Protein phosphatase 2A targets peroxisomes and positively affects fatty acid oxidation.
Abstract
The eukaryotic, highly conserved serine (Ser)/threonine-specific protein phosphatase 2A (PP2A) functions as a heterotrimeric complex composed of a catalytic (C), scaffolding (A), and regulatory (B) subunit. In Arabidopsis (Arabidopsis thaliana), five, three, and 17 genes encode different C, A, and B subunits, respectively. We previously found that a B subunit, B′θ, localized to peroxisomes due to its C-terminal targeting signal Ser-Ser-leucine. This work shows that PP2A C2, C5, andA2 subunits interact and colocalize with B′θ in peroxisomes. C and A subunits lack peroxisomal targeting signals, and their peroxisomal import depends on B′θ and appears to occur by piggybacking transport. B′θ knockout mutants were impaired in peroxisomal β-oxidation as shown by developmental arrest of seedlings germinated without sucrose, accumulation of eicosenoic acid, and resistance to protoauxins indole-butyric acid and 2,4-dichlorophenoxybutyric acid. All of these observations strongly substantiate that a full PP2A complex is present in peroxisomes and positively affects β-oxidation of fatty acids and protoauxins.
Reversible phosphorylation of proteins is one of the most common mechanisms used in the regulation of biological processes and requires both a protein kinase and a protein phosphatase. Protein phosphatase 2A (PP2A) is a conserved Ser/Thr protein phosphatase and belongs to one of the major protein phosphatase families in eukaryotes (Farkas et al., 2007; Eichhorn et al., 2009; Uhrig et al., 2013; Lillo et al., 2014). PP2A plays important roles in plant metabolism, development, stress response, and signal transduction. PP2A holoenzymes are heterotrimeric complexes comprising catalytic (C), scaffolding (A), and regulatory (B) subunits, all encoded by multiple genes. The dimer of the C subunit (36 kD) and A subunit (65 kD) makes up the core enzyme of PP2A. This core enzyme associates with a third subunit, B type, which is essential for subcellular localization and substrate specificity. Through selective incorporation of different B subunits, the complexes are recruited to specific subcellular compartments that define the sphere of active function for the various complexes (Janssens and Goris, 2001; Farkas et al., 2007). In Arabidopsis (Arabidopsis thaliana), three genes (A1 [also called Roots curl in naphthylphthalamic acid1]–A3) code for A subunits, 17 genes for B subunits, and five genes for C subunits (Janssens and Goris, 2001; Farkas et al., 2007). Theoretically, up to 255 different PP2A heterotrimers can be formed. B subunits are classified into B (α and β), B′ (α, β, γ, δ, ε, ζ, η, θ, and κ), and B′′ (α, β, γ, δ, ε, and TONNEAU2/FASS) nonrelated families (Janssens and Goris, 2001; Farkas et al., 2007). The large number of subunits certainly indicates the importance of PP2A in a myriad of processes. For example, the B family members were shown to be important for dephosphorylation and activation of nitrate reductase (Heidari et al., 2011). B′α and B′β from the B′ family were shown to interact with BRASSINOSTEROID INSENSITIVE1, a transcription factor of the brassinosteroid signaling pathway (Tang et al., 2011). Members of the B′′ family were found to interact with a key enzyme of isoprenoid synthesis, 3-hydroxy-3-methylglutaryl-CoA reductase (Leivar et al., 2011). The B′η subfamily from B′ comprises the close homologs B′γ, B′ζ, B′η, and B′θ (Terol et al., 2002; Farkas et al., 2007; Eichhorn et al., 2009), which target the different organelles, nucleus, mitochondria, nucleus and nucleolus, and peroxisomes, respectively (Matre et al., 2009). The most studied member from this subfamily is B′γ, which prevents unnecessary defense reactions under low light in Arabidopsis (Trotta et al., 2011a, 2011b). In addition, b′γ plants display enhanced expression of FLOWERING LOCUS C and flowered later than wild-type plants (Heidari et al., 2013).
Peroxisomes are single-membrane-bound organelles present in all major groups of eukaryotes. They were first discovered as compartments containing hydrogen peroxide-generating oxidases together with catalases that degrade hydrogen peroxide into molecular oxygen and water (De Duve and Baudhuin, 1966; van den Bosch et al., 1992; Kaur et al., 2009). In higher plants, fatty acid β-oxidation takes place in peroxisomes, and all fatty acids can be completely degraded in peroxisomes, whereas in mammalian cells, short-chain fatty acids are transported from peroxisomes to mitochondria for completion of β-oxidation (Mano and Nishimura, 2005). Fatty acid β-oxidation and hydrogen peroxide detoxification are two conserved functions of peroxisomes in all eukaryotes, but specialized functions have also been identified. For example, plant glyoxysomes are specialized peroxisomes in germinating seeds that harbor the glyoxylate cycle, and plant leaf peroxisomes take part in photorespiratory glycolate metabolism and biosynthesis of the plant hormones indole acetic acid, salicylic acid, and jasmonic acid. Peroxisomes are devoid of DNA, and their complete set of matrix proteins are encoded in the nucleus and synthesized in the cytosol before being imported into peroxisomes (Kaur et al., 2009). Peroxisomal matrix proteins contain two types of peroxisome targeting signals, PTS1 and PTS2, by which they are directed to peroxisomes. PTS1 exists in the majority of proteins as a C-terminal conserved tripeptide with the prototype serine-lysine-leucine at C terminus (SKL>). PTS2 is a nonapeptide with RLx5HL as a prototype sequence. Peroxin5 (PEX5) and PEX7 are soluble receptors that recognize proteins with PTS1 and PTS2, respectively, and direct them to the peroxisome membrane (Rehling et al., 1996; Kragler et al., 1998; Woodward and Bartel, 2005; Kaur et al., 2009). There are also examples of peroxisomal proteins with alternative targeting signals and transport by piggybacking through other proteins bearing PTSs (Kaur et al., 2009).
Regulation of peroxisomal metabolism by reversible phosphorylation has not yet been clearly demonstrated, but the presence of protein kinases in peroxisomes has been predicted due to putative targeting signals or implicated from proteome studies (Dammann et al., 2003; Reumann, 2004, 2011). Only one protein kinase (calcium-dependent protein kinase1) has so far been verified to target peroxisomes (Dammann et al., 2003; Coca and San Segundo, 2010). Besides calcium-dependent protein kinase1, three additional protein kinases were reported to have functional PTS1 domains, but the full-length proteins failed to be imported to peroxisomes (Kaur et al., 2009; Lingner et al., 2011; Sørhagen et al., 2013). No protein phosphatase has so far been identified in peroxisomes except for a B subunit (B′θ) of PP2A that targeted peroxisomes when fused N-terminally to enhanced yellow fluorescent protein (EYFP) in transiently transformed cells (Matre et al., 2009). Whereas B′θ has a PTS1 serine-serine-leucine (SSL) in the extreme C-terminal end, none of the C and A subunits have putative peroxisomal targeting signals. In this work, the ability of B′θ to interact with C and A subunits and target them to peroxisomes was investigated. Reverse genetics was applied to gain insight into the effects of B′θ on peroxisomal metabolism and related phenotypes.
