WRKY transcription factors play an essential role in regulating the biosynthesis of plant-specialized metabolites.
Abstract
WRKY transcription factors (TFs) are well known for regulating plant abiotic and biotic stress tolerance. However, much less is known about how WRKY TFs affect plant-specialized metabolism. Analysis of WRKY TFs regulating the production of specialized metabolites emphasizes the values of the family outside of traditionally accepted roles in stress tolerance. WRKYs with conserved roles across plant species seem to be essential in regulating specialized metabolism. Overall, the WRKY family plays an essential role in regulating the biosynthesis of important pharmaceutical, aromatherapy, biofuel, and industrial components, warranting considerable attention in the forthcoming years.
Over the last 20 years, the WRKY transcription factor (TF) family has been well recognized for its role in regulating abiotic and biotic stress tolerance in plants. WRKY domain-containing genes comprise one of the largest TF families in plants and are characterized by a highly conserved WRKYGQK motif and a C2H2 or C2HC zinc finger (Eulgem et al., 2000). Ishiguro and Nakamura (1994) identified the first WRKY TF, SWEET POTATO FACTOR1 (SPF1), as a negative regulator of sporamin and β-amylase expression in sweet potato (Ipomoea batatas). Based on the lack of homologous genes, they speculated that SPF1 may be a new class of DNA-binding proteins. The following year, two additional WRKY TFs, ABF1 and ABF2, from wild oat (Avena fatua), helped solidify these genes as encoding a novel class of plant DNA-binding proteins (Rushton et al., 1995). In 1996, the function of the family was extended to plant defense with the discovery of three parsley (Petroselinum crispum) WRKYs that regulate the PATHOGENESIS RELATED1 promoter (Rushton et al., 1996). That study also clearly identified the core cis-element, (T)TGAC(C), also called the W-box, that is recognized by most WRKY TFs.
A seminal study was published in 2000 characterizing the Arabidopsis (Arabidopsis thaliana) WRKY family (Eulgem et al., 2000). The 72 members of the Arabidopsis WRKY family are phylogenetically classified into three major groups (groups I–III), with group II further divided into five subgroups (groups IIa–IIe). The phylogenetic information led to investigations about the origin of WRKY TFs. Early studies found that WRKYs exist in a limited number of protists, including Giardia lamblia and Dictyostelium discoideum, as well as the green alga Chlamydomonas reinhardtii (Zhang and Wang, 2005). Based on these species, early convention held that the ancestral WRKY gene belonged to the group I WRKY family (Wu et al., 2005; Zhang and Wang, 2005). A more recent study, however, determined that the ancestral WRKY gene most likely encodes a group IIc-like WRKY TF (Brand et al., 2013).
The first structure of the WRKY domain was published in 2005. Since then, the functions of key domain residues have been elucidated (Yamasaki et al., 2005; Duan et al., 2007). The globular WRKY domain is composed of five antiparallel β-strands, a Glu-Trp-Lys triad that stabilizes the structure, and the conserved Cys and His residues binding the Zn2+ ion (Duan et al., 2007). The WRKY domain forms a unique wedge shape that inserts perpendicularly into the major groove of the DNA (Yamasaki et al., 2012) and primarily binds the W-box element through the RKYGQ motif on the second β-strand (Duan et al., 2007; Brand et al., 2013). After elucidation of the WRKY domain structure, the GLIAL CELL MISSING1, FLYWCH, and Mutator-like element transposases from metazoans were identified as having similar protein structures to WRKY TFs (Yamasaki et al., 2005; Babu et al., 2006). These findings suggest that the ancestral WRKY gene encoded a BED finger-type DNA-binding protein that evolved from a C2H2 zinc finger gene. Structural data also led to the identification of another large group of plant TFs, the NO APICAL MERISTEM, ATAF1, and CUP-SHAPED COTYLEDON2 (NAC) family, that are structurally related to WRKYs (Yamasaki et al., 2008).
