Kelch repeat F-box proteins physically interact with phenylpropanoid biosynthetic enzymes, and regulate the production of (poly)phenolics and plant tolerance to ultraviolet irradiation.
Abstract
Phenylpropanoid biosynthesis in plants engenders myriad phenolics with diverse biological functions. Phenylalanine ammonia-lyase (PAL) is the first committed enzyme in the pathway, directing primary metabolic flux into a phenylpropanoid branch. Previously, we demonstrated that the Arabidopsis (Arabidopsis thaliana) Kelch domain-containing F-box proteins, AtKFB01, AtKFB20, and AtKFB50, function as the negative regulators controlling phenylpropanoid biosynthesis via mediating PAL’s ubiquitination and subsequent degradation. Here, we reveal that Arabidopsis KFB39, a close homolog of AtKFB50, also interacts physically with PAL isozymes and modulates PAL stability and activity. Disturbing the expression of KFB39 reciprocally affects the accumulation/deposition of a set of phenylpropanoid end products, suggesting that KFB39 is an additional posttranslational regulator responsible for the turnover of PAL and negatively controlling phenylpropanoid biosynthesis. Furthermore, we discover that exposure of Arabidopsis to ultraviolet (UV)-B radiation suppresses the expression of all four KFB genes while inducing the transcription of PAL isogenes; these data suggest that Arabidopsis consolidates both transcriptional and posttranslational regulation mechanisms to maximize its responses to UV light stress. Simultaneous down-regulation of all four identified KFBs significantly enhances the production of (poly)phenols and the plant’s tolerance to UV irradiation. This study offers a biotechnological approach for engineering the production of useful phenolic chemicals and for increasing a plant’s resistance to environmental stress.
Phenylpropanoid biosynthesis converts l-Phe into diverse aromatic metabolites, collectively termed (poly)phenolics. In excess of 8,000 phenolic structures have been reported, and they are widely dispersed throughout the plant kingdom (Strack, 1997). These diverse phenylpropanoid metabolites are categorized structurally into different classes, such as benzenoids, coumarins, flavonoids/anthocyanins, stilbenes, hydroxycinnamates, lignans, and lignin (Vogt, 2010; Fraser and Chapple, 2011). Many of them have important physiological functions essential for plant growth and development and plant-environment interactions. For example, a variety of small molecule-soluble phenolics, such as hydroxycinnamates and their derivatives, flavonoids, and anthocyanins, serve as the UV light screen, protecting plants from radiation damage, as pigments attracting pollinators, or as signaling molecules mediating plant-microbe interactions (Dixon and Paiva, 1995). Pharmacological studies reveal that many phenolics possess significant antioxidative activity and, therefore, have considerable health-promoting properties; they potentially can be used as nutraceuticals or dietary supplements for chemopreventing human diseases or disorders (Crozier et al., 2009). Owing to their perceivable bioactivities, the methanolic soluble (poly)phenols have been targets of designed metabolic engineering for decades (Winkel-Shirley, 2001; Martin, 2013).
Phenylalanine ammonia-lyase (PAL; EC 4.3.1.5) is the first rate-limiting enzyme in the phenylpropanoid biosynthetic pathway. It channels l-Phe from the primary metabolic pool to the synthesis of trans-cinnamic acid that then is further transformed into different classes of phenolic products (Bate et al., 1994; Cochrane et al., 2004). PAL activity was found in all higher plants analyzed to date and in some fungi and a few bacteria (Hodgins, 1971; Fritz et al., 1976; Xiang and Moore, 2005). PAL is regulated at different levels (Zhang and Liu, 2014). It is well evident in transcriptional regulation; its transcripts readily respond to developmental cues and environmental factors (Dixon and Paiva, 1995). In addition, PAL receives feedback regulation via biosynthetic intermediates or exogenous chemical signals (Bolwell et al., 1986; Blount et al., 2000). Furthermore, our recent study revealed that Arabidopsis (Arabidopsis thaliana) PAL isozymes are (poly)ubiquitinated in vivo, and three Kelch domain-containing F-box proteins (KFBs), the structural component of the S-phase kinase-associated protein1-Cullin-F-box (SCF)-type E3 protein-ubiquitin ligase complex, specifically interact with PAL isozymes and mediate their ubiquitination and subsequent protein degradation. Such KFB-mediated PAL proteolysis acts as an intrinsic regulatory mechanism controlling phenylpropanoid synthesis (Zhang et al., 2013a).
Arabidopsis has around 700 genes encoding the F-box proteins that function as a structural component of the SCF complex (Gagne et al., 2002; Xu et al., 2009). The SCF complex is one type of E3 ubiquitin-protein ligase mediating protein ubiquitylation and subsequent degradation via the 26S proteasome pathway. The role of the F-box proteins in the SCF complex is to interact selectively with target proteins, thereby conferring specificity on the complex (del Pozo and Estelle, 2000). Depending upon the presence of additional protein-protein interaction domains near the C terminus, this large protein family can be divided further into several subfamilies. As one class of F-box protein, the Kelch domain-containing F-box proteins originally were identified from Drosophila melanogaster (Xue and Cooley, 1993; Bork and Doolittle, 1994). KFBs contain a consensus of Kelch motifs characterized by four highly conserved residues, two adjacent Gly residues (G) and a pair of Tyr (Y) and Trp (W) residues, separated by about six amino acids (Adams et al., 2000; Prag and Adams, 2003). In Arabidopsis, more than 90 KFB homologous genes were identified (Sun et al., 2007; Xu et al., 2009), but only a few encoded proteins have been characterized. These include three highly similar Arabidopsis proteins (ZEITLUPE, Flavin-binding kelch domain F-box protein, and LOV KELCH PROTEIN2) involved in flowering timing and/or circadian control (Nelson et al., 2000; Han et al., 2004; Yasuhara et al., 2004; Imaizumi et al., 2005), while recently we identified KFB01, KFB20, and KFB50 as the posttranslational regulators controlling PAL ubiquitination and proteolysis (Zhang et al., 2013a).
In this study, we demonstrate that Arabidopsis KFB39, a close homolog of KFB50, also interacts physically with PAL isozymes and attenuates PAL stability. Up- or down-regulation of KFB39 expression conversely affects the accumulation/deposition of a set of phenylpropanoids, including flavonols, anthocyanins, condensed tannins (CTs), sinapic esters, and lignin, suggesting that KFB39, like its homolog, is an additional negative regulator that acts directly at the enzyme level for phenylpropanoid biosynthesis. Furthermore, we reveal that simultaneous down-regulation of all four identified KFBs in Arabidopsis significantly enhances the concentration of PAL enzyme and its activity and the production of (poly)phenols; consequently, it greatly increases the tolerance of plants to UV radiation. Our study offers an additional biotechnological approach for engineering plants with enhanced production of useful chemicals or increased resistance to environmental stress.
RESULTS
Arabidopsis KFB39 Interacts Physically with PAL Isozymes
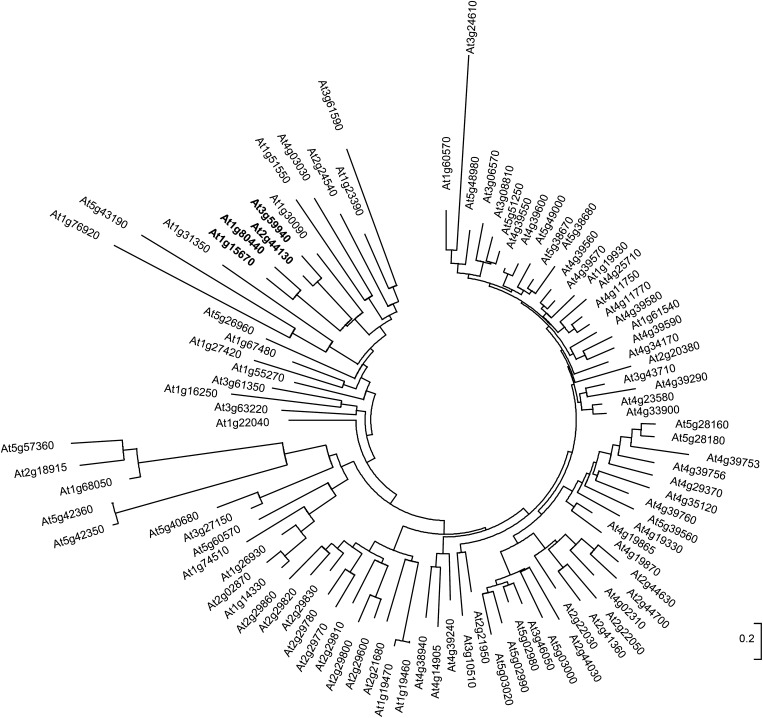
Previously, through in vitro and in vivo assays, we found that Arabidopsis KFB01, KFB20, and KFB50 interact with four PAL isozymes (Zhang et al., 2013a). Using AtKFB01, AtKFB20, and AtKFB50 genes (At1g15670, At1g80440, and At3g59940) as the bait to conduct BLAST searches against genomic sequences of Arabidopsis, and referring to previous reports (Gagne et al., 2002; Xu et al., 2009), we retrieved 98 KFB homolog genes. Among them, we found a close homolog, AtKFB39 (encoded by At2g44130), that shares 62.8% amino acid sequence identity with AtKFB50 and 28% and 26.3%, respectively, with AtKFB01 and AtKFB20 (Supplemental Fig. S1). Phylogenetic analysis indicated that AtKFB39 together with AtKFB01, AtKFB20, and AtKFB50 were classified within the same clade but separated into two different subgroups (Fig. 1). AtKFB39 and AtKFB50 have longer polypeptides (containing 409 and 418 amino acids, respectively) and share a higher similarity at the amino acid level to each other than to AtKFB01 (359 amino acids) or AtKFB20 (354 amino acids); therefore, they are phylogenetically grouped together and distinct from both KFB01 and KFB20 (Fig. 1; Supplemental Fig. S1).
Figure 1.
Phylogenetic analysis of Kelch domain-containing F-box proteins in Arabidopsis. AtKFB01 (At1g15670), AtKFB20 (At1g80440), AtKFB39 (At2g44130), and AtKFB50 (At3g59940) are in boldface.