RESULTS
B′θ Interacts with A2, C2, and C5 Subunits in Peroxisomes
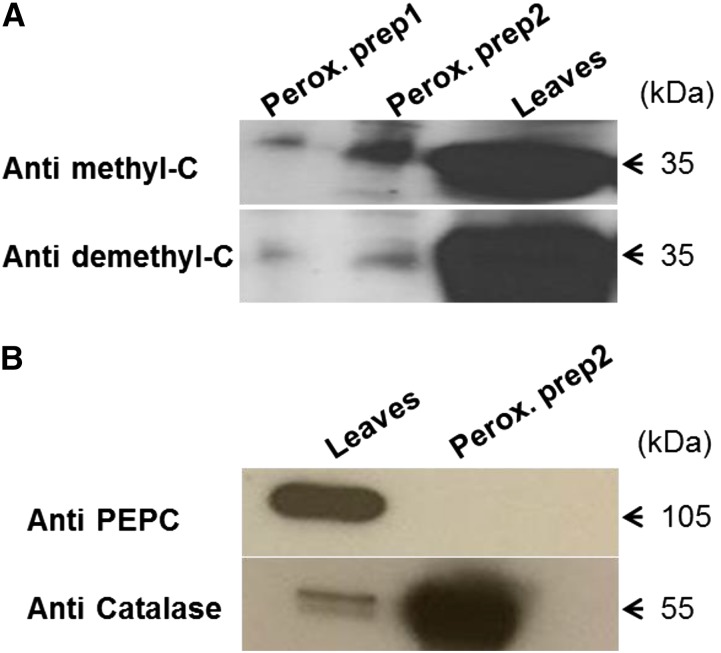
Highly purified peroxisomes were isolated using a Percoll gradient followed by a Suc density gradient (Reumann and Singhal, 2014). In extracts of these peroxisomes, a PP2A C subunit was detected by western blotting using antibodies specific for either methylated or demethylated C subunit (Fig. 1A; Supplemental Fig. S1A). It cannot be excluded that the antibody toward methylated PP2A would not pick up the PP2A-like enzyme PP4, although much less expressed, but to ensure detection of PP2A, another antibody was also used that specifically picked up (demethylated) PP2A. Purity of the peroxisomal extracts was tested by western-blot analysis using antibodies specific for cytosolic phosphoenolpyruvate carboxylase (PEPC) and the peroxisomal catalase (Fig. 1B). Bimolecular fluorescence complementation (BiFC; Waadt et al., 2008) was used to identify C and A subunits that interact with B′θ in peroxisomes. B′θ complementary DNA (cDNA) was subcloned in the vectors Venus Yellow N-terminus enhanced yellow fluorescent protein right [pVYNE(R)] and Venus Yellow C-terminus enhanced yellow fluorescent protein right [pVYCE(R)] to give a free C terminus (Supplemental Fig. S1B). C1 to C5 (except C3, which is highly similar to C4), A1, and A3 were subcloned into vectors to give free N-terminal ends, as were A1 to A3 to give a free C-terminal end (Supplemental Fig. S1B). Microscopy analysis of Arabidopsis mesophyll protoplasts transformed with B′θ::pVYNE(R) together with A1::pVYCE or A3::pVYCE revealed a low background fluorescence that remained mostly cytosolic and was probably caused by some yellow fluorescent protein (YFP; Venus variant) self-assembly. Similar observations were made upon cotransformation by B′θ::pVYCE(R) with A1::pVYNE(R) or A3::pVYNE(R) (24- and 48-h incubation time). In contrast to such weak background fluorescence, use of B′θ::pVYCE(R) with A2::pVYNE(R) revealed strong fluorescence signals, which were observed in organelle-like structures (Fig. 2I). To determine the identity of these structures, the peroxisomal marker glyoxysomal malate dehydrogenase (gMDH)–cyano fluorescent protein (CFP; Kim and Smith, 1994) was coexpressed with B′θ::VYCE and A2::VYNE. The YFP fluorescence from BiFC interaction coincided with CFP fluorescence in the same structures (Fig. 2I). From these results, we conclude that B′θ interacts with A2, and they colocalize to peroxisomes.
Figure 1.
Detection of PP2A catalytic subunit in isolated peroxisomes. A, Catalytic subunit detection in isolated Arabidopsis peroxisomes by western blotting with specific antibodies against methylated (upper) and demethylated (lower) C-terminal end. Two preparations of peroxisomes were used together with Arabidopsis leaf extract (5-week-old plants) as a control. The detected band equals 35 kD. B, Assessment of the purity of isolated peroxisomes using antibodies against cytosolic PEPC and the peroxisomal marker catalase. Perox. prep1, Peroxisomal preparation1; Perox. prep2, peroxisomal preparation2.
Figure 2.
Visualization of B′θ/A2, B′θ/C2, and B′θ/C5 interactions in peroxisomes. Interaction and peroxisomal localization were detected in Arabidopsis mesophyll protoplasts. I, Coexpression of A2::VYNE(R) and B′θ::VYCE(R). II, Coexpression of C2::VYNE and B′θ::VYCE(R). III, Coexpression of C2::VYCE and B′θ::VYNE(R). IV, Coexpression of C5::VYNE and B′θ::VYCE(R) (Leivar et al., 2011). Coexpression of B′θΔSSL::VYCE with C2::VYNE (V) or C5::VYNE (VI) shows cytosolic fluorescence in protoplasts. Peroxisomes were labeled with gMDH-CFP (Fulda et al., 2002). The cyan fluorescence color was converted to red. Scale bars = 5 μM.
C subunits of PP2A are divided into two subgroups in Arabidopsis (group 1: C1, C2, and C5; and group 2: C3 and C4; Farkas et al., 2007). Based on gene expression analyses, C2 and C5 appeared to be good candidates for interacting with B′θ (Zimmermann et al., 2004; Jen et al., 2006; Supplemental Fig. S2). Coexpression of C2::VYNE and B′θ::VYCE(R) (Fig. 2II) and C2::VYCE and B′θ::VYNE(R) (Fig. 2III) in protoplasts showed YFP fluorescence that coincided with gMDH-CFP–labeled peroxisomes (Fig. 2, II and III). Coexpression of C5::VYNE and B′θ::VYCE(R) also revealed interaction in peroxisomes (Fig. 2IV). No YFP fluorescence was detected for BiFC vectors combining C1 or C4 with B′θ. To test the importance of the PTS1 of B′θ, truncated B′θ (ΔSSL) was used. When B′θΔSSL::VYCE(R) was coexpressed with C2::VYNE or C5::VYNE, fluorescence remained in the cytosol (Fig. 2, V and VI), showing that the B′θ PTS1 is necessary to direct the interacting proteins into peroxisomes. We also investigated the BiFC in onion (Allium cepa) epidermal cells after biolistic transformation by B′θ::pVYCE(R) and A2::pVYNE(R) and with C2, C5, C1, or C4::pVYNE for 24 to 48 h. Peroxisomal localization was confirmed in the case of B′θ interacting with A2 and C5 (Supplemental Fig. S3), whereas no fluorescence was observed with C1, C2, or C4. The lack of signal from B′θ::pVYCE(R) combined with C2::pVYNE in onion epidermal cells indicates that the interaction of B′θ and C2 is weaker than the interaction between B′θ and C5, at least under some conditions.
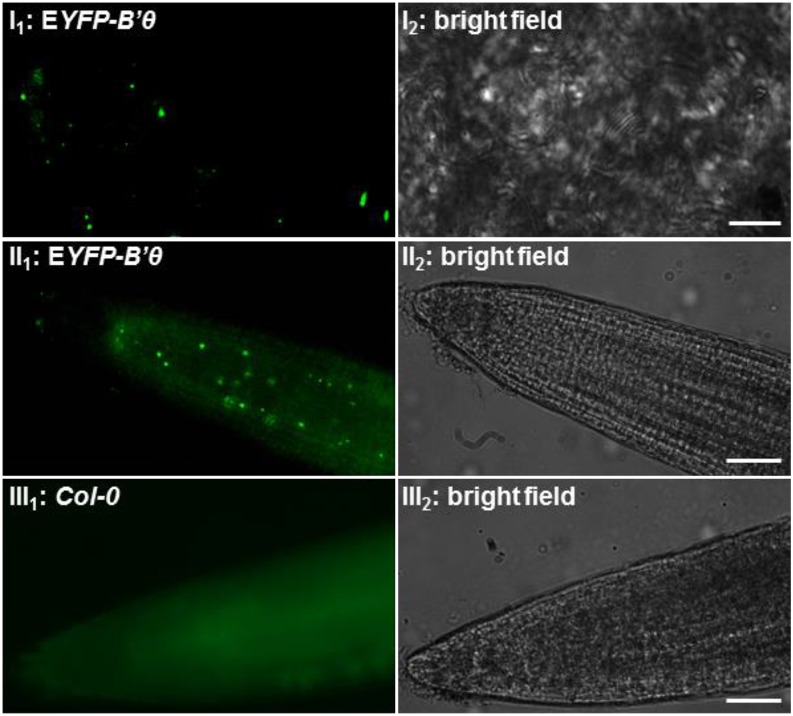
Arabidopsis plants were also stably transformed with a binary vector, and genomic presence of EYFP-B′θ was confirmed by PCR. Fluorescence microscopy revealed the presence of EYFP-B′θ in leaves in punctate structures resembling peroxisomes (Fig. 3I). When the F3 progenies were allowed to grow in liquid one-half-strength Murashige and Skoog medium, fluorescence was also detected in punctate structures in roots (Fig. 3II) and was absent in nontransformed roots (Fig. 3III).
Figure 3.
Expression of EYFP-B′θ in peroxisome-like structures in transgenic stable overexpression plants. The F3 progenies of plants transformed by EYFP-B′θ show EYFP fluorescence in punctate structures with variable levels in mesophyll cells (I). The highest expression was seen in roots of 3-d-old seedlings (meristem and elongation zone; II), whereas no such fluorescence was seen in nontransformed plants (III). Scale bars = 5 μm (I) and 200 μm (II and III).