Since 1994, WRKY TFs have been found to play roles in plant tolerance to a variety of abiotic stresses, including wounding, drought, salt, heat, cold, and osmotic pressure, topics that have been extensively reviewed recently (Chen et al., 2012; Bakshi and Oelmüller, 2014; Tripathi et al., 2014). Arabidopsis WRKY45 mediates the expression of a transporter needed for phosphate acquisition, providing insight into how WRKYs manage plant tolerance to soil abiotic stress (Wang et al., 2014). Additionally, WRKYs function in biotic stress tolerance to numerous pathogens (Zheng et al., 2006; Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012; Chen et al., 2013). WRKYs also have been found to be degraded by the 26S proteasome, divulging the role of proteolysis in regulating WRKY TF functions (Matsushita et al., 2012; Yu et al., 2013). The role of WRKYs in plant development, such as trichome and root hair formation, seed coat color, seed size, plant senescence, and male gametogenesis, is also well documented (Johnson et al., 2002; Robatzek and Somssich, 2002; Luo et al., 2005; Guan et al., 2014).
The involvement of WRKY TFs in numerous phytohormone signaling cascades involving abscisic acid, auxin, brassinosteroids, cytokinin, ethylene, jasmonate, and salicylic acid was recently reviewed (Rushton et al., 2012; Bakshi and Oelmüller, 2014). As salicylic acid and abscisic acid are intricately connected with plant defense and abiotic stress tolerance, respectively, much research has focused on these signaling cascades as regulators of WRKY factors (Yu et al., 2001; Rushton et al., 2012); yet, more recent efforts have focused on other phytohormones (Guo et al., 2014; Li et al., 2014; Schluttenhofer et al., 2014). An Arabidopsis mitogen-activated protein kinase (MAPK) signaling cascade, through MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3)/MPK6 has been shown to regulate WRKY33 (Pecher et al., 2014), which activates the expression of two ethylene biosynthesis genes (Li et al., 2012). Arabidopsis WRKY57 is an important regulator balancing jasmonate and auxin signaling in leaf senescence by interacting with jasmonate zim-domain and auxin/indole-3-acetic acid proteins (Jiang et al., 2014).
THE PRESENT: WRKY TFs AS KEY REGULATORS OF SPECIALIZED METABOLISM
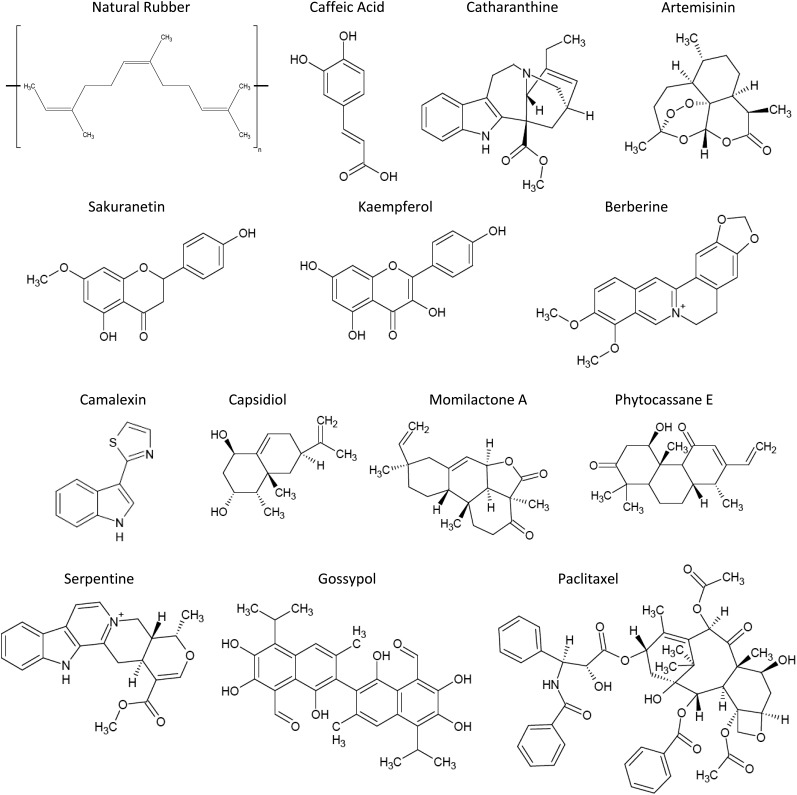
Plants are a rich source of specialized metabolites with antimicrobial and pharmaceutical properties (Fig. 1). However, several key plant-derived drugs are susceptible to shortages, drastically reducing the secure supply for patient treatment (Chabner, 2011). Therefore, developing methods to secure raw material supply and increase yield is essential. One approach is to use TFs to reengineer plants for increased metabolite production. The WRKY TF family is well known for the regulation of stress tolerance based on its association with numerous stress-related phenotypes and our understanding of the underlying mechanisms for stresses (Chen et al., 2012; Bakshi and Oelmüller, 2014; Tripathi et al., 2014). The relative ease of identifying mutant phenotypes in stress-response screens has been a key contributor to establishing the association of WRKYs with stress. In contrast, WRKY TFs have not commonly been considered key regulators of specialized metabolism because of (1) their known association with stress tolerance, (2) the emphasis on other characterized master regulators of specialized metabolism, such as basic helix-loop-helix (bHLH) and ethylene response factors, (3) the difficulty of identifying metabolic mutant phenotypes, (4) the potential redundancy of multiple WRKY TF’s regulating the same biosynthetic genes, and (5) a general lack of mechanistic understanding of the transcriptional regulation of specialized metabolism. Nevertheless, accumulating evidence suggests that certain WRKYs regulate the production of valuable natural products by regulating metabolite biosynthetic genes (Kato et al., 2007; Ma et al., 2009; Suttipanta et al., 2011). WRKYs regulating products from phenylpropanoids, alkaloids, and terpenes, the three major classes of plant metabolites, have been identified (Table I).
