the observed subcellular localizations. Functional overlap with respect to TAG biosynthesis was reported for LACS1 and LACS9, which were localized in different cellular compartments as well (Zhao et al., 2010). It was suggested that fatty acids destined for TAG synthesis are partially activated at plastids by LACS9 and partially activated by LACS1 at the ER. Transport of either free fatty acids or acyl-CoAs could mask the site of synthesis and result in mixed acyl-CoA pools. For the observed cooperation of LACS4 and LACS9, however, a functional overlap based on transport of reactant or product is not convincing given the observed specificity for 18:2, which can, instead, be best explained by substrate channeling between the participating activities. Considering this substrate specificity, it is more intuitive to envision LACS4 and LACS9 enzymes in close proximity to each other. For TGD4, it has been proposed that the protein might act in contact sites between ER and the plastid envelope (Wang et al., 2012). The existence of such contact sites were established by Kjellberg et al. (2000) and Andersson et al. (2007). Because close proximity of the participating membranes might be of vital importance for the lipid transfer between ER and plastid, it seems legitimate to propose that LACS4 and LACS9 are involved in establishing a pool of 18:2-CoA in these contact sites.
lacs4 lacs9 Mutant Lines Are Devoid of TGDGs
Despite many biochemical similarities between tgd mutants and lacs4 lacs9, there is also one important difference. The TGD genes were identified by the detection of TGDGs in the lipid extracts of respective mutant plants (Xu et al., 2003, 2008). Recently, it was shown that galactolipid-galactolipid galactosyltransferase synthesizes TGDG upon exposure to freezing conditions, and for reasons still not completely understood, this activity is constitutively induced in tgd mutant plants (Moellering et al., 2010). In lacs4 lacs9, however, we were unable to detect any TGDG. Currently, it is unclear if this is because of less severe inhibition of ER to plastid lipid trafficking in the double mutant or if the accumulation of TGDG is specifically induced by defects in the TGD machinery.
LACS4 and LACS9 Are Involved in TAG Metabolism and Seed Development
The combined inactivation of LACS4 and LACS9 not only influences ER to plastid lipid trafficking but also, impacts the biosynthesis of seed storage lipids as well as seed development. The TAG content of seeds of the double mutant was about 27% lower than in the wild type (Fig. 3E). This is clearly the strongest effect on seed oil content observed so far for any LACS mutant line. Previously, the most pronounced impact was reported for the double mutant lacs1 lacs9, with TAG levels reduced by about 10% (Zhao et al., 2010). Taking into account the severe morphological phenotype of the lacs4 lacs9 double mutant and the stunted growth, we cannot exclude pleiotropic effects on seed oil content. However, the analysis of the fatty acid profile of TAG revealed highly specific changes in the double mutant, resulting in a significantly lower 18:2 to 18:3 ratio than in the wild type (Supplemental Fig. S3). The modification was especially remarkable, because a comparison of phospholipids between both double mutants and the wild type showed a reciprocal alteration. The TAG-specific changes in fatty acid composition seem to support the hypothesis that limitations of specific acyl-CoA pools are primarily responsible for the lower TAG content of seeds. A recent report on the ATP-binding cassette (ABC) transporter ABCA9 proposed that a special acyl-CoA pool in the ER lumen provides substrate for the biosynthesis of TAG (Kim et al., 2013). The model by Kim et al. (2013) suggests a spatial separation of membrane lipid and TAG biosynthesis based on independent acyl-CoA pools, and it is possible that specific deficiencies in LACS activities could affect these pools differently. The results will require additional consideration, but they may reflect the intricate relationship between phospholipid and TAG biosynthesis. In addition to the reduced TAG content, lacs4 lacs9 was also partially compromised in seed development, resulting in a substantially elevated frequency of undeveloped ovules.