Expression Analysis Indicates the Importance of B′θ in Specific Tissue and under Stressful Conditions
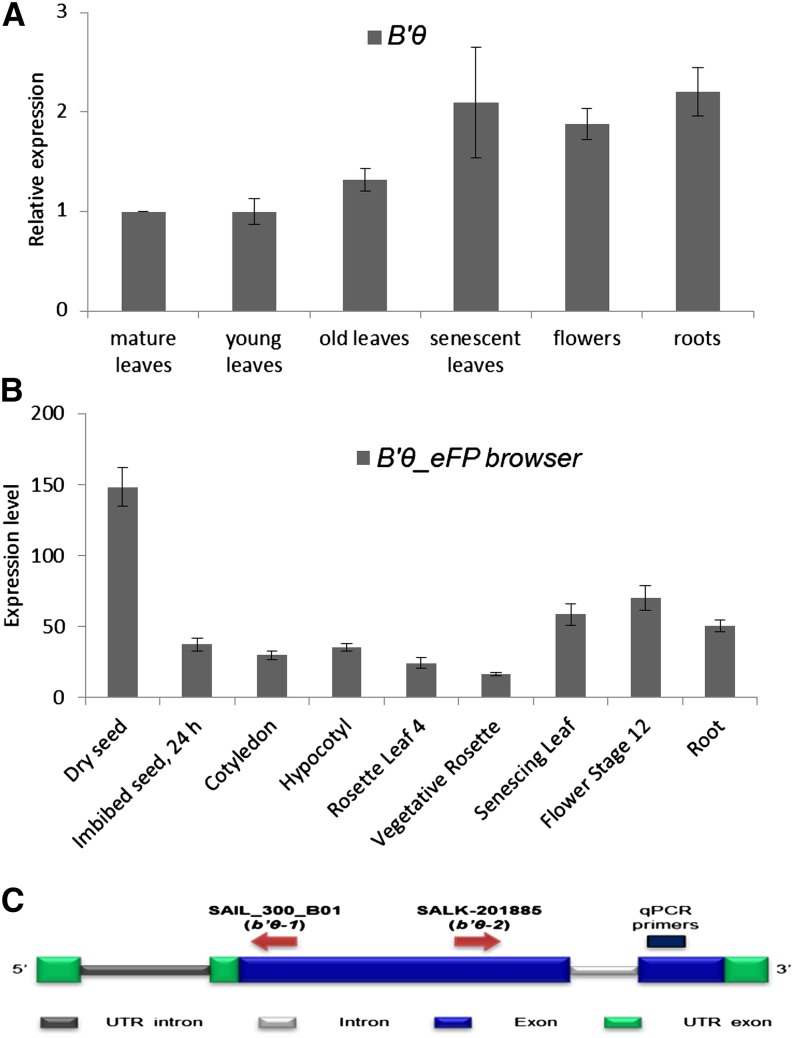
Using quantitative reverse transcription (qRT)-PCR, B′θ expression was found to be higher in roots and reproductive and senescent leaves than in young and mature leaves (Fig. 4A). These results are consistent with the publicly available microarray data (Fig. 4B). It is also seen from the Genevestigator database that expression of B′θ is high in dessicated seeds, decreases in stratified seeds, becomes stable throughout early seedling germination steps, and becomes high during senescence stages (Supplemental Fig. S4). B′θ expression is up-regulated in response to several plant pathogens, such as virulent Pseudomonas syringae pathovars (Psm and Pst), the nematode Meloidogyne incognita, the oomycete Phytophthora parasitica, and fungi (Phytophthora infestans and Blumeria graminis; Supplemental Figs. S4 and S5). Several microarray experiments that include treatment by the bacterial elicitor (flagellin 22) also show up-regulation of B′θ, C2, and C5 expression (Supplemental Figs. S2D and S4). In contrast to its up-regulation due to biotic stress, B′θ expression is down-regulated in response to abiotic stresses (drought, heat, and salt; Supplemental Fig. S4).
Figure 4.
B′θ expression analyses. A, qRT-PCR shows higher levels of B′θ transcript in senescent leaves, roots, and flowers than in young and mature leaves. Expression levels are given relative to the expression in mature leaves (set to one). Vertical bars represent ses. B, Expression levels of B′θ mRNA (absolute values) in different tissues. The graph was made using Arabidopsis electronic fluorescent pictograph (eFP) browser data (Winter et al., 2007). C, Schematic representation of B′θ encoding gene with position of the T-DNA insertions and TaqMan primers used for analyzing expression. UTR, Untranslated region.
B′θ Has a Conserved PTS1 within the Tribe Camelineae
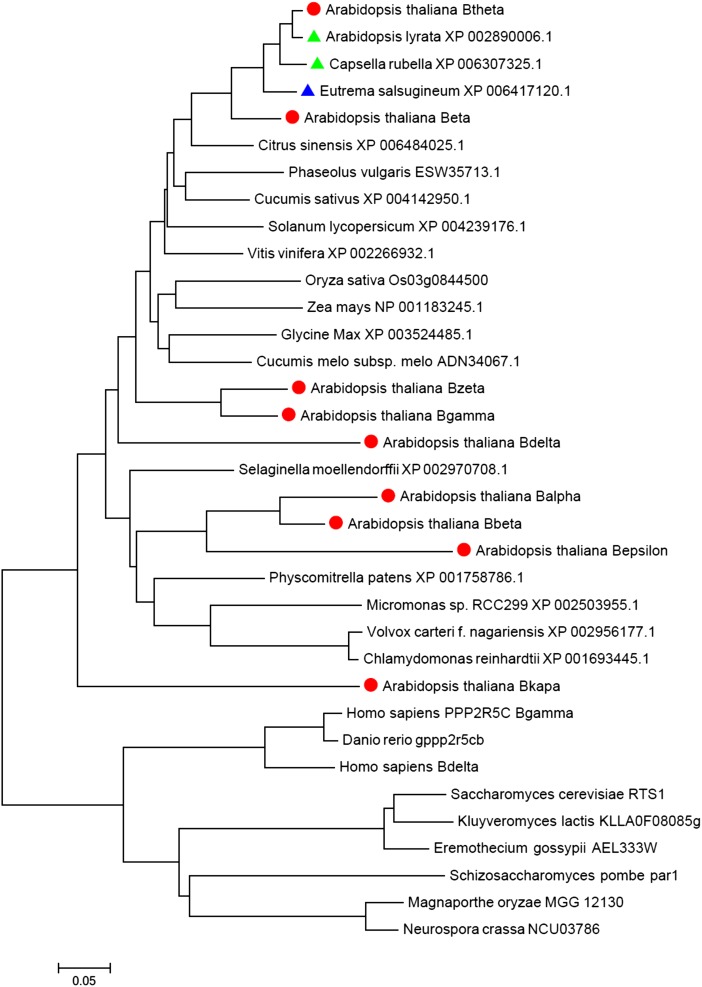
Using the BLAST search tool (protein BLAST, National Center for Biotechnology Information), B′θ was found to be conserved in plants, whereas most of the other eukaryotes had homologs of the B′ family members, not specifically for B′θ (Fig. 5; Supplemental Fig. S6). Although B′θ is conserved in plants, most of its homologs lack the peroxisomal signal SSL> or any other experimentally verified PTS. Two B′θ homologs from the same family/tribe (Brassicaceae/Camelineae) have a conserved SSL>, one from the same genus Arabidopsis (Arabidopsis lyrata) and the other from the genus Capsella (Capsella rubella; Fig. 5). Eutrema salsugineum is a halophytic species in the Brassicaceae family (Yang et al., 2013), and it has a highly similar homolog of Arabidopsis B′θ with a conserved C-terminal domain but with a change of SSL> to SSS> (Fig. 5). This mutation most likely will abolish targeting to peroxisomes, as it lowers the prediction score threshold from 0.368 for SSL> to −0.472 for SSS> based on the latest prediction algorithm of plant PTS1 proteins (Lingner et al., 2011).
Figure 5.
Evolutionary relationship of PP2A-B′θ homologs. B′θ is conserved in plants, whereas mammals and yeast (Saccharomyces cerevisiae) contain homologs of the B′ family. Arabidopsis B′ family members are highlighted by a red circle. The homologs that have a conserved peroxisome PTS1 are highlighted by a green triangle, whereas the blue triangle marks a change in SSL> to SSS>. The phylogram was generated by MEGA6 (Tamura et al., 2013) using the neighbor-joining method of Saitou and Nei (1987). The tree is drawn with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site.