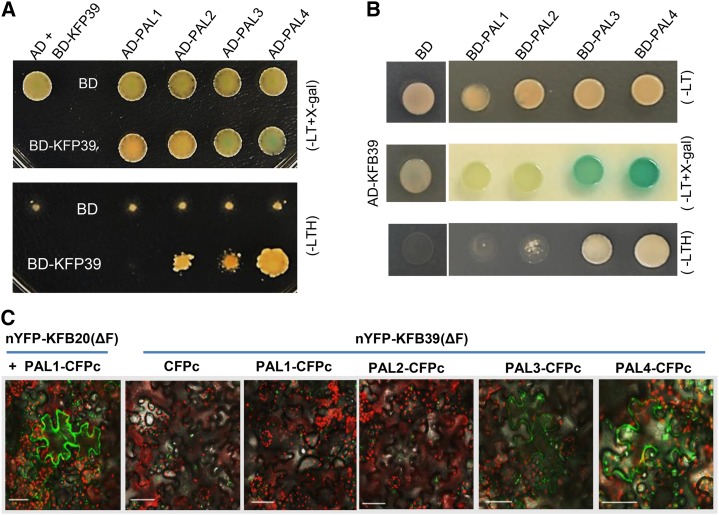
The high sequence identity of AtKFB39 and AtKFB50 inspired us to further examine whether AtKFB39 also has similar functions to AtKFB50 and is involved in controlling PAL stability. We first examined whether it interacts physically with Arabidopsis PAL isozymes via yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) assay. The results showed that the yeast transformants harboring the expressed AtKFB39 and AtPAL4 or AtPAL3 effectively activated the expression of the Lactose Operon gene Z gene that results in a coloration reaction in medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. Also, this coexpression led to a substantial growth rescue of yeast colonies on selective medium, indicating a strong interaction of AtKFB39 with AtPAL4 or AtPAL3 (Fig. 2, A and B). By contrast, AtKFB39 showed only a weak interaction with AtPAL2 and a nearly indiscernible interaction with AtPAL1 in Y2H assay under the same conditions (Fig. 2, A and B). These in vitro data suggest that AtKFB39, similar to AtKFB01, AtKFB20, and AtKFB50, also interacts physically with PAL isozymes, albeit at differential interaction strengths with different PAL isoforms.
Figure 2.
Interaction of PAL isozymes with the KFB39 protein. A, Y2H assay between BD-KFB39 and AD-GADT7-PAL (PAL1–PAL4). Yeasts were grown on two-amino acid dropout (−Leu/Trp; −LT) synthetic defined (SD) medium supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; top) or on three-amino acid dropout (−Leu/Trp/His; −LTH) selective medium (bottom). B, Domain-swapping validation of the interaction between KFB39 and PAL1 to PAL4 in Y2H assays. The assays were conducted between AD-KFB39 and BD-PAL (PAL1–PAL4). Yeasts were grown on SD (−Leu/Trp) medium in the absence (top) and presence (middle) of X-gal and on SD (−Leu/Trp/His) selective medium (bottom). C, BiFC assay for the interactions of PAL isozymes with KFB39 protein in transiently expressed tobacco leaves. PAL1 to PAL4 were fused with CFPc at their C termini, and the truncated KFB39(ΔF) was fused with nYFP at its N terminus. The truncated KFB39(ΔF) fusion construct was coinfiltrated with PAL-CFPc (or CFPc alone) in tobacco leaves. The pair of nYFP-KFB20(ΔF) with PAL1-CFPc served as a positive control. Bars = 50 μm.
Fluorescence complementation assay for each AtPAL isozyme with the truncated KFB39, wherein we removed its conserved F-box domain to prevent its associating with the scaffold component of the SCF complex, in tobacco (Nicotiana tabacum) leaves revealed that coexpression of PAL3- or PAL4-cyan fluorescence protein C-terminus (CFPc) with the N-terminus of yellow fluorescence protein (nYFP)-truncated KFB39 yielded detectable chimeric fluorescence signals in tobacco cells, whereas the coexpression of PAL1- or PAL2-CFPc with nYFP-truncated KFB39 produced no fluorescence (Fig. 2C). In contrast, as the positive control, the coexpression of PAL1-CFPc with nYFP-truncated KFB20, which was demonstrated previously to interact with all four AtPALs (Zhang et al., 2013a), yielded strong chimeric fluorescence (Fig. 2C). These data, consistent with the Y2H assay, imply that AtKFB39 indeed interacts with AtPAL isozymes in vivo, but the affinity for its interaction varies with different PAL isoforms.
In addition to PAL isozymes, the key enzymes in flavonoid-anthocyanin biosynthetic pathways, including chalcone synthase (CHS), dihydroflavonol-4-reductase, flavonol synthase (FLS), and anthocyanidin synthase, also were paired with AtKFB39 and coexpressed in yeast to examine their potential interactions. Among them, the coexpression of AtCHS and AtKFB39 resulted in a partial rescue of the growth of yeast cells on selective medium (Supplemental Fig. S2), indicating that a certain degree of physical interaction occurs between two proteins in vitro.
AtKFB39 Attenuates the Stability of PAL Isozymes
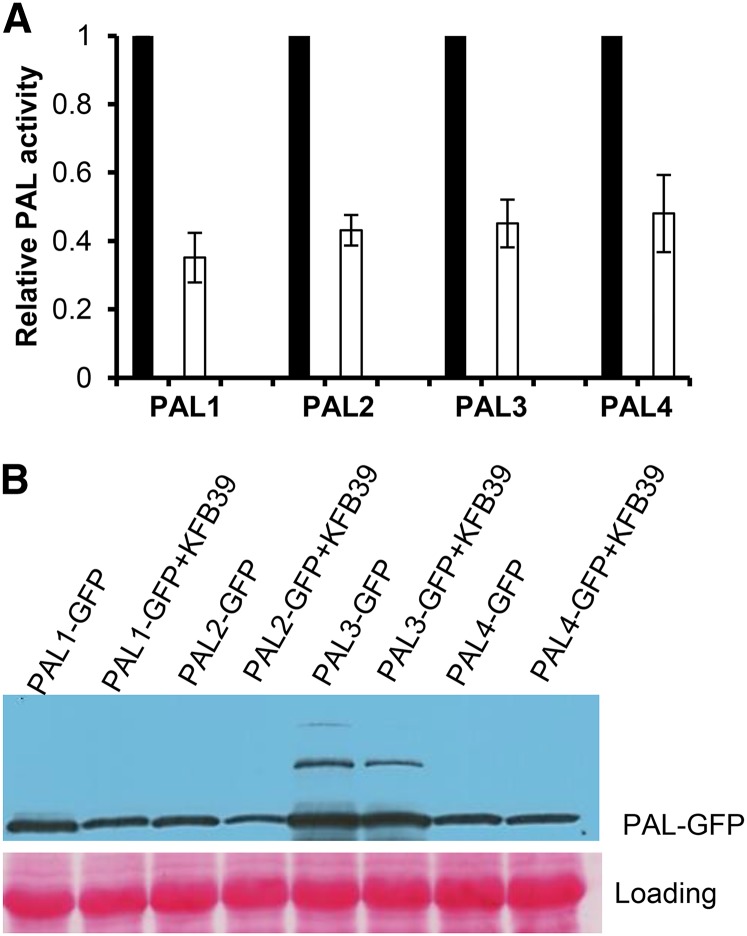
Since AtKFB39 interacts physically with PAL isozymes (particularly with PAL3 and PAL4) in both Y2H and bimolecular fluorescence complementation (BiFC) assays, we investigated whether the interaction also can induce the turnover of PAL enzymes as do AtKFB01, AtKFB20, and AtKFB50 (Zhang et al., 2013a). The full length of AtKFB39 was coexpressed transiently with each of four AtPAL isozymes in tobacco leaves. Surprisingly, we observed that the pairwise coexpression led to an approximately 50% reduction of the total extractable PAL activity, not only in the combination of AtKFB39 with AtPAL3-GFP or PAL4-GFP but also with the AtPAL1-GFP or PAL2-GFP fusion (Fig. 3A). Immunoblots further revealed that the immunosignals for all PAL-GFP fusions (approximately 100 kD), except for AtPAL3, exhibited a readily discernible reduction in the protein extracts from leaves coexpressing PAL with full-length AtKFB39 (Fig. 3B). The 100-kD immunoband of AtPAL3-GFP remained relatively unchanged; however, a couple of upper bands, presumably its incompletely denatured dimer or tetramer, showed an obvious reduction (Fig. 3B). These data suggest that the action of AtKFB39 in planta attenuates the stability of all four PAL isoforms.
Figure 3.
Attenuation of PAL stability and activity by AtKFB39. A, Relative activity of expressed PAL isozymes in crude extracts from tobacco leaves transiently expressing PAL alone (black bars) or coexpressing both PAL and KFB39 (white bars). B, Immunoblot detection of the stability of PAL-GFP proteins using the anti-GFP antibody in tobacco leaves coexpressing PAL-GFP and KFB39 genes. Ponceau S staining served as a control for the amount of protein loading.
When coexpressing AtKFB39 and AtCHS in tobacco leaves, however, we failed to detect the changes in CHS activity and stability, implying that the observation of a physical interaction of AtKFB39 and CHS in the Y2H assay might be artificial or of no biological significance.