Our results proved LACS4 to be a very versatile activity. Recently, we showed that LACS4 has partially overlapping functions with LACS1 in providing substrate for the biosynthesis of very long-chain lipids (Jessen et al., 2011). lacs1 lacs4 double-mutant plants have severely reduced levels of surface wax, and they are conditionally sterile because of low concentrations of tryphine lipids. Here, we present data revealing overlapping functions of LACS4 and LACS9 in glycerolipid synthesis, resulting in obstacles to both TAG metabolism and the retrograde lipid transfer between ER and plastid. In combination, the data suggest a complex network of LACS activities establishing independent pools of acyl-CoA within the cell, which can be assigned to specific pathways.
In conclusion, our genetic, cell biological, physiological, and biochemical approaches consistently show that LACS activity is essentially required for ER to plastid lipid trafficking. Our data support a model postulating deacylation and subsequent reacylation of PC concomitant to the lipid transport. LACS4 and LACS9 are both involved in establishing a very special pool of 18:2-CoA, which is tightly connected to the transfer of PC. Despite that ER to plastid lipid trafficking was shown to be essential in Arabidopsis, lacs4 lacs9 double-mutant plants showed diverse phenotypes but were perfectly viable. However, the additional inactivation of LACS8 in the lacs4 lacs9 background resulted in lethality and revealed overlapping functions of all three LACS proteins.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of the T-DNA insertion lines lacs4-1 (At4g23850; SALK_101543), lacs4-2 (SALK_126610), lacs9-2 (At2g47240; SALK_111835), lacs9-7 (SALK_040810), and lacs9-8 (SALK_124615) were obtained from the European Arabidopsis Stock Center (Alonso et al., 2003). To produce lacs triple-mutant lines, the following T-DNA insertion lines obtained from the same source have been used: lacs1-1 (At2g47240; SALK_127191), lacs2-3 (At1g49430; GK_368C02), lacs3-1 (At1g64400; SALK_084299), lacs5-1 (At4g11030; GK_567F06), lacs8-1 (At2g04350; SALK_136060), and lacs8-5 (SAIL_439_H08). All Arabidopsis (Arabidopsis thaliana) lines under investigation were of the ecotype Columbia-0 (Col-0). Seeds were stratified for 2 d at 4°C, and plants were grown on soil in environmental chambers under either short- (8-h photoperiod) or long-day conditions (16-h photoperiod) at 21°C, 60% humidity, and a light intensity of 150 μmol m−2 s−1.
Nicotiana benthamiana plants were grown on soil in a greenhouse at 22°C under a 16-h photoperiod. Plants with six to eight leaves were used for transient transformation by infiltration with Agrobacterium spp.
Isolation of Single and Double T-DNA Insertion Mutants
Homozygous plants were identified by PCR-based genotyping. For both mutant alleles of LACS4, a single pair of gene-specific primers was used and designated as lacs4F and lacs4R. A complete list with all oligonucleotides is given in Supplemental Table S2. For lacs9-2 and lacs9-7, the primer pair lacs9F and lacs9R was used, whereas lacs9-8 was analyzed by the primer combination lacs9F2 and lacs9R2. For lacs8-1, the primers lacs8F and lacs8R were used, whereas lacs8-5 was analyzed by the combination of lacs8F2 and lacs8R2. For all other LACS genes under investigation, only a single-mutant allele was used to generate triple-mutant lines, and the corresponding primer pairs were lacs1F, lacs1R; lacs2F, lacs2R; lacs3F, lacs3R; and lacs5F, lacs5R. The presence of T-DNA insertions was verified using appropriate gene-specific primers combined with the individual T-DNA left border primer SALK_LB, SAIL_LB3, or GABI_LB. All lacs4 lacs9 double-knockout lines (lacs4-1 lacs9-2, lacs4-1 lacs9-7, lacs4-1 lacs9-8, lacs4-2 lacs9-2, lacs4-2 lacs9-7, and lacs4-2 lacs9-8) were obtained by crossing the corresponding single-mutant lines. Homozygous double-knockout lines were identified by screening plants of the F2 generation for obvious phenotypes, and the genotypes were confirmed by PCR.