The b′θ Mutant Seedlings Show a Sugar Dependence Phenotype
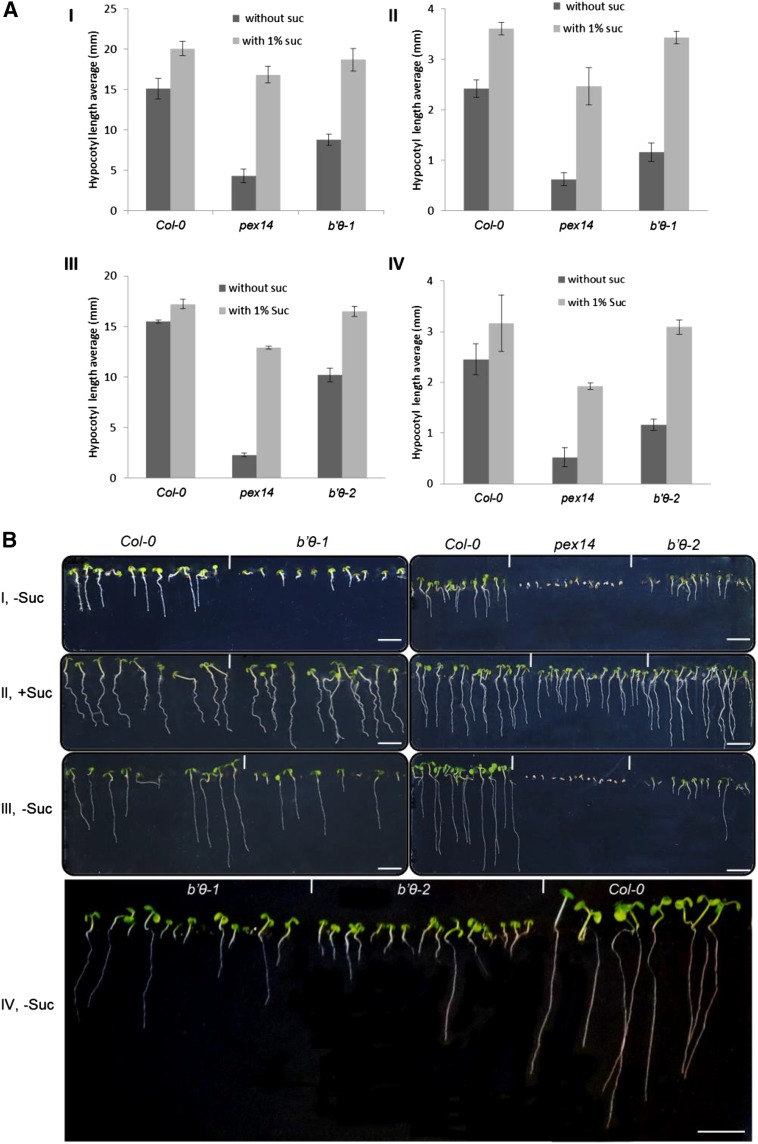
Oilseed plants, such as Arabidopsis, rely on fatty acid catabolism during the early seedling stage. In mutants that do not properly catabolize fatty acids, growth is hampered unless an alternative energy source is provided (Mano and Nishimura, 2005). Two homozygous transfer DNA (T-DNA) insertion lines, SAIL_300_ B01 (b′θ-1; Matre et al., 2009) and SALK_201885 (b′θ-2), with inserts in the first coding exon (Fig. 4C), were used to study the effects of B′θ inactivation. PEX14 is a peroxisomal membrane protein involved in protein import, and the pex14 mutant was used as a sugar dependence control (Orth et al., 2007; Zhang and Hu, 2010). Hypocotyl length of seedlings grown in continuous darkness (Fig. 6A, I) and in short days (Fig. 6A, II) in the presence and absence of Suc was measured 6 d after germination. On Suc-free medium, hypocotyl elongation was strongly inhibited in pex14 and b′θ-1 seedlings as compared to Suc-containing medium, whereas only a small effect was seen for the wild type Columbia (Col-0; Fig. 6A, I and II). The hypocotyl-length phenotype was confirmed by b′θ-2 seedlings (Fig. 6A, III and IV). Both b′θ-1 and b′θ-2 mutants showed seedling development defects on Suc-free medium 6 and 12 d after germination (Fig. 6B, I, III, and IV), whereas this was not the case on Suc-containing medium (Fig. 6B, II). However, some seedlings were able to recover after their photosynthesis had started but still were not comparable with the wild type (Fig. 6B, III and IV). These data indicate that b′θ seedlings are impaired in triacylglycerol (TAG) metabolism. Moreover, obtaining the same phenotype for both b′θ-1 and b′θ-2 lines confirms that the disruption of B′θ is the cause of the observed phenotypes.
Figure 6.
Suc dependence assay of b′θ mutants. A, Hypocotyl length of seedlings grown for 6 d in the dark (I and III) or 8 h of light/16 h of dark (II and IV) on one-half-strength Linsmeier and Skoog (LS) medium with or without 1% Suc. The homozygous lines b′θ-1 (I and II) and b′θ-2 (III and IV) were tested. The experiment was repeated three times, and error bars represent sd. B, Representative images showing developmental defects in b′θ-1, b′θ-2, and pex14 (sugar dependence control) seedlings 6 d after germination in short days without Suc (I). Impaired growth was not seen for medium with Suc (II). After 12 d on Suc-free medium, the differences between mutants and the wild type were still clear, but some mutant seedlings started to recover (III and IV). Scale bars = 5 mm.
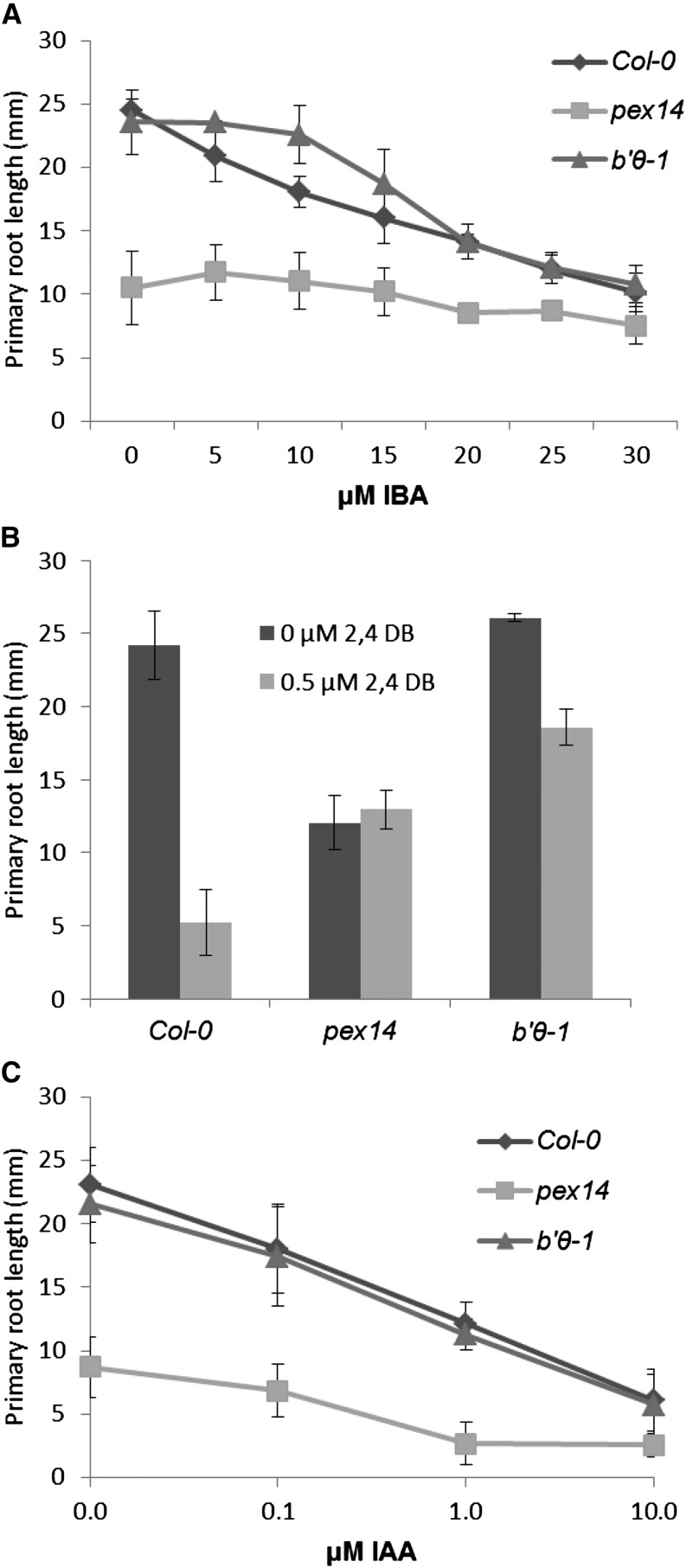
The b′θ Mutant Seedlings Are Impaired in Their Response to Protoauxins
Indole-3-butyric acid (IBA) and 2,4-dichlorophenoxybutyric acid (2,4-DB) are protoauxins that can be transformed by peroxisomal β-oxidation to the bioactive auxins indole-3-acetic acid (IAA) and 2,4-dichlorophenoxyacetic acid, respectively. Mutants impaired in β-oxidation show resistance to the inhibitory effect of these compounds on primary root elongation (Zolman et al., 2001; Mano and Nishimura, 2005). In the presence of low levels of IBA, b′θ-1 seedlings showed significant resistance to auxin in comparison with the wild type Col-0 (Fig. 7A). Moreover, b′θ-1 seedlings showed resistance to 2,4-DB (Fig. 7B). The pex14 mutant was also resistant to the inhibition of root elongation by IBA and 2,4-DB, consistent with previous reports (Zhang and Hu, 2010). These results support the hypothesis that the b′θ mutant seedlings are impaired in transforming protoauxins into active auxins, especially since the seedlings responded as the wild type upon treatment with IAA (Fig. 7C).
Figure 7.
Effects of IBA and 2,4-DB on primary root elongation of b′θ-1 seedlings. Plants were grown for 7 d in light on one-half-strength LS medium supplemented with 0.5% Suc and different concentrations of IBA (A), 2,4-DB (B), and IAA (C). The experiment was repeated three times; error bars represent sd.