The Up-Regulation of AtKFB39 in Arabidopsis Impairs the Synthesis of Phenylpropanoids
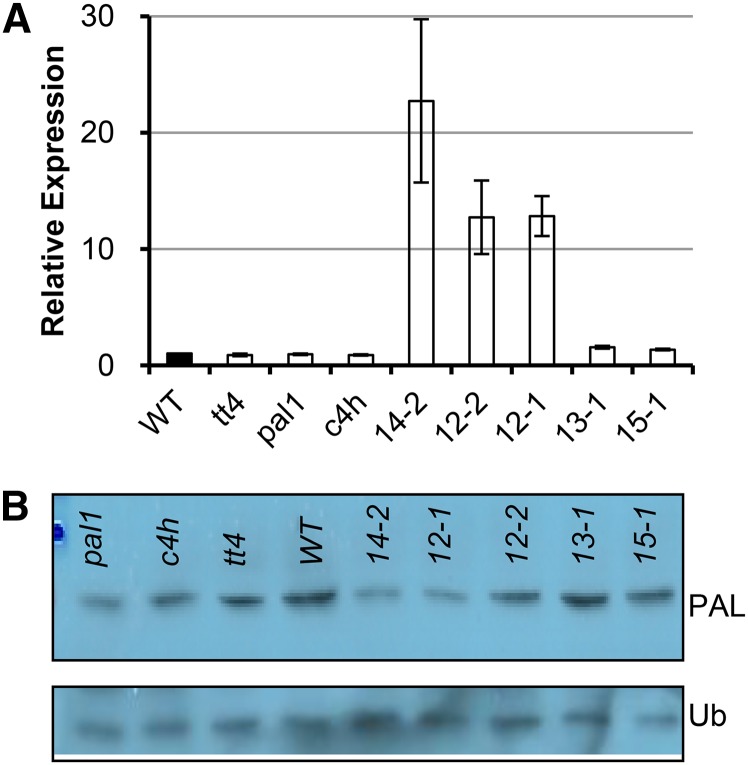
To further examine the roles of AtKFB39 in vivo for their potential in regulating phenylpropanoid biosynthesis, we stably overexpressed KFB39 in Arabidopsis driven by a constitutive 35S promoter. The expression level of AtKFB39 was higher in most of the generated transgenic lines than in the wild type, albeit with a large variation among the independent transgenic events (Fig. 4A). Overexpression of AtKFB39 in Arabidopsis did not significantly affect the transcript levels of the endogenous PAL isogenes, nor for AtKFB01, AtKFB20, and AtKFB50 (Supplemental Fig. S3A), whereas the level of the PAL protein, detected with an anti-PAL peptide antibody, was reduced substantially in the transgenic lines bearing highly expressed AtKFB39 (Fig. 4B; Supplemental Fig. S3B).
Figure 4.
Overexpression of KFB39 and reduction of PAL protein level in Arabidopsis. A, Relative expression level of KFB39 in 5-DAG seedlings of the selected KFB39 overexpression lines. Its level in transgenic lines, after normalization to that of the UBIQUITIN10 control gene, was expressed relative to its level in an empty-vector control line. tt4, pal1, and c4h were the controls. WT, Wild type. B, Level of endogenous PAL protein in the KFB39 overexpression lines. The PAL proteins were detected with anti-PAL peptide antibody. The monoubiquitin immunoblot against the anti-ubiquitin antibody (Ub) served as a control for the amount of protein loading.
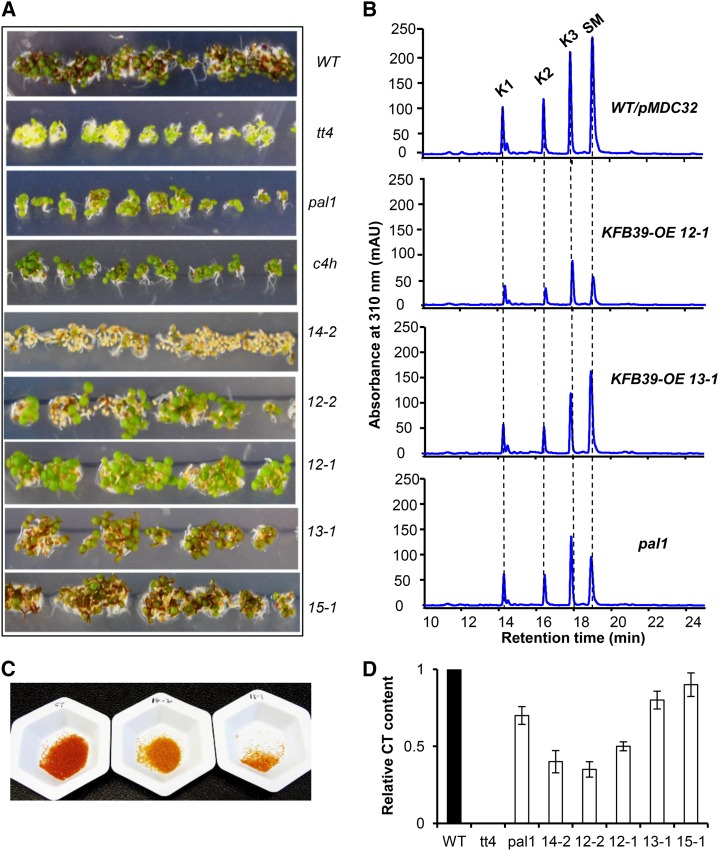
Growing Arabidopsis seedlings in one-half-strength Murashige and Skoog (MS) medium containing 4% (w/v) Suc, the wild-type ones (5 d after germination [DAG]) developed a perceivable violet-red anthocyanin pigmentation (Fig. 5A); in contrast, the purple pigmentation in the emerging seedlings harboring the highly overexpressed AtKFB39 was clearly faded, and the development of purple color was reciprocally proportional to the expression level of KFB39 transgene (Figs. 4A and 5A). In seedlings of lines 12-1, 12-2, and 14-2 that have strongly expressed KFB39, the purple coloration was hardly discernible, much resembling or comparable to those of the transfer DNA insertion mutant lines deficient in PAL1 or cinnamate 4-hydroxylase (C4H; Fig. 5A). However, in the weak expression lines of AtKFB39 (i.e. lines 13-1 and 15-1), the purple color was clearly visible and its intensity was similar to that of wild-type plants (Figs. 4A and 5A). These data suggest that the expression level of AtKFB39 is directly correlated to its effect on the synthesis of anthocyanins. Moreover, most seeds of line 14-2, which has the strongest expression of AtKFB39, and some seeds of lines 12-1 and 12-2 failed to germinate under the same growth conditions as the wild type (Fig. 5A; Supplemental Fig. S4), reminiscent of a previous report by Rohde at al. (2004) of the deleterious effect on seed viability when total PAL level is lowered to a certain threshold. Quantifying flavonoids and phenolic esters accumulated in the emerging seedlings confirmed that overexpression of AtKFB39 resulted in up to a 50% reduction of both anthocyanins and sinapoyl esters and up to an 80% decrease in flavonol content in some transgenic lines (Fig. 5B; Table I). After growing plants for 4 weeks in soil, we collected their rosette leaves and measured the level of anthocyanin accumulated. The content in the rosette leaves of transgenic lines 12-1 and 12-2 was less than half of that in the wild-type plants (Supplemental Fig. S3C). Together, these data suggest that up-regulation of KFB39 reduces the cellular level of the PAL enzyme, thereby lowering the accumulation of soluble phenolics.
Figure 5.
Alteration of the accumulation of flavonoids, anthocyanins, and CTs in KFB39 overexpression plants. A, The 5-DAG seedlings of the KFB39 overexpression lines with control and tt4, pal1, and c4h mutants grown on one-half-strength MS medium containing 4% (w/v) Suc. Note the change in their pigmentation. B, Representative UV-HPLC profiles of methanolic soluble phenolics in the KFB39 overexpression lines. K1, Kaempferol 3-O-[6′′-O-(rhamnosyl)glucoside] 7-O-rhamnoside; K2, kaempferol 3-O-glucoside 7-O-rhamnoside; K3, kaempferol 3-O-rhamnoside 7-O-rhamnoside; SM, sinapoyl malate. Metabolites were extracted from 0.1 g fresh weight of seedlings with 1 mL of 80% methanol, and 10-µL extracts were injected for HPLC profiling. C, Seed coat brown pigmentation of the control (left), KFB39 overexpression line 14-2 (middle), and KFB39 overexpression line 12-1 (right). D, Relative content of CTs in the mature seeds of KFB39-OE lines. Data represent means ± sd from three biological repeats. WT, Wild type.
Table I. Accumulation levels of anthocyanins, flavonols, and sinapoyl esters in the T3 generation of KFB39 overexpression, kfb triple, and kfb triple/KFB39-RNAi Arabidopsis seedlings (5 DAG).
Data are means ± se from three biological repeats. Since line 14-2 exhibited a severe defect in seed germination, there was not enough material for analysis. n/a, Not detectable; ND, not determined.
| Lines | Anthocyanins | Kaempferol Flavonols | Sinapic Acid Esters |
|---|---|---|---|
| pmol g−1 fresh wt | pmol mg−1 fresh wt | pmol mg−1 fresh wt | |
| c4h | 15.7 ± 0.69 | 80.9 ± 5.4 | 713 ± 16.3 |
| pal1 | 25.9 ± 1.02 | 130 ± 5.6 | 1,293 ± 16.3 |
| tt4 | n/a | n/a | 2,877 ± 80.5 |
| Wild type/pMDC32 | 70.5 ± 3.24 | 298 ± 3.9 | 2,145 ± 18.9 |
| KFB39/OE(14-2) | ND | ND | ND |
| KFB39/OE(12-1) | 30.1 ± 1.53 | 54.2 ± 4.8 | 1,019 ± 12.7 |
| KFB39/OE(12-2) | 35.8 ± 1.73 | 166 ± 5.3 | 1,716 ± 16.7 |
| KFB39/OE(13-1) | 40.5 ± 3.48 | 133 ± 7.9 | 1,602 ± 15.8 |
| KFB39/OE(15-1) | 51.4 ± 1.96 | 156 ± 9.8 | 1,864 ± 20.6 |
| Triple | 200.9 ± 5.68 | 200 ± 6.7 | 4,993 ± 15.3 |
| Triple/KFB39RNAi-1 | 240.1 ± 14.32 | 220 ± 4.8 | 5,790 ± 26.7 |
| Triple/KFB39RNAi-3 | 278.6 ± 13.23 | 264 ± 5.8 | 6,300 ± 37.8 |
| Triple/KFB39RNAi-6 | 260.9 ± 15.18 | 278 ± 4.7 | 5,237 ± 12.3 |
| Triple/KFB39RNAi-7 | 250.5 ± 13.18 | 260 ± 3.9 | 5,055 ± 21.3 |
In Arabidopsis, anthocyanin and CTs (or proanthocyanidins) share common intermediates from the phenylpropanoid pathway. Thus, we expected that an altered level of the PAL enzyme also would affect the accumulation of CTs in the seed coats. Indeed, we observed a clear reduction in both the brown color that appeared on the seed coats and the measured CT content from the mature seeds of the KFB39 overexpression lines (Fig. 5, C and D); with the strong expression of KFB39, the transgenic lines 14-2 and 12-2 contained only about 40% of the wild-type CT content. The reduction was even more severe than what resulted from a single pal1 disruption (Fig. 5D).