In addition, the absence of corresponding transcripts was verified by RT-PCR. Total RNA was extracted from young leaves using the Invisorb Spin Plant RNA Mini Kit (Invitek). The RNA was transcribed into complementary DNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. For RT-PCR, gene-specific primers flanking the T-DNA insertion were used. For LACS9, an additional reverse primer according to the work by Zhao et al. (2010) was tested. The Actin2 gene served as a constitutive control, and the following primers were used for the amplifications: LACS9_RTf, LACS9_RTr; LACS9_RTf_Zhao, LACS9_RTr_Zhao (Zhao et al., 2010); LACS4_RTf, LACS4_RTr; and Actin2_RTf, Actin2_RTr.
Lipid Analysis
For lipid extraction, rosette leaves of 6-week-old plants were frozen in liquid nitrogen and extracted with hexane:isopropanol (3:2, v/v) with 0.0025% (w/v) 2-butyl-6-hydroxytoluene (Sigma). As internal standard for total fatty acid analysis, pentadecanoic acid was added, whereas for the analyses of the fatty acid profiles of distinct lipid classes, diheptadecanoyl-glycerolipids were added. Extracts were analyzed on TLC plates by using different solvent systems. Phospholipids were separated in chloroform:methanol:acetic acid (65:25:8, v/v/v), whereas for galactolipids, chloroform:methanol (85:20, v/v) was used. Lipids were identified by comigration of appropriate standards.
For methylation, lipids were resuspended in methanol:sulfuric acid:2,2-dimethoxypropane (50:1:1, v/v/v) and incubated for 1 h at 80°C. After stopping the reaction with 200 µL of 5 m NaCl solution, fatty acid methyl esters were extracted with hexane and analyzed by gas chromatography. Free fatty acids were methylated in 400 µL of a solution of 10 mg of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide in 4 mL of MeOH and incubated for 2 h at room temperature. The methylated fatty acids were extracted with 2 mL of hexane, dried under a stream of nitrogen, and resuspended in 15 µL of acetonitrile (Stumpe et al., 2001). The analysis of fatty acid methyl esters was carried out using an Agilent 6890 Series GC System (Agilent) equipped with a flame ionization detector. For this analysis, the column DB-23 (30 m × 0.25 mm, 0.25-µm coating thickness; J&W Scientific; Agilent) was used with helium as carrier gas (1 mL min−1). The quantification was based on the integration of peak areas relative to the internal standard.
For analysis of storage lipids, 10 seeds were transferred into a glass vial, and 2 mL of extraction solution composed of methanol:toluene:sulfuric acid:2,2-dimethoxypropane (30:20:1:1, v/v/v/v) was added. Vials were incubated for 1 h at 80°C, and the reaction was terminated by adding 200 µL of 5 m NaCl solution. Fatty acids methyl esters were extracted with hexane. Gas chromatography analysis was performed as described above.
Acyl-CoAs were extracted and analyzed by HPLC exactly as described by Larson and Graham (2001).
Molecular species compositions of galactolipids were analyzed using Rhizopus sp. lipase according to the protocol established by Siebertz and Heinz (1977) with modifications introduced later (Miquel et al., 1998).
Labeling of Lipids
In vivo labeling with [1-14C]acetic acid sodium salt (specific activity of 55 mCi mmol−1) for short time-course experiments was performed as previously described (Bates et al., 2007) For each time point, about 0.3 g of detached Arabidopsis leaves from 4- to 5-week-old plants were incubated in the light at 22°C in labeling solution (20 mm MES, pH 5.5, 0.1-strength Murashige and Skoog medium salts, and 0.01% [w/v] Tween 20). After preincubation for 30 min in the light in 20 mL of solution without radioisotope, 250 µCi of [1-14C]acetic acid was added (0.225 mm), and incubation was continued. After 3, 6, and 9 min, leaves were removed, washed quickly in water, and then frozen in liquid nitrogen. Lipids were extracted as described before.