Figure 1.
Phytochemicals synthesized by metabolic pathways regulated by WRKY TFs.
Table I. A list of reported WRKY TFs regulating specialized metabolite production.
–, Information not available.
| Species | WRKY | GenBank No. | Metabolic Pathways Affected | Metabolite Class | Reference |
|---|---|---|---|---|---|
| A. thaliana | WRKY12 | AF404857 | Lignin | Phenylpropanoid | Wang et al. (2010) |
| A. thaliana | WRKY23 | AY052647 | Flavonol | Phenylpropanoid | Grunewald et al. (2012) |
| A. thaliana | WRKY33 | AK226301 | Camalexin | Indole alkaloid | Mao et al. (2011) |
| A. thaliana | WRKY44 | NM_129282 | Tannin, mucilage | Phenylpropanoids | Johnson et al. (2002) |
| A. annua | WRKY1 | FJ390842, KC118517 | Artemisinin | Sesquiterpene | Ma et al. (2009); Han et al. (2014) |
| C. roseus | WRKY1 | HQ646368 | Catharanthine, serpentine | TIAs | Suttipanta et al. (2011) |
| C. japonica | WRKY1 | AB267401 | Berberine | BIA | Kato et al. (2007) |
| E. californica | AtWRKY1 | AF442389 | Sanguinarine, chelirubine | BIAs | Apuya et al. (2008) |
| G. arboreum | WRKY1 | AY507929 | Gossypol | Sesquiterpene | Xu et al. (2004) |
| H. brasiliensis | WRKY41 | GU372969 | Rubber | Polyterpene | Zhang et al. (2012) |
| H. brasiliensis | WRKY1 | JF742559 | Rubber | Polyterpene | Wang et al. (2013b) |
| H. brasiliensis | WRKY33 | AJJZ010966566 | Rubber | Polyterpene | Li et al. (2014) |
| H. brasiliensis | WRKY14 | AJJZ010236935 | Rubber | Polyterpene | Li et al. (2014) |
| H. brasiliensis | WRKY55 | AJJZ010198483 | Rubber | Polyterpene | Li et al. (2014) |
| M. truncatula | WRKY100577 | EU526033 | Rutin, kaempferol, caffeic acid, lignin | Phenylpropanoids | Naoumkina et al. (2008) |
| M. truncatula | WRKY100630 | EU526034 | Rutin, kaempferol, caffeic acid, lignin | Phenylpropanoids | Naoumkina et al. (2008) |
| M. truncatula | WRKY108715 | EU526035 | Rutin, kaempferol, caffeic acid, lignin | Phenylpropanoids | Naoumkina et al. (2008) |
| M. truncatula | WRKY109669 | EU526036 | Rutin, kaempferol, caffeic acid, lignin | Phenylpropanoids | Naoumkina et al. (2008) |
| M. truncatula | STP | HM622066 | Lignin | Phenylpropanoid | Wang et al. (2010) |
| N. benthamiana | WRKY8 | AB445392 | Capsidiol | Sesquiterpene | Ishihama et al. (2011) |
| O. sativa | WRKY13 | EF143611 | Momilactone A | Diterpene | Qiu et al. (2008a) |
| O. sativa | WRKY45 | AK066255 | Momilactone A, oryzalexin, phytocassane | Diterpenes | Akagi et al. (2014) |
| O. sativa | WRKY53 | AB190436 | Momilactone A | Diterpene | Chujo et al. (2014) |
| O. sativa | WRKY76 | AK068337 | Momilactone A, phytocassane, sakuranetin | Diterpenes and phenylpropanoid | Yokotani et al. (2013) |
| O. sativa | WRKY89 | AY781112 | Lignin | Phenylpropanoid | Wang et al. (2007) |
| P. quinquefolius | WRKY1 | JF508376 | – | Triterpene | Sun et al. (2013) |
| P. somniferum | WRKY1 | JQ775582 | – | BIA | Mishra et al. (2013) |
| S. lycopersicum | WRKY73 | NM_001247873 | – | Monoterpenes | Spyropoulou et al. (2014) |
| T. chinensis | WRKY1 | JQ250831 | Paclitaxel | Diterpene | Li et al. (2013) |
| V. vinifera | WRKY2 | AY596466 | Lignin | Phenylpropanoid | Guillaumie et al. (2010) |