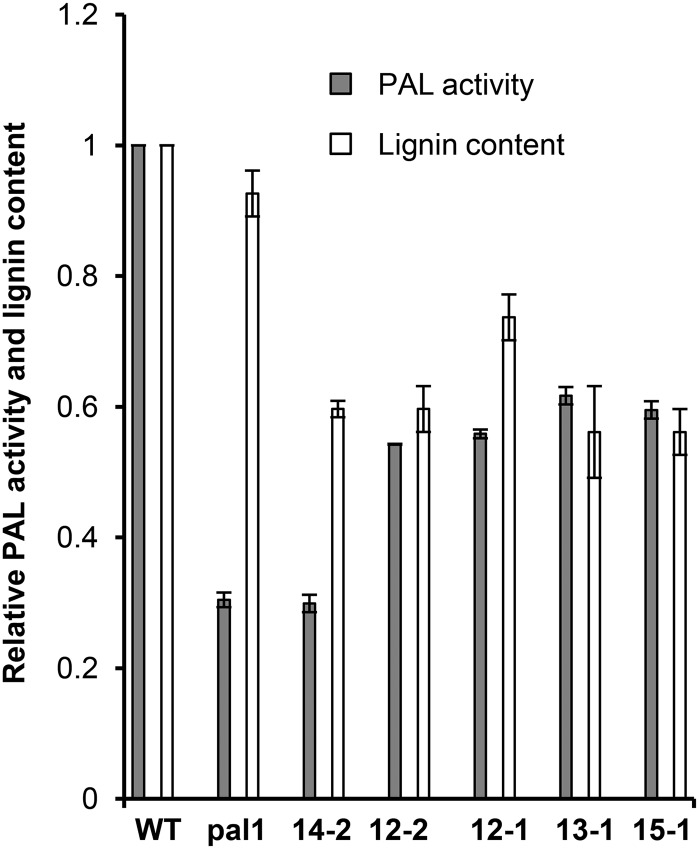
Similarly, the measured acetyl-bromide lignin content in the stems of 12-week-old transgenic lines was also reduced by up to 40% of the level of the wild-type controls (Fig. 6), although when plants approached the fully mature stage, the difference of lignin content in the cell walls of KFB39 transgenic and control lines became small. Notably, the lignin reductions did not correlate well with the decrease of total PAL activity in transgenic plants. Line 14-2, which had the most severe reduction in PAL activity (Fig. 6) and a severe alteration in the color of the seed coat and CT content (Fig. 5), did not exhibit the strongest reduction in lignin content (Fig. 6). Conversely, lines 13-1 and 15-1, which have a lower level of FKB39 overexpression (Fig. 4) and lesser effects on the accumulation of anthocyanins and flavonols in seedlings and CT deposition in seed coats (Table I; Fig. 5), did show a similar level of lignin reduction to line 14-2.
Figure 6.
Reduction of total PAL activity and lignin content in KFB39 overexpression lines. PAL activity was measured using protein crude extracts from leaves of 4-week-old transgenic plants. Acetyl-bromide lignin was determined using the cell walls of 12-week-old Arabidopsis stems. Data represent means ± sd with three biological repeats. The enzyme activity and lignin content of the control plants were set at 1. WT, Wild type.
The Down-Regulation of AtKFB39 Conversely Enhances the Synthesis of Phenylpropanoids
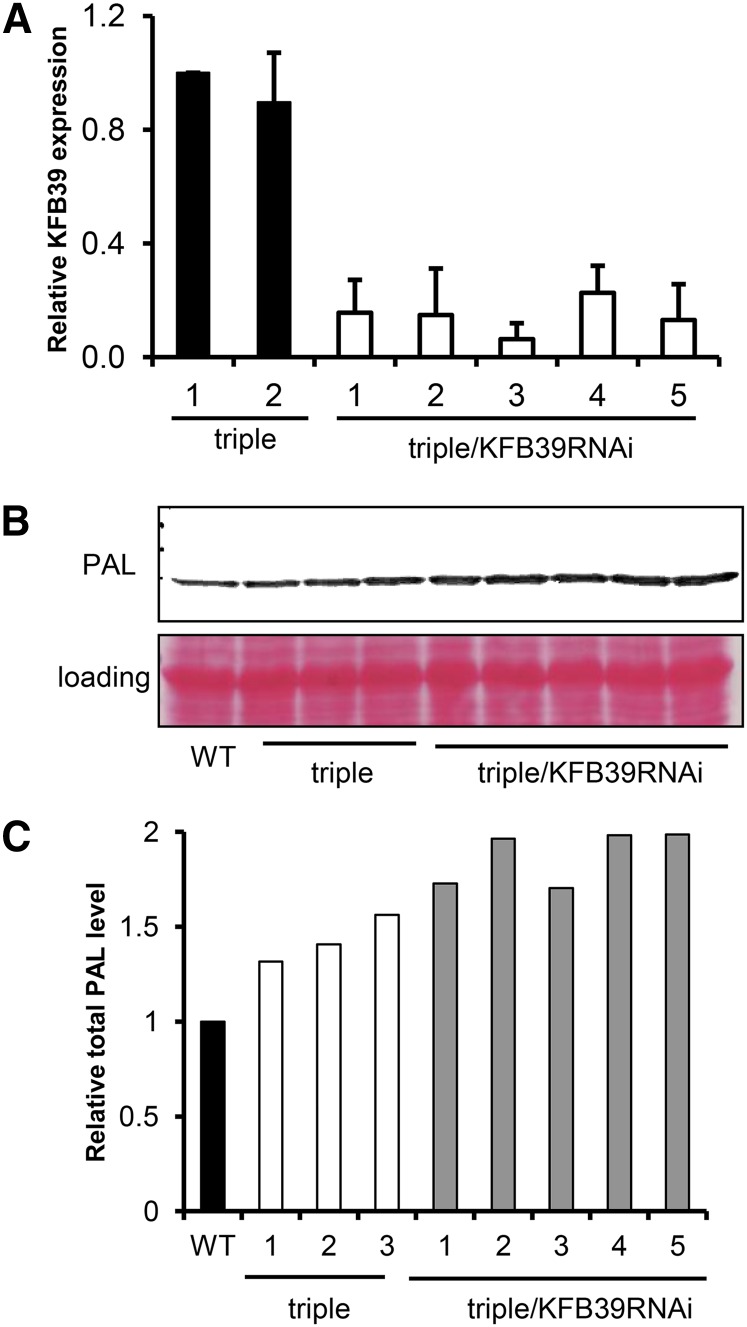
Previously, we discovered that AtKFB01, AtKFB20, and AtKFB50 exhibit partially redundant activity in controlling PAL stability; the high-order atkfb01/20/50 triple mutant increased the total protein level and activity of PAL nearly three times compared with the single mutant (Zhang et al., 2013a). Since there are no available mutant lines for AtKFB39, to further evaluate KFB39’s in vivo functions, we created transgenic lines that suppress AtKFB39 expression by 80% of its level in the atkfb01/20/50 triple mutant via RNA interference (RNAi)-mediated gene silencing in the triple mutant background (Fig. 7A). Immunoblot analysis showed that the accumulated levels of PAL protein in the atkfb01/20/50/KFB39-RNAi lines could further increase by about 30% the level of the atkfb01/20/50 triple mutant (Fig. 7, B and C), suggesting that KFB39 has an additive effect on PAL stability.
Figure 7.
Suppression of KFB39 and alteration of PAL protein level in KFB39 RNAi-mediated gene-silencing lines. A, qRT-PCR analysis of the expression level of KFB39 in RNAi silencing lines. Data represent means ± sd with three biological replicates. B, Immunoblot detection of the cellular concentration of the PAL protein using anti-PAL antibody. C, Relative PAL protein level in RNAi silencing lines calculated based on B. WT, Wild type.
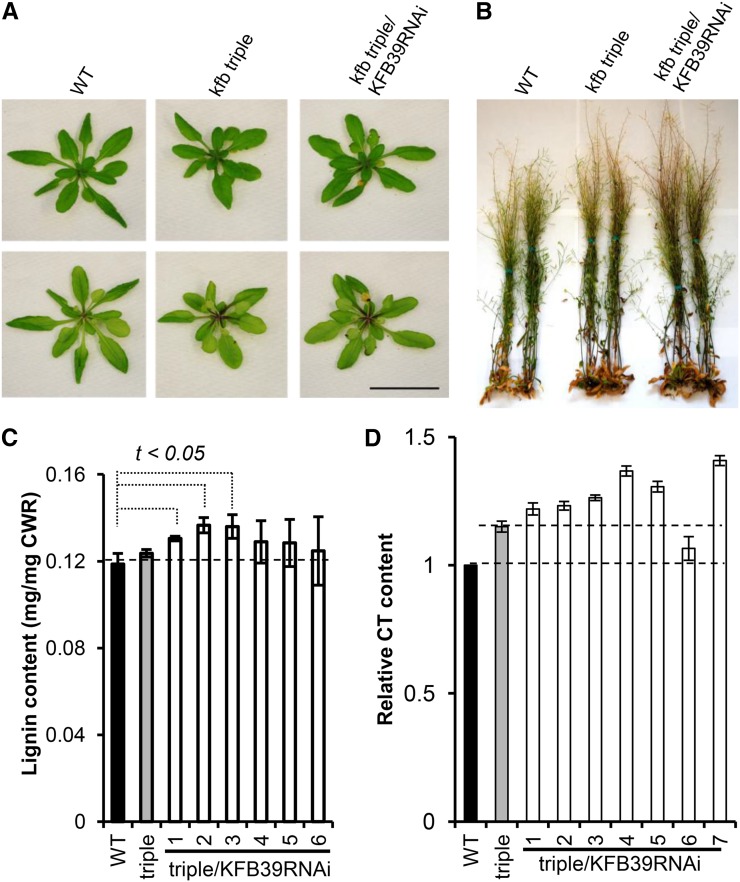
Measuring the contents of accumulated flavonoids/anthocyanins and soluble phenolic esters in atkfb01/20/50/KFB39-RNAi seedlings, we found that the level of accumulated anthocyanins increased nearly 4-fold, reaching 280 pmol g−1 fresh weight, and that the sinapoyl ester content rose 3-fold to 6.3 nmol mg−1 fresh weight compared with the level in the wild-type control plants; down-regulation of KFB39 further raised the accumulation of both anthocyanins and phenolic esters compared with those in the atkfb01/20/50 triple mutant (Table I). Consistently, the atkfb01/20/50/KFB39-RNAi lines at 4 to 5 weeks of age developed much more intense purple pigmentation on the adaxial and abaxial surfaces of their rosette leaves and in the leaf petioles than the wild-type control (Fig. 8A). When plants approached the mature stage (approximately 10 weeks), both the triple mutant and atkfb01/20/50/KFB39-RNAi lines grew stronger and taller than the control lines, and their inflorescence stems accumulated more intense purple pigmentation (Fig. 8B). Measuring total lignin content in the harvested mature stems revealed a slight but statistically significant increase in the atkfb01/20/50/KFB39-RNAi plants (approximately 15% increase compared with the control plants; Fig. 8C). The CT content in mature seeds of most RNAi transgenic lines increased 20% to 30% compared with the control plants; silencing KFB39 further enhanced the CT accumulation compared with the atkfb01/20/50 triple mutant (Fig. 8D).