Pulse-chase experiments with [1-14C]acetic acid sodium salt were performed with the same setup as described above. The detached leaves were incubated for 1 h in the labeling solution before they were removed, washed three times in water, and chased in solution without radioisotope for 1, 4, or 10 h. The leaf material was frozen in liquid nitrogen, and the lipids were extracted as described before.
Labeling with [1-14C]oleic acid (specific activity of 50 mCi mmol−1) was performed according to a previously described procedure (Xu et al., 2003). Briefly, detached leaves were incubated in 20 mL of labeling solution containing 100 µCi of [1-14C]oleic acid in the light for 1 h at 22°C under gentle shaking. Afterward, the leaves were washed three times in water and then, further incubated in solution without radioisotope for 5 and 24 h. Finally, the leaf material was collected and frozen in liquid nitrogen, and the lipids were extracted as described before.
The lipid extracts were dissolved in hexane:isopropanol (3:2, v/v) with 0.0025% (w/v) 2-butyl-6-hydroxytoluen (Sigma) and further analyzed by two-dimensional TLC using chloroform:methanol:water (65:35:4, v/v/v) in the first dimension and chloroform:methanol:ammonia hydroxide (62:25:5 v/v/v) in the second dimension. The developed TLC plates were exposed to imaging plates, and the radiolabeled lipids were quantified after 5 d of exposure using a phosphorimager (Fuji FLA-3000; Raytest).
Promoter-GUS Reporter Gene Fusions and GUS Histochemical Assay
Promoter activity was analyzed by using the GUS reporter gene. Promoter fragments of 2,305 and 1,108 bp of LACS4 and LACS9, respectively, were amplified from Arabidopsis genomic DNA using the primer pairs LACS4P_fSseI/LACS4P_rBamHI and LACS9Pf_SseI/LACS9P_rBamHI. The fragments were inserted into the SseI/BamHI sites of the vector pBI121 (Jefferson et al., 1987). The constructs were transformed into Agrobacterium tumefaciens EHA105. Confirmed clones were used for transformation of wild-type Arabidopsis Col-0 plants using the floral dip method (Clough and Bent, 1998). For each construct, at least 18 transgenic lines were isolated, and a representative plant was chosen for additional analyses as described earlier (Malamy and Benfey, 1997).
Subcellular Localization
Subcellular localization was investigated by generating stable transgenic Arabidopsis lines with endogenous levels of tagged LACS proteins. For C-terminal epitope tagging, three repeats of the HA tag were fused to genomic fragments of LACS4 and LACS9 and cloned into a modified version of pCAMBIA3300. The vector pCAMBIA3300 was adapted by removing the 35S promoter, extending the polylinker, and inserting the 3HA tag. In detail, the 35S promoter was excised by PstI and HindIII and subsequently religated in the presence of a small linker generated by annealing of the two oligonucleotides LinkerF and LinkerR. The fragment encoding 3HA was amplified from the vector pYM-N20 (Janke et al., 2004) by the primer combination 3HA_fBamHIAsisI and 3HA_rSacISseI, digested by BamHI and SacI, and inserted into the modified pCAMBIA3300, resulting in the new vector pC33-3HA. Genomic fragments of LACS4 (6,602 bp) and LACS9 (4,900 bp) were amplified using the primer combinations LACS4_fNotIAscI/LACS4_rAsiSI and LACS9_fNcoIAscI/LACS9_rAsiSI, respectively. Both fragments were digested by AscI and AsiSI and ligated into pC33-3HA digested accordingly, resulting in the constructs pC33-LACS4g-3HA and pC33-LACS9g-3HA. In the final constructs, the coding sequence of the LACS genes and the 3HA tag were separated by a small fragment encoding the polypeptide AIAPGAG, because fusing EYFP directly to the C terminus of LACS4 resulted in improper results.