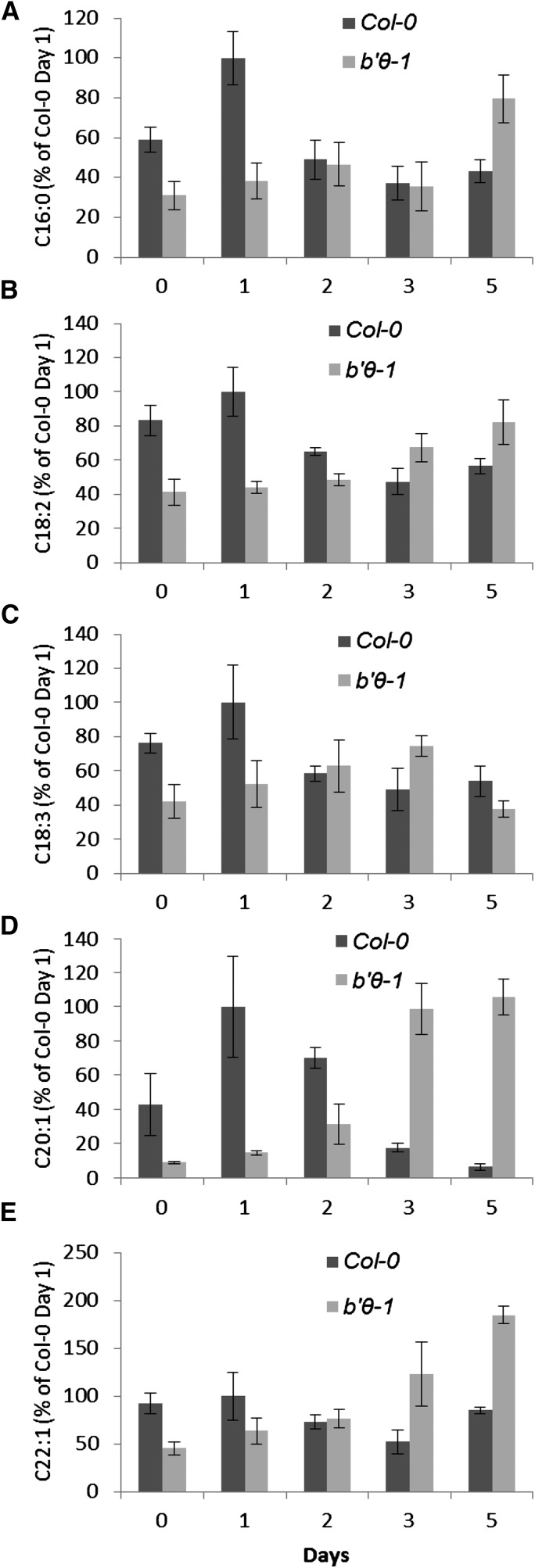
The b′θ Mutant Seedlings Lack Normal TAG Mobilization
In Arabidopsis, eicosenoic acid (C20:1), which is a component of TAG, is considered as a convenient reporter of TAG breakdown in seeds (Lemieux et al., 1990; Fulda et al., 2004). Normally, C20:1 is rapidly degraded during early seedling growth and is almost absent 4 to 5 d after imbibition of the seeds (Fulda et al., 2004). Mutants blocked in β-oxidation failed to degrade TAG and suffered during early seedling growth (Zolman et al., 2001). The levels of fatty acids in the wild type and b′θ-1 seedlings were followed and showed that C20:1 in the wild type was high during the first days, then declined rapidly as expected, whereas in the mutant, the level was lower during the first days, then increased (Fig. 8D). Not only C20:1 but also other long-chain fatty acids were noticeably lower in b′θ-1 than in the wild type during the first 2 d of testing (Fig. 8, A–E). The data indicate that b′θ-1 cannot mobilize TAG normally and underpin that B′θ has an influence on peroxisomal fatty acid β-oxidation.
Figure 8.
Delayed lipid mobilization in b′θ-1 seedlings. Seeds were sown on one-half-strength LS medium without sugar and stratified for 2 d before being transferred to a growth chamber with continuous light at d 0. The relative level of individual fatty acids (A–E) in the wild type Col-0 and b′θ-1 are given as percentages and standardized to the wild type Col-0 levels at d 1. Error bars represent ses (n = 3) of three batches, each with 39 seedlings.
The b′θ Mutant Seedlings Have Unique Phosphorylated Proteins Involved in TAG Metabolism
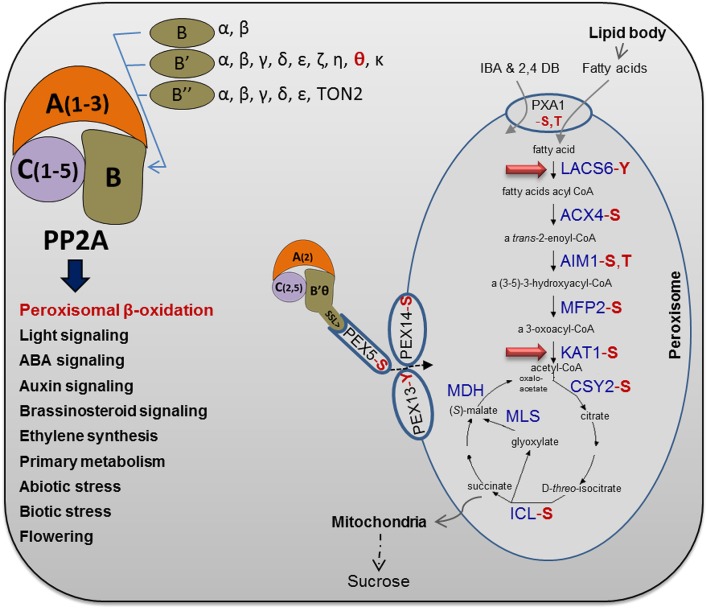
Very recent studies experimentally identified phosphopeptides related to peroxisomal β-oxidation, glyoxysomal cycle, and PTS1 import proteins and showed that several of the proteins involved in β-oxidation are in fact phosphorylated (Fig. 9; Supplemental Table S1). Most of the identified proteins are phosphorylated at Ser or Thr residues and, hence, can be potential substrates for PP2A. In an initial attempt to identify peroxisomal targets of PP2A, total protein extracts were prepared from 7-d-old b′θ-1 mutant and wild-type Col-0 seedlings grown in Suc-free medium on short days. Tryptic digests were enriched on a titanium dioxide column and subjected to liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis on an LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific). The analysis identified nine phosphopeptides from eight enzymes involved in TAG metabolism. Importantly, these phosphopeptides were uniquely detected in the b’θ-1 mutant (Table I). All except one of the peptides were phosphorylated at Ser or Thr residues, thus constituting potential substrates for PP2A. Three of the phosphopeptides were previously unidentified, two of which matched acyl-CoA synthetase (LACS5), which is involved in the first step of peroxisomal β-oxidation, and one of which matched peroxisomal ketoacyl-CoA thiolase (KAT1; Fig. 9; Table I). These two peroxisomal β-oxidation-related proteins show high potential to be PP2A substrates in peroxisomes (Fig. 9).
Figure 9.
Brief overview of Arabidopsis PP2A heterotrimer complexity and functions. The PP2A heterotrimer subunits are presented together with functions of PP2A in Arabidopsis (Farkas et al., 2007; Uhrig et al., 2013). Regulation of peroxisomal β-oxidation is a unique function of PP2A that was approved in this study. Several peroxisomal proteins, involved in fatty acid β-oxidation and the glyoxylate cycle, have been shown experimentally to be phosphorylated (Supplemental Table S1), as highlighted in red for the type of phosphorylated amino acid (S, Ser; T, Thr; and Y, Tyr). As highlighted by the red arrows, β-oxidation steps including LACS5 and KAT1 were found to be potential substrates for a peroxisomal PP2A heterotrimer based on the differences between phosphoproteomes of the b’θ mutant and the wild type (this work). For more details, see Table I. The protein abbreviations are as follows: ACX, acyl-CoA oxidase; AIM, enoyl-CoA hydratase; CSY, citrate synthase; ICL, isocitrate lyase; MDH, malate dehydrogenase; MFP, multifunctional protein; and MLS, malate synthase. In glyoxysomes, the acetyl-CoA produced by fatty acid β-oxidation is used as a substrate for the glyoxylate cycle, where succinate is produced and transferred to mitochondria to be metabolized for energy production. Moreover, the peroxisomal ATP-binding cassette transporter (PXA1) and the PEX5, PEX13, and PEX14 were found to be phosphorylated (Supplemental Table S1). The pathway of fatty acid β-oxidation and glyoxylate cycle is modified from the online database (Karp et al., 2005).
Table I. List of identified β-oxidation–related enzymes uniquely detected in the b′θ-1 phosphoproteome.