Figure 8.
Alteration of the accumulation of anthocyanins, CTs, and lignin in KFB39-RNAi transgenic lines. A, Effect of KFB39 silencing on anthocyanin pigmentation. The top row shows the front view of 4-week-old control (left), kfb01/20/50 triple mutant (middle), and kfb01/20/50/KFB39-RNAi transgenic (right) rosettes; the bottom row shows the opposite view of control, mutant, and transgenic rosettes, illustrating the enhanced anthocyanin pigment in the petioles of the triple mutant and RNAi transgenic lines. Bar = 5 cm. B, Approximately 10-week-old kfb01/20/50/KFB39-RNAi transgenic plants showing pigmentation in the inflorescence stems. C, Change in the total lignin content in kfb01/20/50/KFB39-RNAi transgenic Arabidopsis. The acetyl-bromide lignin was measured in independent T2 lines. The data represent means ± sd with three biological replicates. CWR, Cell wall residue. D, Relative content of CTs in mature seeds of kfb01/20/50 triple mutant and kfb01/20/50/KFB39-RNAi transgenic plants. The data represent means ± sd from four biological repeats. WT, Wild type.
Suppression of KFB Mitigates Damage from UV-B Irradiation in Plants
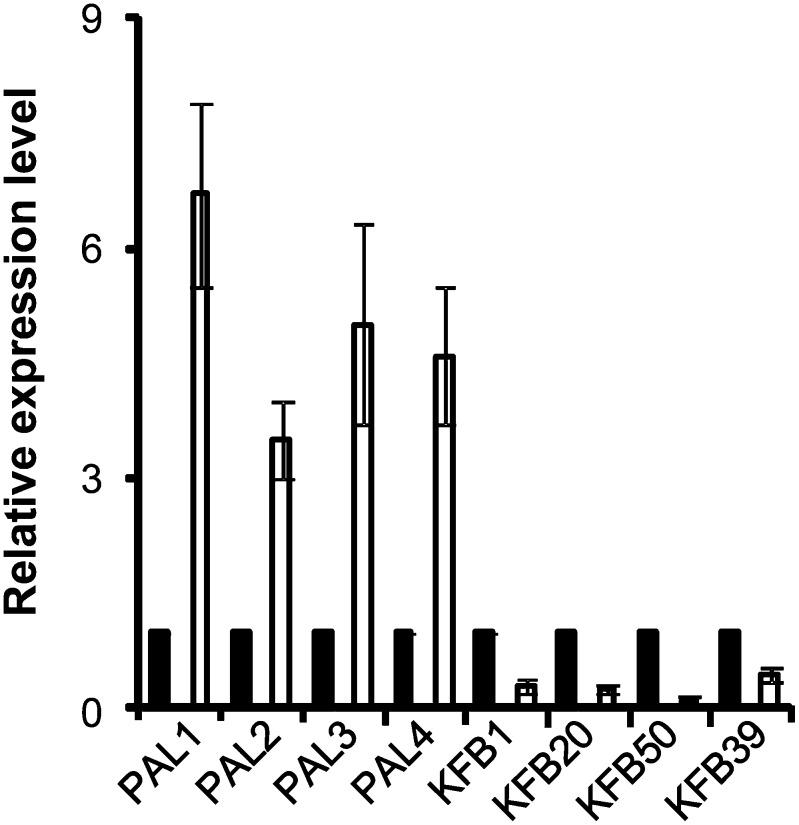
The plant’s UV-B protection process involves the up-regulation of key genes in the phenylpropanoid-flavonoid pathway, including PAL and CHS, and the consequent accumulation of a set of phenylpropanoid metabolites, including flavonoids and sinapic esters, that are well established as being the sunscreen photoprotecting plants from UV-B radiation (Li et al., 1993; Jansen et al., 1998). To evaluate whether KFBs are involved in sensing/transducing UV light signals, which, in turn, can regulate PAL and phenylpropanoid biosynthesis, we quantified the transcription changes of AtKFB01, AtKFB20, AtKFB39, and AtKFB50 and of the PAL isogenes in 5-week-old Arabidopsis growing under normal full-range light supplemented with UV-B radiation. As expected, PAL expression was sharply up-regulated by 3- to 6-fold after 12 h of exposure. By contrast, the expression levels of all four KFBs were suppressed substantially (Fig. 9); AtKFB50 showed the strongest suppression, with its levels of expression decreased by more than 5-fold compared with that in the control plants. These data imply that during the process of protection against UV-B light, both transcriptional and posttranslational controls are implemented in plants (i.e. by up-regulating the transcription of the key biosynthetic gene PAL and reducing the level of KFBs, thus modulating the stability of PAL protein, to maximize the synthesis of phenylpropanoids as a defense mechanism). Among four identified KFB proteins, KFB50 might play a dominant role in regulating plant responses to UV-B light stress (Fig. 9).
Figure 9.
Relative expression levels of PALs and KFBs in Arabidopsis exposed to UV light. Five-week-old Arabidopsis was illuminated with cool-white fluorescent light supplemented with UV-B radiation from an FS20-UVB lamp (0.2 J m−2 s−1) for 12 h. The error bars represent sd from three biological repeats. The expression level in untreated samples (black bars) was set at 1.
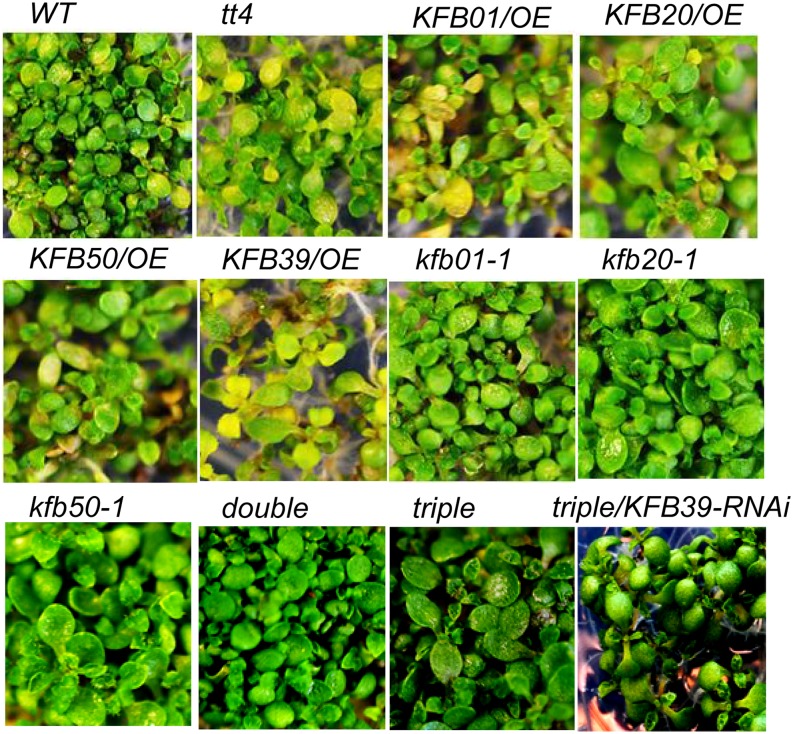
To further assess the significance of KFBs in affecting the capacity of plant UV-B light protection, we exposed 5-DAG seedlings of the KFB overexpression plants and the knockout/RNAi lines to UV-B light for 8 h. After they recovered for 2 d, we examined their morphology. As Figure 10 (and Supplemental Fig. S4) shows, the wild-type control and the pal1, c4h, and transparent testa4 (tt4; chs) mutants in which phenylpropanoid biosynthesis is impaired showed obvious sensitivity to UV-B light exposure; a portion of their seedlings wilted or turned yellowish in color. Overexpression of any of the four KFBs made plants more sensitive to UV-B irradiation; a larger population of seedlings showed foliar bleaching or lesions compared with the wild-type control plants (Fig. 10; Supplemental Fig. S4). In sharp contrast, the seedlings of KFB disruption lines, particularly the double and triple mutant lines and the triple mutant with KFB39 RNAi, grew normally and did not exhibit obvious foliar injury under the same conditions as those for the wild-type and overexpression plants.
Figure 10.
Growth and injury of Arabidopsis seedlings under UV-B exposure. Seedlings at 5 DAG were grown on one-half-strength MS agar medium and illuminated with full-wavelength light supplemented with UV-B light from an FS20-UVB lamp (0.2 J m−2 s−1) for 8 h, then left to recover for 2 d. WT, Wild type.