The phytohormone jasmonate is a key elicitor of specialized metabolism (Aerts et al., 1994; Singh et al., 1998; Qi et al., 2011; Lenka et al., 2012), primarily through the induction of key TFs, such as MYC2 and ethylene response factors, that regulate the biosynthetic genes (van der Fits and Memelink, 2000; Shoji et al., 2010; Shoji and Hashimoto, 2011; Zhang et al., 2011; Patra et al., 2013; Schweizer et al., 2013; Sears et al., 2014). Study of the grape (Vitis vinifera) WRKY family found that 80% of WRKY genes are jasmonate responsive (Guo et al., 2014). Recently, bioinformatics and comparative genomics approaches to identify jasmonate-inducible WRKY transcriptions in Arabidopsis and Catharanthus roseus uncovered the association of WRKY TFs with jasmonate in the production of specialized metabolites (Schluttenhofer et al., 2014).
WRKYs Regulating Phenylpropanoid Pathways
WRKY TFs have been found to regulate the production of a variety of phenolic compounds, including lignin (Naoumkina et al., 2008; Guillaumie et al., 2010; Wang et al., 2010). As lignin is derived from the same phenylpropanoid pathway as other specialized metabolites, it is likely that WRKYs regulating lignin production or deposition may also directly or indirectly affect flux through the phenylpropanoid pathway, resulting in altered biosynthesis of other phenolic-based compounds (e.g. flavonoids, lignans, etc.; Besseau et al., 2007). This is supported by the observation that orthologs of WRKYs regulating lignin production also control the biosynthesis of other specialized metabolites (Table I; Supplemental Table S1). Four Medicago truncatula WRKY TFs, each individually overexpressed in tobacco (Nicotiana tabacum), demonstrate that multiple WRKY TFs can affect the accumulation of the same metabolites (Naoumkina et al., 2008). M. truncatula SECONDARY WALL THICKENING IN PITH (STP) and its Arabidopsis ortholog, WRKY12, illustrate that WRKY TFs involved in plant metabolism can have conserved roles across species (Wang et al., 2010). Studies on OsWRKY89 and VvWRKY2 indicate how modulating specialized metabolism regulation may also alter plant physiology and crop performance (Wang et al., 2007; Guillaumie et al., 2010).
In addition to lignin, WRKYs control the production of flavonol and tannin compounds, also derived from the phenylpropanoid pathway. Flavonols, particularly kaempferol and quercetin, can function as polar auxin transport inhibitors (Brown et al., 2001; Peer et al., 2004), thus connecting specialized metabolism to phytohormone signaling. Arabidopsis WRKY23 regulates the production of flavonols in an auxin-inducible manner and illustrates how metabolites can have a negative feedback on phytohormone signaling (Grunewald et al., 2012). The first WRKY characterized in regulating specialized metabolism was AtWRKY44, also called TRANSPARENT TESTA GLABRA2. AtWRKY44 regulates the production of proanthocyanins, a subset of tannin compounds, and pectinaceous seed mucilage (Johnson et al., 2002). The expression of AtWRKY44 is governed by a MYB-bHLH-WD40 TF complex that is a key regulator of anthocyanin production (Ishida et al., 2007), indicating the interconnections of WRKY TFs with other networks regulating specialized metabolism. Together, these findings indicate the incorporation of WRKY TFs into multiple TF and hormone signaling networks.