LC-MS/MS analysis of phosphoproteome differences between b′θ-1 and wild-type seedlings grown in Suc-free media. The phosphopeptides related to peroxisomal β-oxidation enzymes were found once in three replicates. The detected peptide and its q value are presented. The phosphorylated residue is shown in boldface for the type of amino acid (S, Ser; T, Thr; and Y, Tyr). AGI, Arabidopsis Genome Initiative; GDSL, glycine, aspartate, serine, and leucine around the active site serine.
| AGI | Annotation | Acronym | Detected Peptide | q Value |
|---|---|---|---|---|
| AT4G11030 | Long-chain acyl-CoA synthetase5 | LACS5 | GQSHVAASPFCDK | 0.025 |
| GVMISNESIVTITTGVMHFLGNVNASLSEK | 0.037 | |||
| AT1G04710 | Peroxisomal 3-ketoacyl-coa thiolase1 | KAT1 | INVNGGAIAIGHPLGATGAR | 0.049 |
| AT1G73610 | GDSL esterase/lipase | ACELFVNQGAAMFNQQLSADIDNLGATFPGAK | 0.03 | |
| AT3G14820 | GDSL-like lipase/acylhydrolase superfamily protein | LNNMALHFNSKLSSSLDTLK | 0.015 | |
| AT4G28570 | Long-chain fatty alcohol dehydrogenase family protein | LHPVLMTWGYFPEKDSEFSGKMYEGGIITSVHHMNDTESGCK | 0.045 | |
| AT2G19450 | Diacylglycerol O-acyltransferase1 | DGAT | SLANAADKANPEVSYYVSLKSLAYFMVAPTLCYQPSYPR | 0.013 |
| AT1G31480 | Phosphatidic acid-preferring phospholipase A1 | NPDYVGKISIYGHSLGSVLSYDILCHQHNLSSPFPMDSVYK | 0.017 | |
| AT1G10670 | ATP-citrate lyase A-1 | MRALGDDIGVPIEVYGPEATMTGICK | 0.036 |
DISCUSSION
Inspection of stably transformed plants confirmed the presence of B′θ in peroxisomes, as was seen in both leaves and roots (Fig. 3). No PP2A subunits had been reported as part of the proteome of isolated peroxisomes when analyzed by mass spectrometry (MS; Fukao et al., 2002; Reumann et al., 2007, 2009; Eubel et al., 2008; Quan et al., 2013), but sensitive immunological methods used in this work showed the presence of the PP2A C subunits in extracts of highly purified peroxisomes (Fig. 1A).
Localization of PP2A C and A subunits to peroxisomes was further confirmed by BiFC analysis of B′θ in combination with each of the two other types of subunits. Out of the five C subunits present in Arabidopsis, only C2 and C5 gave positive BiFC results (Fig. 2, II–IV). Interactions of B′θ with C5 were more persistent than with C2 under some conditions, indicating that C5 was the more important C subunit in peroxisomes. We also concluded that C2 interacts with B′θ but probably under special conditions because they failed to interact in onion epidermal cells.
PP2A C and A subunits are constitutively expressed in many organisms; interestingly, however, the Arabidopsis C5 subunit has very variable expression levels in different tissues and under different conditions. In accordance with the BiFC results, microarray expression data also pointed to C5 as the more important candidate compared with C2 for interacting with B′θ (Supplemental Figs. S2, S4, and S5). Of the three A subunits, only A2 targeted peroxisomes (Fig. 2I). Altogether, the experiments strongly support the hypothesis that a full PP2A complex is present in peroxisomes. Peroxisomes are distinguished from other organelles by their capacity to import fully folded proteins. It has been shown for some proteins that, although they do not have a PTS, they are able to enter peroxisomes by being carried or piggybacked by proteins that do possess a PTS (Lee et al., 1997; Kaur et al., 2009). A and C subunits lack endogenous PTSs and are generally found throughout the cell (Koroleva et al., 2005; Michniewicz et al., 2007; Tran et al., 2012). Their import into peroxisomes clearly depended on the B′θ PTS1 because truncation of the B′θ C terminus abolished their peroxisomal targeting (Fig. 2, V and VI). Taken together, we conclude that A2, C2, and C5 interact with B′θ in the cytosol and are imported by a piggybacking process into peroxisomes.
Although B′θ is conserved among plants, a peroxisomal targeting signal was only predicted in very close relatives of Arabidopsis (Fig. 5). It is not clear why these plants in particular have evolved a PP2A specialized for peroxisomes. When speculating on a function for PP2A in Arabidopsis peroxisomes, it became clear that expression data as well as features of b′θ knockout plants were compatible with PP2A influencing fatty acid β-oxidation. Peroxisomal β-oxidation is activated during senescence, where it is thought to have a role in the turnover of lipids derived from membranes of various organelles (Mano and Nishimura, 2005). The expression of most genes encoding peroxisomal proteins involved in β-oxidation is up-regulated during senescence (Supplemental Fig. S5). Furthermore, as an oilseed plant, Arabidopsis needs to rapidly metabolize TAGs into sugars for translocation to growing tissues during the early seedling stage. B′θ loss-of-function mutants showed a sugar dependence phenotype typically of impaired fatty acid breakdown (Fig. 6). Plant peroxisomes are also the site of some steps in hormone synthesis, which requires a cycle of β-oxidation. It is known that the effect of the protohormones IBA and 2,4-DB on primary root length will be weakened in β-oxidation–defective mutants because of the inability of such mutants to metabolize these compounds into IAA and 2,4-dichlorophenoxyacetic acid (Mano and Nishimura, 2005). The b′θ mutant seedlings showed resistance to both IBA and 2,4-DB (Fig. 7). The defect in β-oxidation was further confirmed by following the level of C20:1 after germination, which was clearly different in the b′θ mutant and the wild type (Fig. 8D). This aligns with the reported defect in peroxisome function in kat2 mutant (Fig. 9; Supplemental Table S1). In the kat2 mutant, lipid body degradation and TAG mobilization during seedling growth were severely hampered, and it was suggested that the metabolism of peroxisomes would also influence the metabolism in the lipid bodies (Germain et al., 2001).
Transport of fatty acids and protoauxins into peroxisomes has been suggested to occur by the same transporter, PXA1 (Zolman et al., 2001), which has five known Ser/Thr phosphorylation sites (Supplemental Table S1); hence, a possible function of PP2A would be to activate PXA1. Another possibility is that PP2A acts directly on enzymes in the peroxisomal matrix that are involved in β-oxidation. This possibility is supported in this study by comparing phosphoproteomes of the b′θ-1 mutant and the wild type during seedling growth. Our experiments revealed two peroxisomal matrix proteins (LACS5 and KAT1) involved in β-oxidation that were phosphorylated in the mutant but not in the wild type. Finally, other possibilities, such as PP2A effects on import of enzymes of β-oxidation, cannot be excluded.
Overall, this work shows that Arabidopsis and some close relatives have developed a PP2A B subunit (B′θ) with a PTS1 peroxisomal targeting signal. Using Arabidopsis, we have shown that this signal is essential for targeting all three types of PP2A building blocks, C, A, and B subunits, to peroxisomes. Examination of b′θ mutant lines strongly supports a function of PP2A in peroxisomal processing of protoauxins and fatty acids.
MATERIALS AND METHODS
Gene Cloning for in Planta Expression
Arabidopsis (Arabidopsis thaliana) cDNAs for PP2A-B′θ (At1g13460 [U85787]), C subunits (At1g59830 [U1180], At1g10430 [U16346], At3g58500 [U14624], and At1g69960 [U82420]), and all A subunits (At1g25490 [U16272], At3g25800 [U24842], and At1g13320 [U25062]) were obtained from the Arabidopsis Biological Resource Center. The coding sequences of the genes were cloned by PCR and subcloned via SpeI and XhoI restriction sites into the multicolor BiFC vectors, pVYCE, pVYCE(R), pVYNE, and pVYNE(R) (Waadt et al., 2008), to create protein fusions with the N- or C-terminal fragments of YFP (Venus variant) at the N-terminal end or C-terminal end of the gene (Supplemental Fig. S1B).
Transformation and Microscopy
Arabidopsis (Col-0) wild-type seeds were sown on soil and stratified for 2 d, then transferred to short days (8 h of light) and irrigated once a week with complete Hoagland solution. Plants 3 to 4 weeks old were used for protoplast isolation. For BiFC analysis in Arabidopsis mesophyll protoplasts, plasmids for PP2A subunits and peroxisomal marker proteins were cotransformed into protoplasts using a polyethylene glycol transformation protocol (Yoo et al., 2007). For BiFC analysis in epidermal cells, plasmids were transiently introduced into onion (Allium cepa ‘Kepaløk’) by a helium-driven particle accelerator (PDS/1000; Bio-Rad) with all basic adjustments set according to the manufacturer’s recommendations. The bombarded onion cells were incubated for 1 to 2 d in the dark at room temperature and then observed under the microscope. Peroxisomal markers used were gMDH-CFP that contains the PTS2 sequence of gMDH from cucumber (Cucumis sativus; Kim and Smith, 1994) and Red fluorescent protein-SKL (Matre et al., 2009). Microscopy was carried out on a Nikon TE-2000U inverted fluorescence microscope equipped with an Exfo X-Cite 120 fluorescence illumination system and filters for CFP (exciter, S436/10; and emitter, S470/30), YFP (exciter, HQ500/20; and emitter, S535/30), and a special red chlorophyll autofluorescence filter set (exciter, HQ630/39; and emitter, HQ680/40; Chroma Technology). Images were captured using a Hamamatsu Orca ER 1394 cooled CCD camera. Volocity II software (Improvision, Coventry, UK) was used to capture 0.5 Z-sections to generate extended focus images. Images were subsequently processed for optimal presentation with Adobe Photoshop version 9.0 (Adobe Systems).