DISCUSSION
AtKFB39 Is an Additional Negative Regulator Directly Controlling PAL Stability and Phenolic Synthesis
As the first rate-limiting enzyme in the phenylpropanoid pathway, PAL receives multiple levels of regulation in response to developmental cues and environmental challenges (Zhang and Liu, 2015). Previously, we found that a group of Arabidopsis KFB proteins (AtKFB01, AtKFB20, and AtKFB50) interact with PAL, thereby mediating PAL ubiquitination and subsequent degradation, by which plants negatively regulate phenylpropanoid biosynthesis (Zhang et al., 2013a). AtKFB39 shares 62.8% sequence identity with AtKFB50 at the amino acid level, and they are phylogenetically clustered (Fig. 1), indicating that the pair of genes encoding these two KFB proteins might arise from gene duplication. In both Y2H and BiFC assays, KFB39 exhibits a perceivable physical interaction with PAL isozymes, suggesting that this KFB member, like its homolog KFB50, is involved in mediating PAL proteolysis. Manipulating KFB39 expression indeed engenders a notable and reciprocal alteration of the cellular concentration of PAL protein and its activity (Figs. 3, 4, and 7) and affects the synthesis or accumulation of an array of phenylpropanoid end products, including flavonoids/anthocyanins, CTs, phenolic esters, and lignin (Table I; Figs. 5, 6, and 8). These data are evidence that AtKFB39 represents an additional member of the F-box protein family that functions as the structural component of the canonic SCF-type protein-ubiquitin ligase specific for regulating PAL turnover and, thus, negatively controlling phenylpropanoid synthesis.
Similar to AtKFB50, AtKFB39 exhibits a preferential interaction with AtPAL4 (and AtPAL3) but has little or no detectable interaction with AtPAL1 or PAL2 in both Y2H and BiFC assays (Fig. 2). This biochemical property contrasts with those of AtKFB01 and KFB20, both of which interact with all AtPAL isoforms in vitro and in vivo (Zhang et al., 2013a). Amino acid sequence alignment reveals a large difference at the C termini of AtKFB39 (and AtKFB50) compared with AtKFB01 or AtKFB20 (Supplemental Fig. S1). The C termini of F-box proteins are well known for specifying substrate specificity (del Pozo and Estelle, 2000; Lechner et al., 2006). However, interestingly, when AtKFB39 was coexpressed with AtPAL1 or PAL2 isoforms in tobacco leaves, the overexpressed PAL was degraded and total PAL activity was reduced (Fig. 3). This apparent discrepancy of the physical interaction assay and the in vivo action of AtKFB39 can be interpreted in different ways. One simple explanation is that the detection limitation of both Y2H and BiFC assays for physical interaction may not be able to completely mirror the weak interaction (with AtPAL1 or PAL2) in planta, but when AtKFB39 is overexpressed in cells, the hyperaccumulation (i.e. the high abundance) of the KFB39 protein may functionally compensate for its weak interaction with PAL1 or PAL2, thereby leading to the ubiquitination and turnover of those PAL isoforms. Another possibility is that both PAL1 and PAL2 in plant cells may undergo appropriate modification or be specifically folded, thus creating the better interface required for a tighter association with KFB39. However, the failure to detect any interaction of KFB39 with PAL1 or PAL2 in tobacco cells via BiFC assay (Fig. 2) seemingly negates this explanation. Alternatively, PAL is known to be able to form homotetramers or heterotetramers for its deamination functionality (Reichert et al., 2009). Tetramerization may change the conformation of PAL1 or PAL2, thus facilitating the interaction with AtKFB39; or heterotetramerization may include the PAL3 or PAL4 unit with which KFB39 preferentially interacts, and the actions of KFB39 with PAL3 or PAL4 might turn over the entire PAL complex in vivo, including the PAL1 or PAL2 unit.
Arabidopsis has four PAL homologous genes (Wanner et al., 1995; Raes et al., 2003). AtPAL1, AtPAL2, and AtPAL4 are highly expressed in different tissues (Rohde et al., 2004; Zhang et al., 2013a), and their encoded isozymes exhibit a high binding affinity for Phe (Cochrane et al., 2004); PAL3, in contrast, has only a base-level expression in plants (Rohde et al., 2004; Zhang et al., 2013a), and the encoded enzyme has lower affinity for Phe (Cochrane et al., 2004). Purportedly, these PAL isozymes may have overlapping but distinct functions in phenylpropanoid production. Disruption of both PAL1 and PAL2 in Arabidopsis nearly completely eliminates flavonoid accumulation, although PAL activity in pal1 and pal2 double mutant lines remains up to 50% of the wild-type level (Rohde et al., 2004). Disrupting both PAL1 and PAL2 also leads to changes in lignin deposition and the ultrastructure of the secondary cell wall (Rohde et al., 2004). These studies suggest that AtPAL1 and PAL2 might have functional specification for flavonoids/anthocyanin biosynthesis; meanwhile, they may also work with AtPAL4, which is primarily expressed in vascular tissues of stem, responsible for the synthesis of both soluble phenolics and tissue-specific lignin (Rohde et al., 2004; Huang et al., 2010). When AtKFB39 is stably transformed into Arabidopsis, the influence on the accumulation of flavonoids/anthocyanins and CTs is highly correlated with its expression level; the higher the KFB39 transcript level (e.g. in lines 14-2 and 12-1), the greater the reduction of flavonoid/anthocyanin synthesis (Table I; Figs. 4 and 5). In contrast, the effect of overexpressed KFB39 on lignin deposition in the cell wall does not exhibit such a clear correlation with its expression level. The alteration of lignin content in the cell walls of different transgenic lines is nearly the same, irrespective of the levels of KFB39 expression and the decrease in total PAL activity (Figs. 4 and 6). These data imply that, depending on the overexpression level of the KFB39 transgene, and thus the abundance of accumulated KFB39 protein, it has imposed differential effects on the stability of different PAL isozymes in different transgenic lines. At a high level of expression, the highly abundant KFB39, besides modulating the stability and activity of the PAL isozymes that are primarily involved in lignin synthesis (e.g. AtPAL4), also impairs those isoforms dominant in flavonoid/anthocyanin synthesis (e.g. AtPAL1 and/or PAL2); consequently, this impairs the synthesis of different sets of phenylpropanoid products. Since AtKFB39 can strongly interact with the isoform AtPAL4 that is primarily associated with lignin synthesis (Davin et al., 2008), a limited accumulation of KFB39 may already reach the functional threshold for modulating the stability and activity of PAL4 (or PAL3). Therefore, transgenic plants even with low-level expression of KFB39 can display discernible effects on the synthesis and deposition of lignin. On the other hand, lignin has a large pool size, and its accumulation prolongs throughout developmental stages after the onset of lignification; therefore, it is understandable that the decrease in lignin content is inconsistent with the reduction of the measured steady-state PAL activity in KFB39 overexpression lines (Fig. 6). Taken together, these results support the concept that AtPAL isozymes have subtle but distinguishable functional specifications: AtKFB39 in vivo might exhibit different interactions with four PAL isoforms, as is displayed in Y2H and BiFC assays (i.e. be more preferable for PAL3 and PAL4 than for PAL1 and PAL2), although this subtle substrate preference can be easily wiped out due to the abundant accumulation of KFB39 protein in planta.
KFBs as Targets for Engineering the Production of Desired (Poly)phenols
Many phenylpropanoid metabolites have high economic value and can be used as flavors or fragrance ingredients or as pigments in beverages, processed foods, perfumes, and cosmetics. The dietary flavonoids and hydroxycinnamates exhibit possible potential in preventing or treating different cancers, cardiovascular disorders, and/or obesity or in increasing life span and improving spatial working memory (Boudet, 2007; Del Rio et al., 2013). In particular, anthocyanins, the water-soluble pigments in many colored fruits and flowers, can inhibit the initiation and progression of tumor development, reduce inflammatory inducers of tumor initiation, suppress angiogenesis, and minimize cancer-induced DNA damage (Martin, 2013). Preclinical studies with models of animal disease also suggest that the consumption of anthocyanins confers cardioprotection, inhibits weight gain on a high-fat diet, and extends the life span of cancer-prone mice (Glover and Martin, 2012). In addition, the antioxidative activity of anthocyanins/flavonoids also confers enhanced resistance of fruits to the rot fungi and slows the processes of overripening, therefore increasing the shelf life of fruits (Zhang et al., 2013b). CTs, the oligomers or polymers of flavonoid units (flavan-3-ols), on the other hand, serve as a pest deterrent, protecting plants against herbivores (McMahon et al., 2000; Peters and Constabel, 2002; Xie et al., 2003). CTs bind reversibly to proteins, and their presence in the leaves of forage plants can slow the rate of protein degradation in the rumen and thus protect cattle and sheep from pasture bloat (McMahon et al., 2000). CTs are also powerful antioxidants. CTs in fruits, fruit juice, red wine, and green tea have been claimed to reduce the incidence and progression of cancer (particularly prostate cancer) and cardiovascular disease (Bagchi et al., 2000).
Considering the significance of phenylpropanoid products to the well-being of plants, animals, and humans, there is growing interest in the metabolic engineering of plants with higher levels of desirable (poly)phenols (Martin, 2013; Zhang et al., 2014). PAL, as the entry point enzyme, controls the partitioning of primary metabolic flux into a variety of desirable phenolics; therefore, it is an ideal target for enhancing the pathway’s activity. However, it is difficult to boost PAL’s activity by genetically manipulating the level of gene expression in plants (Elkind et al., 1990; Howles et al., 1996). Overexpression of PAL often causes an unexpected severe reduction of both endogenous and transgenic PAL at the levels of RNA and enzyme activity (Elkind et al., 1990; X. Zhang and C.-J. Liu, unpublished data). This high, frequent sense suppression of PAL represses the overall biosynthesis activity and, consequently, decreases the accumulation of phenolics. Meanwhile, PAL transgenic plants are also phenotypically abnormal, displaying stunted growth (Elkind et al., 1990; Howles et al., 1996; X. Zhang and C.-J. Liu, unpublished data). Our study here demonstrates that knocking out or silencing KFB genes significantly enhanced PAL stability and thus the cellular concentration and activity. Consequently, it led to severalfold increases in the production of anthocyanins and phenolic esters and to considerable enhancement of the formation and deposition of CTs in the seed coats of plants (Figs. 7 and 8; Table I). This feature offers a unique opportunity to overcome the obstacle in directly manipulating PAL to produce the desired (poly)phenols.