To establish stably transformed Arabidopsis lines expressing LACS-EYFP fusion proteins, the fragment encoding 3HA was replaced by a sequence encoding EYFP (amplified by the primer combination XFP_fAsiSI and XFP_rSseI) using the restriction sites AsiSI and SseI. The resulting constructs pC33-LACS4g-EYFP and pC33-LACS9g-EYFP were transformed into A. tumefaciens EHA105 and used for the transformation of Arabidopsis wild-type plants as described above.
For the transient expression of LACS4 in N. benthamiana, a complementary DNA clone was amplified using the primer pair LACS4_fKpn and LACS4_rSpe. The resulting fragment was digested by KpnI/SpeI and inserted into the vector pEG104 (Earley et al., 2006) digested by SpeI and partially digested by KpnI. The resulting plasmid pEG104LACS4 encoding an EYFP-LACS4 fusion protein was transformed into A. tumefaciens EHA105. For transient transformation of N. benthamiana, 5 mL of Agrobacterium spp. overnight cultures were harvested by centrifugation (10 min at 4,000 rpm at 22°C), the cell pellet was resuspended in infiltration medium (10 mm MgCl2, 10 mm MES, pH 5.5, and 150 µm acetosyringone), and the optical density at 600 nm was adjusted to 0.4. The bacterial mixture was incubated for 2 h at 22°C before it was used to infiltrate leaves of 4- to 6-week-old N. benthamiana plants (Voinnet et al., 2003). The leaves were analyzed by confocal microscopy after 2 to 3 d. As control for localization in the ER, Agrobacterium spp. cells transformed with the vector ER-ck (Nelson et al., 2007) were used. Confocal laser-scanning microscopy after transient expression in N. benthamiana and of stable transgenic Arabidopsis plants was performed on a Leica SP5-DM6000 (Leica GmbH) equipped with Leica LAS AF software (v.2.6.7266.0) using excitation wavelengths of 514 nm for EYFP (detection at 525–600 nm), 485 nm for ECFP (detection at 465–485 nm), and 561 nm for chlorophyll autofluorescence (detection at 680–700 nm). Images were processed using Photoshop CS6 (Adobe).
Intact chloroplasts were purified by density gradient centrifugation using Percoll (GE Healthcare) as described earlier (Aronsson and Jarvis, 2002) and analyzed by SDS-PAGE and immunoblotting. The samples were mixed with 2 times Laemmli buffer and separated on 10% (w/v) SDS-PAGE. The proteins were transferred to nitrocellulose membrane, blocked with 2% (w/v) dried milk in buffer of 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.25% (v/v) Tween 20 (TBS-T) for 1 h, and then incubated with primary antibodies at various dilutions overnight at 4°C. After four washes in TBS-T for 10 min each, the membranes were exposed to polyhorseradish peroxidase-conjugated anti-rabbit (diluted 1:5,000) or anti-mouse (diluted 1:5,000) sera for 2 h at 20°C. After five washes with TBS-T, the signals were detected by a 3:1 mixture of SuperSignal West Pico:Femto Chemiluminescent Substrates (Pierce). HA antibodies (diluted 1:5,000) were purchased from Roche, TOC75 (diluted 1:2,000) was a gift of John Froehlich, and TPR7 (diluted 1:2,000) was provided by Katrin Philippar and Serena Schwenkert.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At4g23850 (LACS4), At2g04350 (LACS8), and At2g47240 (LACS9).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Leaf area per plant.
Supplemental Figure S2. Detection of truncated LACS9 transcript in the lacs9-7 mutant line.
Supplemental Figure S3. Fatty acid profiles of seeds.
Supplemental Figure S4. Mutant leaf polar lipid composition is comparable to the wild type.
Supplemental Figure S5. Relative expression levels of FAD3 and FAD7 in double mutant plants.
Supplemental Figure S6. Detailed analysis of leaf lipids from plants grown under long-day conditions.
Supplemental Figure S7. Detailed analysis of leaf lipids from plants grown under short-day conditions.
Supplemental Figure S8. Composition of the acyl-CoA pools.
Supplemental Figure S9. In vivo short-term acetate labeling of lipids.