Generation of Transgenic Lines
To generate an overexpressor line of N-terminally fused protein, gene-specific primers were used to amplify full-length Arabidopsis cDNA of B′θ, and were cloned in the pGEMT-EYFP vector. Subsequently, the available EYFP-fused fragment was excised and subcloned into the 35S promoter-containing binary vector pBA002. The construct was transformed into Agrobacterium tumefaciens nopalin strain from A208 via the freeze-thaw method. Arabidopsis Col-0 was transformed by the floral dip method (Clough and Bent, 1998). Seeds were screened on one-half-strength Murashige and Skoog agar plates containing 10 µg mL−1 phosphinotricin. Phosphinotricin-resistant seedlings were selected 10 to 14 d after germination. Successful transformation was validated by isolation of genomic DNA of the primary transformants and using primers upstream (forward) and downstream (reverse) of the EYFP-B′θ insertion sites in the vector.
Peroxisome Isolation and Western-Blot Analysis
Leaves were harvested from 4- to 6-week-old Arabidopsis plants grown in a 12-h photoperiod, and peroxisomes were isolated as described previously (Reumann et al., 2007). Leaf extracts were obtained by direct grinding of leaf tissue in Laemmli buffer. Leaf extracts or peroxisomes were mixed with loading buffer (Laemmli buffer) and resolved in SDS-PAGE. Western-blot detection of PP2A C subunit was performed with the specified monoclonal antibodies for the methylated and demethylated C-terminal end of PP2A C subunits (Santa Cruz Biotechnology; sc-81603 and sc-80990). Western-blot comparison between isolated peroxisomes and total protein extract was performed with polyclonal anti-Catalase and anti-PEPC (Agrisera AB). Equal amounts of protein from total leaf extract and precipitated peroxisomal protein were used for the comparison. Total protein extraction from 6-week-old leaves was performed using the TCA-acetone method. Proteins from isolated peroxisomal preparations were precipitated by the chloroform-methanol method (Wessel and Flügge, 1984). Protein concentrations were measured using the Bradford protein assay (Bradford, 1976).
Plant Growth
Arabidopsis mutant lines SAIL_300_B01 (Sessions et al., 2002), SALK_201885 (Alonso et al., 2003), SAIL_890_C10 (Sessions et al., 2002), and FLAG_185E04 (Samson et al., 2002), for B′θ, were obtained from the European Arabidopsis Stock Centre. Mutant selection was done by PCR using primers for T-DNA insertion lines recommended at the Salk Institute Web site SIGnAL (http://signal.salk.edu/tdnaprimers.2.html). The pex14 mutant (Orth et al., 2007; Zhang and Hu, 2010) seeds were kindly provided by Jianping Hu (Michigan State University). For plant material grown on soil, seeds were sown directly in a regular soil plant mix. Seeds were stratified at 4°C for 2 d and then transferred to standard growth conditions. During germination and growth, plants were placed at 22°C under artificial light in short-day (8 h of light/16 h of dark), 12 h of light/12 h of dark, or long-day (16 h of light/8 h of dark) regimens.
Sugar Dependence and 2,4-DB/IBA Response Assays
For sugar dependence analysis, seeds of the wild-type Col-0 and mutants were sown on one-half-strength LS medium with vitamins (Caisson Labs) with or without 1% (w/v) Suc and stratified in the dark at 4°C for 2 d before being transferred to darkness or short-day conditions (8 h of light/16 h of dark) at 20°C. Six-day-old seedlings were scanned using an EPSON scanner (http://www.epson.com), and hypocotyl length was measured using ImageJ (Schneider et al., 2012; http://rsb.info.nih.gov/ij/). To study the response to protoauxins, 2,4-DB or IBA was added to one-half-strength LS agar medium with 0.5% (w/v) Suc. Seeds were sown and stratified for 2 d, then kept in continuous light for 7 d. Length of the primary root was measured using ImageJ (Zolman et al., 2001; Zhang and Hu, 2010).
qRT-PCR
Total RNA was isolated using the RNeasy Plant Mini Kit and treated with on-column DNaseI digestion (Qiagen). Afterward, RNA was treated in solution with DNaseI (Life Technologies) and precipitated by ammonium acetate (7.5 m) and ethanol. The cDNA was synthesized using the High Capacity cDNA Archive Kit (Applied Biosystems). Real-time PCR reactions were assayed using the ABI 7300 Fast Real-Time PCR System. The reaction volume was 25 µL containing 12.5 µL of TaqMan buffer (Applied Biosystems; includes 6-Carboxyl-X-Rhodamine as a passive reference dye), 8.75 µL of water, 2.5 µL of cDNA, and 1.25 µL of primers. Primers were predesigned TaqMan Gene Expressions for B′θ At1g13460 (At02780124; Applied Biosystems identification no.). The TaqMan assay is based on a light signal from each transcript copy formed and allows the comparison of expression levels between the various genes. Standard cycling conditions (2 min at 50°C, 10 min at 95°C, and 40 cycles altering between 15 s at 95°C and 1 min at 60°C) were used. Real-time PCR products were analyzed by Sequence Detection Software version 1.3. The comparative threshold cycle method for relative quantification was used with ACTIN8 At1g49240 (At02270958).
Fatty Acid Analysis
Total fatty acids were extracted (Browse et al., 1986) and quantified by gas chromatography. Heptadecanoic acid methyl ester was obtained from Sigma-Aldrich and served as the internal standard. In brief, the plant materials were treated by 1 mL of 1 m methanolic HCL reagent (dilution 3 m methanolic HCL [Sigma-Aldrich] by methanol + 5% [v/v] 2,2-dimethoxypropane) and heated (80°C) for 1 h in glass tubes sealed with Teflon. After the samples were allowed to cool down, 0.3 and 1 mL of hexane and 0.9% (w/v) sodium chloride, respectively, were added and vigorously vortexed to release fatty acid methyl esters. Centrifugation at 1000g for 30 s was applied to allow transfer of the upper hexane layer for gas chromatography-mass spectrometry/flame ionization detector analysis. An Agilent GC6890 connected to an Agilent 5975 mass selective detector (Agilent Technologies) and equipped with a J&W HP-5MS column (19091S-433; Agilent Technologies) was used. Flame ionization detector quantification using the column type (J&W HP-5, 19091J-413; Agilent Technologies) was applied. The obtained chromatograms were compared with different databases, and the individual fatty acids were monitored based on retention time and quantified by internal standard calibration.
Phosphoproteomics
Total protein from 7-d-old seedlings of the wild type and b′θ-1 (grown on one-half-strength LS medium without Suc) was extracted using TCA-acetone precipitation. The precipitate was dissolved in 100 µL of 50 mm NH4HCO3 and 20 mm dithiothreitol and heated for 30 min at 60°C. The sample was cooled to room temperature, and 20 µL of 1 m iodoacetamide was added and incubated for 20 min in the dark at room temperature. The reduced and alkylated sample was treated with trypsin (1:50 trypsin:protein ratio), and the proteins were digested overnight at 37°C. For total protein analysis, the digested sample was made acidic, and a C18 purification step was performed (Rappsilber et al., 2003). For phosphopeptide enrichment, the sample was treated with titanium dioxide (Engholm-Keller and Larsen, 2011). For both total protein and phosphopeptide analyses, the dried peptides were dissolved in 60 µL of 0.1% (v/v) formic acid prior to LC-MS/MS.
MS Analysis
The peptides were analyzed on a LC-MS/MS platform consisting of an Easy-nLC 1000 ultra high performance liquid chromatography system (Thermo Scientific/Proxeon) interfaced with an LTQ-Orbitrap Elite hybrid eastrometer (Thermo Scientific) via a nanospray ESI ion source (Proxeon). Peptides were injected onto a C-18 trap column (Acclaim PepMap100, 75-μm × 2-cm i.d., C18, 5 μm, 100 Å; Thermo Scientific) and further separated on a C-18 analytical column (Acclaim PepMap100, 75-μm × 50-cm i.d., C18, 3 μm, 100 Å; Thermo Scientific) using a 240-min gradient (total protein) from 7% to 32% CH3CN, 0.1% formic acid, or 120-min gradient (phosphopeptides) from 0% to 30% CH3CN and 0.1% formic acid at a flow rate of 250 nL min−1.