It is noteworthy that disrupting KFBs significantly enhanced the metabolic flux into anthocyanins, proanthocyanidins, sinapic esters, and lignin but did not further promote the accumulation level of flavonols (e.g. kaempferol) in Arabidopsis (Table I). This implies that the enzymatic step leading to the flavonol branch, most likely the FLS or the following glycosyltransferase reaction, might be the additional rate-limiting point. This information helps to define the regulatory architecture of the flavonol branch. It remains to be determined whether the limitation of the FLS reaction (or the glycosyltransferase reaction) occurs at the transcriptional or enzyme level.
Manipulating KFB Expression to Enhance the Tolerance of Plants to UV-B Radiation
In response to UV-B stress, flowering plants produce a variety of UV light-absorptive secondary products primarily derived from Phe. Arabidopsis mutants with defects in the synthesis of flavonoids, like tt4, tt5, and tt6, or of hydroxycinnamate esters (e.g. sinapoyl esters), like ferulic acid hydroxylase1, are more sensitive to UV-B light than the wild type when grown under high UV-B irradiance, demonstrating that both flavonoids and hydroxycinnamates play critical roles in protecting plants against UV-B irradiation (Li et al., 1993; Landry et al., 1995). Manipulating their accumulation could significantly affect a plant’s photoprotection ability (Landry et al., 1995; Milkowski et al., 2004; Nair et al., 2004). In Brassicaceae, hydroxycinnamate esters are synthesized via the phenylpropanoid-lignin biosynthetic pathway and are presumably stored in the vacuoles in epidermal and mesophyll cells (Milkowski et al., 2004), while flavonoids were detected in the vacuoles, endoplasmic reticulum, chloroplast outer membranes, and nuclear locations in epidermal and mesophyll cells, where they serve as UV light filters and/or scavengers of singlet oxygen or reactive oxygen species for photoprotection (Agati and Tattini, 2010; Agati et al., 2012). It was also proposed that hydroxycinnamates may be incorporated into the secondary cell wall, offering additional protection against the penetration of UV light in leaves through the anticlinal cell walls (Strack et al., 1988).
Exposure of Arabidopsis to UV irradiation noticeably suppressed the expression of the identified KFB genes, along with increasing the expression of PALs, suggesting that KFBs, as the posttranslational regulators of PAL, can sense UV-B light signal to mitigate the turnover of PAL enzyme. In conjunction with the up-regulation of PAL at the mRNA level, the UV light-responsive behavior of KFB synergistically maximizes the activity of the PAL-catalyzed reaction, by which it coordinately regulates phenolic synthesis to build up photoprotection ability.
Knocking out or silencing KFB gene expression, including KFB39, increased the accumulation of hydroxycinnamates (sinapic acid ester) and anthocyanins 3- to 4-fold in Arabidopsis seedlings (Table I); consequently, the kfb mutant or RNAi silencing seedlings exhibited a noted enhancement in resistance to UV radiation damage. The mutant or RNAi plants displayed largely improved viability (Fig. 9). This property could be important in agricultural applications to protect crops from UV stress, since the release of man-made chlorine and bromine compounds increases the depletion of stratospheric ozone, leading to an increase of solar UV-B radiation reaching the surface of the earth (Xu and Sullivan, 2010). It could be a useful strategy to tailor the UV-B light stress tolerance in bioenergy crops by manipulating KFB expression, thereby fostering their adaptability in a high-UV-irradiance environment.
MATERIALS AND METHODS
Phylogenetic and Sequence Analyses
Arabidopsis (Arabidopsis thaliana) KFB homologs were retrieved using AtKFB01, AtKFB20, and AtKFB50 genes (At1G15670, At1G80440, and At3G59940) as the bait through BLAST searches in The Arabidopsis Information Resource based on previous reports (Gagne et al., 2002; Xu et al., 2009). Phylogenetic analysis was conducted by using translated protein full-length sequences. Full-length amino acid sequences were first aligned by the integrated ClustalW program with default parameters in the MEGA package version 5.1 (Kumar et al., 2004). Phylogenetic analyses were conducted using the neighbor-joining method (Saitou and Nei, 1987) implemented in MEGA, with the pairwise deletion option for handling alignment gaps and with the Poisson correction model for computing distance.
Isolation of AtKFB39 and Y2H Assay
The full-length complementary DNA (cDNA) of AtKFB39 (AT2G44130) first was amplified by reverse transcription-PCR from Arabidopsis seedlings with primers At2g44130-attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGACTATGGAAGTGTCGAAAAAGAA-3′) and At2g44130-attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAACATAGATTGAAGCATGCGAAA-3′) and ligated into a pDONR207 entry vector by the BP reaction Gateway cloning technique (Invitrogen). After confirming the sequence, the cDNA was subcloned into pDEST-GADT7 (Arabidopsis Biological Resource Center no. CD3-763) or pDEST-GBKT7 (for domain swap) to generate an activation domain (AD)- or a binding domain (BD)-KFB39 fusion protein, driven by an alcohol dehydrogenase I constitutive promoter. Expression vectors of BD- (or AD-) PAL1, PAL2, PAL3, or PAL4 (Zhang et al., 2013a) and AD- (or BD-) KFB39 were cotransformed into the yeast cell AH109 and then spread onto SD-Leu/Trp medium to select positive yeast transformants. Thereafter, Y2H assays were conducted as described (Zhang et al., 2013a) on three-amino acid dropout medium (−His/Leu/Trp) and on medium containing 5-bromo-4-chloro-3-indoyl-α-d-galactopyranoside (Gold Biotechnology). For a more precise assay, the transformants were also dropped onto four-amino acid dropout medium (−adenine/His/Leu/Trp) to assess the activation of the additional nutrient reporter gene (adenine synthase).
BiFC Assay
KFB39 was amplified with the deletion of its F-box domain (N-terminal 50 amino acids) with PCR using the primers AT2G44130(ΔF)-attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGTAGCTTACTCTCCGAC-3′) and AT2G44130-attB2, as described above. The amplified truncated KFB39 gene first was cloned into the pDONR207 vector by BP reactions (Invitrogen) and then subcloned into pDEST-GW VYNE(R) (Gehl et al., 2009) to obtain the nYFP-KFB39(ΔF) constructs. After the sequence was confirmed, the construct was transferred into Agrobacterium tumefaciens strain GV3101 by the electroporation method. Pairwise interaction examinations were done by mixing A. tumefaciens strains carrying the nYFP-KFB39(ΔF) and PAL(s)-CFPc constructs (Zhang et al., 2013a) with the gene-silencing inhibitor strain pBA-HcPro (Menke et al., 2005) at 1:1:1 ratio and coinfiltrating onto the abaxial surface side of 4- to 6-week-old tobacco (Nicotiana tabacum) leaves. The fluorescence of the expressed fusion proteins was examined 2 to 4 d after infiltration with a Leica TCS SP5 Laser Scan Confocal Microscope as described (Zhang et al., 2013a).
Assays for the Transient Expression of PAL in Tobacco Leaves and PAL Activity
The procedures for the transient expression of native and GFP-tagged versions of PAL and KFB39 proteins in tobacco leaves, the total PAL activity assay, and western-blot analysis were essentially the same as described (Zhang et al., 2013a).
Arabidopsis Growth Conditions, Transformation, and UV Light Treatment
All wild-type (Columbia-0) and mutant Arabidopsis seeds were germinated on agar plates and then either directly used for analysis or transferred into soil for further studies according to standard protocols. Briefly, seeds were first surface sterilized with 10% (v/v) commercial bleach for 10 min, washed three times with 0.1% (v/v) Triton X-100 water, followed by a brief wash in 70% (v/v) ethanol. The sterilized seeds were spread on one-half-strength MS medium (Murashige and Skoog, 1962; Sigma) containing 0.8% (w/v) agar and 1% (w/v) Suc (except for inducing anthocyanin; we added 4% [w/v] Suc). The plates were stratified at 4°C for 2 to 3 d in the dark. The seeds were germinated under long-day conditions with 16 h of light and 8 h of dark. Seven-day-old seedlings were transferred into soil and grown under photoperiodic cycles of 16 h of light and 8 h of dark at 22°C in a growth chamber.
To overexpress the F-box protein genes, their full-length cDNAs were subcloned into binary vector pMDC32 (Curtis and Grossniklaus, 2003) from the entry clones in pDONR207 via Gateway cloning (Invitrogen) as described above. The resulting constructs then were transferred into Arabidopsis (Columbia-0) by the floral dip method (Clough and Bent, 1998), mediated by A. tumefaciens strain GV3101. The presence of the transgene was confirmed by PCR. Homozygous transgenic plants were obtained from a T3 generation plant based on antibiotic resistance and PCR confirmation.
For UV light treatment, 5-DAG seedlings grown on MS medium-filled petri dishes under cool-white fluorescent light (approximately 140 µmol m−2 s−1 photosynthetically active radiation) were exposed to UV light from a UVB-FS20 fluorescent lamp (Westinghouse) with irradiation intensity of 0.2 J m−2 s−1 for 8 h. After treatment, the petri dishes were resealed with Parafilm, and the seedlings were left to recover in the growth room for 2 d before their images were taken.
To quantify PAL and KFB gene expression in response to UV light exposure, 5-week-old soil-grown plants were illuminated under cool-white fluorescent light (approximately 140 µmol m−2 s−1 photosynthetically active radiation) supplemented with UV-B light from an FS20-UVB fluorescent lamp at an intensity of 0.2 J m−2 s−1 for 12 h. Thereafter, the rosette leaves were immediately collected and the total RNAs were extracted as described below.