Supplemental Table S1. Fatty acid composition at the sn-2 position of the two major galactoglycerolipids in the wild type and lacs4-1 lacs9-2.
Supplemental Table S2. List of oligonucleotides.
Supplementary Material
Acknowledgments
We thank Ronald Scholz, Corinna Thurow, and Christiane Gatz for the real-time PCR analysis; Volker Lipka for providing access to the microscope; John Froehlich for provision of TOC75 antibody; Katrin Philippar and Serena Schwenkert for provision of TPR7 antibody; and Jay Shockey for critical reading of the article.
Glossary
- acyl-ACP
acyl-acyl carrier protein
- Col-0
Columbia-0
- DAG
diacylglycerol
- DGDG
digalactosyldiacylglycerol
- ER
endoplasmic reticulum
- MGDG
monogalactosyldiacylglycerol
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- RT
reverse transcription
- TBS-T
10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.25% (v/v) Tween 20
- TGDG
trigalactosyldiacylglcyerol
- TLC
thin-layer chromatography
- T-DNA
transfer DNA
Footnotes
This work was supported by the International Max Planck Research School (PhD program “Molecular Biology” to D.J.), the Göttingen Graduate School for Neurosciences and Molecular Biosciences at the Georg-August-University (Deutsche Forschungsgemeinschaft [DFG] grant no. GSC 226/1 to D.J.), the Evangelisches Studienwerk e.V. Villigst (scholarship to D.J.), and the DFG (grant nos. WI 3208/4–1 and WI 3208/5–1 to M.W., and FU 430/4–1 to M.F.).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Goksör M, Sandelius AS (2007) Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J Biol Chem 282: 1170–1174 [DOI] [PubMed] [Google Scholar]
- Andrews J, Keegstra K (1983) Acyl-CoA synthetase is located in the outer membrane and acyl-CoA thioesterase in the inner membrane of pea chloroplast envelopes. Plant Physiol 72: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Awai K, Xu C, Tamot B, Benning C (2006) A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc Natl Acad Sci USA 103: 10817–10822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Ohlrogge JB, Pollard M (2007) Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J Biol Chem 282: 31206–31216 [DOI] [PubMed] [Google Scholar]
- Bertrams M, Heinz E (1981) Positional specificity and fatty acid selectivity of purified sn-glycerol 3-phosphate acyltransferases from chloroplasts. Plant Physiol 68: 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoule JJ, Testet E, Cassagne C (1995) Synthesis of phosphatidylcholine in the chloroplast envelope after import of lysophosphatidylcholine from endoplasmic reticulum membranes. Eur J Biochem 228: 490–497 [PubMed] [Google Scholar]
- Block MA, Dorne AJ, Joyard J, Douce R (1983) Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J Biol Chem 258: 13281–13286 [PubMed] [Google Scholar]
- Browse J, McConn M, James D Jr, Miquel M (1993) Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J Biol Chem 268: 16345–16351 [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem J 235: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassagne C, Juguelin H, Boiron F (1991) Phospholipid acylation by mouse sciatic nerve microsomes. Biochim Biophys Acta 1070: 119–126 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Heinz E, Roughan PG (1983) Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol 72: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AT. (1963) The biosynthesis of long-chain saturated and unsaturated fatty acids in isolated plant leaves. Biochim Biophys Acta 70: 9–19 [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen D, Olbrich A, Knüfer J, Krüger A, Hoppert M, Polle A, Fulda M (2011) Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J 68: 715–726 [DOI] [PubMed] [Google Scholar]
- Kim S, Yamaoka Y, Ono H, Kim H, Shim D, Maeshima M, Martinoia E, Cahoon EB, Nishida I, Lee Y (2013) AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc Natl Acad Sci USA 110: 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellberg JM, Trimborn M, Andersson M, Sandelius AS (2000) Acyl-CoA dependent acylation of phospholipids in the chloroplast envelope. Biochim Biophys Acta 1485: 100–110 [DOI] [PubMed] [Google Scholar]
- Lands WE. (1958) Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J Biol Chem 231: 883–888 [PubMed] [Google Scholar]