Eluted peptides were analyzed on the LTQ-Orbitrap Elite hybrid mass spectrometer operating in positive ion and data-dependent acquisition mode using the following parameters: electrospray voltage, 1.9 kV; collision-induced dissociation fragmentation with normalized collision energy, 35; and automatic gain control target value, 1E6 for Orbitrap MS and 1E3 for MS/MS scans. Each MS scan (mass-to-charge ratio 400:1,600) was acquired at a resolution of 120,000 full width at half maximum, followed by 20 MS/MS scans triggered for intensities above 500, at a maximum ion injection time of 200 ms for MS and 50 ms for MS/MS scans. For phosphopeptide analysis, a multistage activation was used. Neutral loss masses specified for multistage activation fragmentation were 32.67 and 49 D.
Raw data files of spectra obtained were analyzed in the Proteome Discoverer version 1.4 (Thermo Scientific) platform using the SEQUEST HT search engine (Thermo Scientific). Files were searched against the UniProt Arabidopsis protein database using the following search parameters: Enzyme was specified as trypsin with a maximum of three miscleavages allowed; precursor mass tolerance was 20 parts per million, and fragment mass tolerance was 0.5 Daltons. False discovery rate was 0.05. Carbamidomethylated Cys (57.021 Daltons) was set as fixed modification, whereas phosphorylations at Ser/Thr/Tyr as well as Met oxidation were set as variable modifications.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of PP2A in peroxisomes and constructing BiFC recombinant genes.
Supplemental Figure S2. Clustering and coexpression of Arabidopsis PP2A subunits C and A with B′θ.
Supplemental Figure S3. Identification of PP2A complexes in peroxisomes of onion epidermal cells.
Supplemental Figure S4. Gene expression profiles of Arabidopsis PP2A subunits C, A, and B′θ in response to various stimuli.
Supplemental Figure S5. PP2A subunits C5 and B′θ show high expression during senescence.
Supplemental Figure S6. PP2A B’θ homolog sequence alignments.
Supplemental Table S1. Summary of phosphorylated peroxisomal proteins involved in β-oxidation, glyoxylate cycle, and PTS1 import.
Supplementary Material
Glossary
- PP2A
protein phosphatase 2A
- C
catalytic
- A
scaffolding
- B
regulatory
- EYFP
enhanced yellow fluorescent protein
- PEPC
phosphoenolpyruvate carboxylase
- BiFC
bimolecular fluorescence complementation
- cDNA
complementary DNA
- YFP
yellow fluorescent protein
- gMDH
glyoxysomal malate dehydrogenase
- CFP
cyano fluorescent protein
- RT
reverse transcription
- T-DNA
transfer DNA
- Col-0
Columbia
- TAG
triacylglycerol
- IBA
indole-3-butyric acid
- 2,4-DB
2,4-dichlorophenoxybutyric acid
- IAA
indole-3-acetic acid
- C20:1
eicosenoic acid
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- LACS5
acyl-CoA synthetase
- KAT1
ketoacyl-CoA thiolase
- MS
mass spectrometry
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the Norwegian Research Council (grant no. 213853/F20 to C.L.).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coca M, San Segundo B (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63: 526–540 [DOI] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Baudhuin P (1966) Peroxisomes (microbodies and related particles). Physiol Rev 46: 323–357 [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Creyghton MP, Bernards R (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 1795: 1–15 [DOI] [PubMed] [Google Scholar]
- Engholm-Keller K, Larsen MR (2011) Titanium dioxide as chemo-affinity chromatographic sorbent of biomolecular compounds: applications in acidic modification-specific proteomics. J Proteomics 75: 317–328 [DOI] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696 [DOI] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J (2004) Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M, Shockey J, Werber M, Wolter FP, Heinz E (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J 32: 93–103 [DOI] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28: 1–12 [DOI] [PubMed] [Google Scholar]
- Heidari B, Matre P, Nemie-Feyissa D, Meyer C, Rognli OA, Møller SG, Lillo C (2011) Protein phosphatase 2A B55 and A regulatory subunits interact with nitrate reductase and are essential for nitrate reductase activation. Plant Physiol 156: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari B, Nemie-Feyissa D, Kangasjärvi S, Lillo C (2013) Antagonistic regulation of flowering time through distinct regulatory subunits of protein phosphatase 2A. PLoS ONE 8: e67987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353: 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen CH, Manfield IW, Michalopoulos I, Pinney JW, Willats WGT, Gilmartin PM, Westhead DR (2006) The Arabidopsis co-expression tool (ACT): a WWW-based tool and database for microarray-based gene expression analysis. Plant J 46: 336–348 [DOI] [PubMed] [Google Scholar]
- Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahrén D, Tsoka S, Darzentas N, Kunin V, López-Bigas N (2005) Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res 33: 6083–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Reumann S, Hu J (2009) Peroxisome Biogenesis and Function. The Arabidopsis Book 7: e0123, doi/10.1199/tab.0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Smith SM (1994) Expression of a single gene encoding microbody NAD-malate dehydrogenase during glyoxysome and peroxisome development in cucumber. Plant Mol Biol 26: 1833–1841 [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH (2005) High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J 41: 162–174 [DOI] [PubMed] [Google Scholar]
- Kragler F, Lametschwandtner G, Christmann J, Hartig A, Harada JJ (1998) Identification and analysis of the plant peroxisomal targeting signal 1 receptor NtPEX5. Proc Natl Acad Sci USA 95: 13336–13341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Mullen RT, Trelease RN (1997) Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Antolín-Llovera M, Ferrero S, Closa M, Arró M, Ferrer A, Boronat A, Campos N (2011) Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 23: 1494–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B, Miquel M, Somerville C, Browse J (1990) Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet 80: 234–240 [DOI] [PubMed] [Google Scholar]
- Lillo C, Kataya AR, Heidari B, Creighton MT, Nemie-Feyissa D, Ginbot Z, Jonassen EM (2014) Protein phosphatases PP2A, PP4 and PP6: mediators and regulators in development and responses to environmental cues. Plant Cell Environ 37: 2631–2648 [DOI] [PubMed] [Google Scholar]
- Lingner T, Kataya AR, Antonicelli GE, Benichou A, Nilssen K, Chen XY, Siemsen T, Morgenstern B, Meinicke P, Reumann S (2011) Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23: 1556–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Nishimura M (2005) Plant peroxisomes. Vitam Horm 72: 111–154 [DOI] [PubMed] [Google Scholar]
- Matre P, Meyer C, Lillo C (2009) Diversity in subcellular targeting of the PP2A B’eta subfamily members. Planta 230: 935–945 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J (2007) The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Yang P, Cassin-Ross G, Kaur N, Switzenberg R, Aung K, Li J, Hu J (2013) Proteome analysis of peroxisomes from etiolated Arabidopsis seedlings identifies a peroxisomal protease involved in β-oxidation and development. Plant Physiol 163: 1518–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rehling P, Albertini M, Kunau WH (1996) Protein import into peroxisomes: new developments. Ann N Y Acad Sci 804: 34–46 [DOI] [PubMed] [Google Scholar]
- Reumann S. (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. (2011) Toward a definition of the complete proteome of plant peroxisomes: Where experimental proteomics must be complemented by bioinformatics. Proteomics 11: 1764–1779 [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, Linka N, Switzenberg R, Wilkerson CG, Weber AP, et al. (2009) In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Singhal R (2014) Isolation of Leaf Peroxisomes from Arabidopsis for Organelle Proteome Analyses. In JV Jorrin-Novo, S Komatsu, W Weckwerth, S Wienkoop, eds, Plant Proteomics, Ed 2, Vol 1072. Humana Press, pp 541–552 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørhagen K, Laxa M, Peterhänsel C, Reumann S (2013) The emerging role of photorespiration and non-photorespiratory peroxisomal metabolism in pathogen defence. Plant Biol (Stuttg) 15: 723–736 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Bargues M, Carrasco P, Pérez-Alonso M, Paricio N (2002) Molecular characterization and evolution of the protein phosphatase 2A B’ regulatory subunit family in plants. Plant Physiol 129: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A, Moorhead GB (2012) Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J 71: 263–272 [DOI] [PubMed] [Google Scholar]
- Trotta A, Konert G, Rahikainen M, Aro EM, Kangasjärvi S (2011b) Knock-down of protein phosphatase 2A subunit B’γ promotes phosphorylation of CALRETICULIN 1 in Arabidopsis thaliana. Plant Signal Behav 6: 1665–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A, Wrzaczek M, Scharte J, Tikkanen M, Konert G, Rahikainen M, Holmström M, Hiltunen HM, Rips S, Sipari N, et al. (2011a) Regulatory subunit B’gamma of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol 156: 1464–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Labandera AM, Moorhead GB (2013) Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci 18: 505–513 [DOI] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RB, Wanders RJ, Tager JM (1992) Biochemistry of peroxisomes. Annu Rev Biochem 61: 157–197 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, et al. (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang X, Hu J (2010) The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.