Generation of kfb Triple/KFB39-RNAi Lines
The isolation and characterization of the transfer DNA insertion lines for KFB1, KFB20, and KFB50 and the generation of their double and triple mutant lines were described previously (Zhang et al., 2013a). To create KFB39-RNAi in the kfb triple mutant background, full-length KFB39 cDNA was subcloned into an RNAi construct of pB7GWIWG(II) (Karimi et al., 2002), and the construct was transformed to the kfb triple mutant via the floral dip method (Clough and Bent, 1998) mediated by A. tumefaciens strain GV3101. Knocking down KFB39 expression was confirmed by quantitative reverse transcription (qRT)-PCR using primers At2g44130_RTF (5′-TACGGAACGGAATCTCAAGG-3′) and At2g44130_RTR (5′-GTTTCCCATCGACGTTCACT-3′), with the RNA extracted from 5-DAG seedlings of the T2 generation. The qRT-PCR device was set with SsoAdvanced SYBR Supermix (Bio-Rad) following the formalized standard procedure in the manufacturer’s instructions. The delta-cycle threshold method was used to quantify the expression of individual genes. Arabidopsis UBIQUITIN10 (At4g05320) was used as the control for normalization. Homozygous transgenic plants were obtained from T3 generation plants based on antibiotic resistance and PCR confirmation.
Examining PAL Activity and Stability in Transgenic Plants
Protein extraction from transgenic plants and PAL activity assays were carried out according to the description of Edwards and Kessmann (1992). To examine the protein levels of endogenous PALs in transgenic lines, western blots were run against with the generated PAL peptide antibody that cross-reacts with all the Escherichia coli-expressed PAL proteins (AtPAL1–AtPAL4; Zhang et al., 2013a).
Analyses and Quantification of Flavonoids/Anthocyanins, Soluble Phenolics, CTs, and Cell Wall Lignin
To extract flavonoids and soluble phenolics, we used 5-DAG T2 or T3 plants. Extraction and analysis of flavonoids and soluble phenolics by liquid chromatography-mass spectrometry were as described (Zhang et al., 2012). Briefly, 0.1 g fresh weight of seedlings was ground into powder in liquid N2 and then extracted with 1 mL of 80% (v/v) methanol at 4°C overnight, and 10 µL of extract was injected for HPLC profiling. Anthocyanin was extracted overnight from the 5-DAG seedlings or 4-week-old T2 plants as described by Zhang et al. (2013a). For measurements of acetyl-bromide lignin, we used the mature stems of 12-week-old KFB39 overexpression lines or of 10-week-old kfb01/20/50/KFB39-RNAi lines. For CT measurements, 30 mg of dry mature seeds was weighed and ground in a mortar with 1 mL of acetonitrile:water (75:25, v/v) for 5 min on ice, then sonicated for 20 min. Following centrifugation, the pellet was extracted again with 1 mL of acetonitrile:water (75:25, v/v) overnight at 4°C. The extracts were pooled and concentrated under a flow of nitrogen, and the dry extract was dissolved in 200 μL of acetonitrile:water (75:25, v/v). Fifty-microliter aliquots of the final extracts were used to analyze soluble CTs. After adding 3 mL of butanol-HCl reagent (butanol-concentrated HCl, 95:5, v/v) and 0.1 mL of ferric reagent (2% [w/v] ferric ammonium sulfate in 2 n HCl), the tubes were put into a heating block adjusted to 98°C for 60 min. Thereafter, the tubes were cooled on ice, and CT-related absorbance was recorded at 550 nm.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: At1g15670 (KFB01), At1g80440 (KFB20), At2g44130 (KFB39), At3g59940 (KFB50), At2g37040 (PAL1), At3g53260 (PAL2), At5g04230 (PAL3), and At3g10340 (PAL4).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence alignment of Arabidopsis KFB01 (At1g15670), KFB20 (At1g80440), KFB39 (At2g44130), and KFB50 (At3g59940).
Supplemental Figure S2. Interaction of KFB39 protein with flavonoid biosynthetic enzymes in vitro.
Supplemental Figure S3. Expression of KFB01, KFB20, KFB39, KFB50, and PAL isogenes and accumulation of anthocyanins in KFB39 overexpression lines.
Supplemental Figure S4. The growth and injury of KFB39 overexpression seedlings, and kfb01/20/50/KFB39-RNAi transgenic seedlings under UV-B exposure.
Supplementary Material
Glossary
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- DAG
days after germination
- MS
Murashige and Skoog
- CT
condensed tannin
- RNAi
RNA interference
- cDNA
complementary DNA
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy (grant no. DEAC0298CH10886 to C.-J.L.); H.Y. was supported by the National Science Foundation (grant no. MCB–1051675 to C.-J.L.). The use of the confocal microscope in the Center for Functional Nanomaterials, Brookhaven National Laboratory was supported by the Office of Basic Energy Sciences, U.S. Department of Energy (contract no. DEAC0298CH10886).
Articles can be viewed without a subscription.
References
- Adams J, Kelso R, Cooley L (2000) The Kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 10: 17–24 [DOI] [PubMed] [Google Scholar]
- Agati G, Azzarello E, Pollastri S, Tattini M (2012) Flavonoids as antioxidants in plants: location and functional significance. Plant Sci 196: 67–76 [DOI] [PubMed] [Google Scholar]
- Agati G, Tattini M (2010) Multiple functional roles of flavonoids in photoprotection. New Phytol 186: 786–793 [DOI] [PubMed] [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148: 187–197 [DOI] [PubMed] [Google Scholar]
- Bate NJ, Orr J, Ni W, Meromi A, Nadler-Hassar T, Doerner PW, Dixon RA, Lamb CJ, Elkind Y (1994) Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc Natl Acad Sci USA 91: 7608–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Cramer CL, Lamb CJ, Schuch W, Dixon RA (1986) L-Phenylalanine ammonia-lyase from Phaseolus vulgaris: modulation of the levels of active enzyme by trans-cinnamic acid. Planta 169: 97–107 [DOI] [PubMed] [Google Scholar]
- Bork P, Doolittle RF (1994) Drosophila Kelch motif is derived from a common enzyme fold. J Mol Biol 236: 1277–1282 [DOI] [PubMed] [Google Scholar]
- Boudet AM. (2007) Evolution and current status of research in phenolic compounds. Phytochemistry 68: 2722–2735 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cochrane FC, Davin LB, Lewis NG (2004) The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65: 1557–1564 [DOI] [PubMed] [Google Scholar]
- Crozier A, Jaganath IB, Clifford MN (2009) Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep 26: 1001–1043 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Jourdes M, Patten AM, Kim KW, Vassão DG, Lewis NG (2008) Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat Prod Rep 25: 1015–1090 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol 44: 123–128 [DOI] [PubMed] [Google Scholar]
- Del Rio D, Rodriguez-Mateos A, Spencer JP, Tognolini M, Borges G, Crozier A (2013) Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid Redox Signal 18: 1818–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Kessmann H (1992) Isoflavonoid phytoalexins and their biosynthetic enzymes. InGurr SJ, McPherson MJ, Bowles DJ, eds, Molecular Plant Pathology: A Practical Approach, Vol 2. IRL Press, Oxford, pp 45–62 [Google Scholar]
- Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ (1990) Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87: 9057–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152, doi/10.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz RR, Hodgins DS, Abell CW (1976) Phenylalanine ammonia-lyase: induction and purification from yeast and clearance in mammals. J Biol Chem 251: 4646–4650 [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New Gateway vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Glover BJ, Martin C (2012) Anthocyanins. Curr Biol 22: R147–R150 [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE (2004) Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Hodgins DS. (1971) Yeast phenylalanine ammonia-lyase: purification, properties, and the identification of catalytically essential dehydroalanine. J Biol Chem 246: 2977–2985 [PubMed] [Google Scholar]
- Howles PA, Sewalt V, Paiva NL, Elkind Y, Bate NJ, Lamb C, Dixon RA (1996) Overexpression of l-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol 112: 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Jansen MAK, Gaba V, Greenberg BM (1998) Higher plants and UV-B radiation: balancing damage, repair and acclimation. Trends Plant Sci 3: 131–135 [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CC, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9: 631–638 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. (2013) The interface between plant metabolic engineering and human health. Curr Opin Biotechnol 24: 344–353 [DOI] [PubMed] [Google Scholar]
- McMahon LR, McAllister TA, Berg BP, Majak W, Acharya SN, Popp JD, Coulman BE, Wang Y, Cheng KJ (2000) A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Can J Plant Sci 80: 469–485 [Google Scholar]
- Menke FL, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Baumert A, Schmidt D, Nehlin L, Strack D (2004) Molecular regulation of sinapate ester metabolism in Brassica napus: expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant J 38: 80–92 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701–712 [DOI] [PubMed] [Google Scholar]
- Prag S, Adams JC (2003) Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert AI, He XZ, Dixon RA (2009) Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem J 424: 233–242 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Strack D. (1997) Phenolic metabolism. InDey PM, Harborne JB, eds, Plant Biochemistry. Academic Press, London, pp 387–416 [Google Scholar]
- Strack D, Heilemann J, Mömken M, Wray V (1988) Cell wall-conjugated phenolics from Coniferae leaves. Phytochemistry 27: 3517–3521 [Google Scholar]
- Sun Y, Zhou X, Ma H (2007) Genome-wide analysis of Kelch repeat containing F-box family. J Integr Plant Biol 49: 940–952 [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wanner LA, Li G, Ware D, Somssich IE, Davis KR (1995) The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol Biol 27: 327–338 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Moore BS (2005) Biochemical characterization of a prokaryotic phenylalanine ammonia lyase. J Bacteriol 187: 4286–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Xu C, Sullivan JH (2010) Reviewing the technical designs for experiments with ultraviolet-B radiation and impact on photosynthesis, DNA and secondary metabolism. J Integr Plant Biol 52: 377–387 [DOI] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Cooley L (1993) Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693 [DOI] [PubMed] [Google Scholar]
- Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T (2004) Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot 55: 2015–2027 [DOI] [PubMed] [Google Scholar]
- Zhang KW, Bhuiya MW, Pazo JR, Miao Y, Kim H, Ralph J, Liu CJ (2012) An engineered monolignol 4-O-methyltransferase (MOMT4) represses lignin polymerization and confers novel metabolic capability in Arabidopsis. Plant Cell 24: 3122–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gou M, Liu CJ (2013a) Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25: 4994–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu CJ (2015) Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant 8: 17–27 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, De Stefano R, Schoonbeek HJ, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, et al. (2013b) Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol 23: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19: 81–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.