- Larson TR, Graham IA (2001) Technical advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Li Z, Gao J, Benning C, Sharkey TD (2012) Characterization of photosynthesis in Arabidopsis ER-to-plastid lipid trafficking mutants. Photosynth Res 112: 49–61 [DOI] [PubMed] [Google Scholar]
- Lu B, Xu C, Awai K, Jones AD, Benning C (2007) A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J Biol Chem 282: 35945–35953 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J 12: 957–963 [DOI] [PubMed] [Google Scholar]
- McLean LR, Phillips MC (1984) Kinetics of phosphatidylcholine and lysophosphatidylcholine exchange between unilamellar vesicles. Biochemistry 23: 4624–4630 [DOI] [PubMed] [Google Scholar]
- Miquel M, Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267: 1502–1509 [PubMed] [Google Scholar]
- Miquel M, Cassagne C, Browse J (1998) A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol 117: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Cassagne C, Bessoule JJ (2000) Import of lyso-phosphatidylcholine into chloroplasts likely at the origin of eukaryotic plastidial lipids. Plant Physiol 122: 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Kuhn DN, Stumpf PK (1979) Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc Natl Acad Sci USA 76: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Shine WE, Stumpf PK (1978) Fat metabolism in higher plants. Characterization of plant acyl-ACP and acyl-CoA hydrolases. Arch Biochem Biophys 189: 382–391 [DOI] [PubMed] [Google Scholar]
- Roughan PG, Holland R, Slack CR (1980) The role of chloroplasts and microsomal fractions in polar-lipid synthesis from [1-14C]acetate by cell-free preparations from spinach (Spinacia oleracea) leaves. Biochem J 188: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Thompson GA Jr, Cho SH (1987) Metabolism of exogenous long-chain fatty acids by spinach leaves. Arch Biochem Biophys 259: 481–496 [DOI] [PubMed] [Google Scholar]
- Scharnewski M, Pongdontri P, Mora G, Hoppert M, Fulda M (2008) Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J 275: 2765–2778 [DOI] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer GJ, Browse JA (2002) Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol 129: 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger R, Müller NC, Schmitt MJ, Soll J, Schwenkert S (2012) AtTPR7 is a chaperone-docking protein of the Sec translocon in Arabidopsis. J Cell Sci 125: 5196–5207 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebertz M, Heinz E (1977) Galactosylation of different monogalactosyldiacylglycerols by cell-free preparations from pea leaves. Hoppe Seylers Z Physiol Chem 358: 27–34 [DOI] [PubMed] [Google Scholar]
- Slack CR, Roughan PG, Balasingham N (1977) Labelling studies in vivo on the metabolism of the acyl and glycerol moieties of the glycerolipids in the developing maize leaf. Biochem J 162: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe M, Kandzia R, Göbel C, Rosahl S, Feussner I (2001) A pathogen-inducible divinyl ether synthase (CYP74D) from elicitor-treated potato suspension cells. FEBS Lett 507: 371–376 [DOI] [PubMed] [Google Scholar]
- Testet E, Verdoni N, Cassagne C, Bessoule J (1999) Transfer and subsequent metabolism of lysolipids studied by immobilizing subcellular compartments in alginate beads. Biochim Biophys Acta 1440: 73–80 [DOI] [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang Z, Xu C, Benning C (2012) TGD4 involved in endoplasmic reticulum-to-chloroplast lipid trafficking is a phosphatidic acid binding protein. Plant J 70: 614–623 [DOI] [PubMed] [Google Scholar]
- Xu C, Fan J, Cornish AJ, Benning C (2008) Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20: 2190–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Froehlich JE, Awai K, Benning C (2005) Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Riekhof W, Froehlich JE, Benning C (2003) A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J 22: 2370–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Katavic V, Li F, Haughn GW, Kunst L (2010) Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. Plant J 64: 1048–1058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.