WRKYs Regulating Alkaloid Pathways
WRKYs have emerged as key regulators of alkaloid metabolism. Analysis of Catharanthus spp. terpene indole alkaloid (TIA) biosynthetic gene promoters revealed that W-boxes are a commonly present cis-element, suggesting that WRKY TFs are important in regulating alkaloid production (Suttipanta et al., 2011). At least 25% of WRKY TFs in Catharanthus spp. are jasmonate response factors and could potentially regulate TIA biosynthesis (Schluttenhofer et al., 2014). CrWRKY1 from C. roseus directly regulates the expression of TRYPTOPHAN DECARBOXYLASE, the enzyme synthesizing the indolic tryptamine precursor (Suttipanta et al., 2011). Additionally, metabolites from two branches of the TIA pathway, catharanthine and serpentine, accumulate differently in CrWRKY1 RNA interference lines of hairy root cultures, indicating that CrWRKY1 regulates genes governing metabolite flux within the pathway. Analysis of the CrWRKY1 promoter reveals cis-elements for bHLH, DNA binding with one finger, MYB, and TGACG sequence-specific binding protein TFs, suggesting that additional transfactors with greater pathway hierarchy may regulate this gene (Yang et al., 2013).
Similar to TIA, benzylisoquinoline alkaloids (BIAs) also are regulated by WRKY TFs. Coptis japonica WRKY1 dictates the expression of nine genes involved in berberine biosynthesis but does not appear to have an effect on primary metabolism (Kato et al., 2007). Importantly, this finding illustrates that WRKY TFs are capable of up-regulating entire specialized metabolic pathways. More recently, a WRKY from opium poppy (Papaver somniferum) was found to bind to the TYROSINE DECARBOXYLASE promoter and possibly to regulate BIA production (Mishra et al., 2013). That study also found that W-boxes are present in the seven available BIA gene promoters. Heterologous expression of Arabidopsis AtWRKY1 in California poppy (Eschscholzia californica) affects the accumulation of sanguinarine and chelirubine (Apuya et al., 2008). Therefore, one way to engineer a plant, which synthesizes valuable compounds but is hindered by the limited availability of pathway information, is to express WRKY TF genes with known roles in governing specialized metabolism.
Camalexin is a primary defense metabolite in Arabidopsis and is governed by a set of MAPK cascades through MPK3/MPK6 and MPK4. MPK3/MPK6 directly phosphorylates WRKY33, which proceeds to activate PHYTOALEXIN DEFICIENT3, a key gene in camalexin biosynthesis (Ren et al., 2008; Mao et al., 2011). In the MPK4-WRKY33 cascade, MPK4 interacts with VQ MOTIF-CONTAINING PROTEIN21, and phosphorylation of the VQ protein destabilizes the complex, releasing WRKY33 (Qiu et al., 2008b; Petersen et al., 2010). These results emphasize the role of phosphorylation and protein complexes in regulating WRKY TF activity. VQ proteins, which interact with the WRKY domain, thus inhibiting TF-DNA binding, appear to be key regulators of group I and group IIc WRKYs activities (Petersen et al., 2010; Cheng et al., 2012; Chi et al., 2013) and also are likely to be important regulators of specialized metabolism. In Arabidopsis, MPK3 and MPK6 are important in regulating VQ-WRKY protein interactions (Lai et al., 2011; Pecher et al., 2014). In Catharanthus spp., phosphorylation regulates TIA biosynthesis, part of which occurs through the MAPK CrMPK3, an ortholog of Arabidopsis MPK3 (Raina et al., 2012). The work on CrMPK3 suggests that additional WRKY TFs, and probably VQ proteins, are involved in regulation of the TIA pathway.
WRKYs Regulating Terpene Pathways
While considerably less is known about the transcriptional regulatory networks governing terpene synthesis (Patra et al., 2013), the majority of regulators identified are WRKY TFs. Other than in Arabidopsis, early work established the role of WRKY TFs in the regulation of specialized metabolism in cotton (Gossypium arboreum; Xu et al., 2004). GaWRKY1 regulates a sesquiterpene cyclase at a pathway branch point leading to the production of gossypol, an antifeedant phytoalexin. Recently, the Gossypium raimondii WRKY family was characterized, which identified seven WRKY TFs primarily expressed in fiber trichomes (Cai et al., 2014). In plants, trichomes are a major site of specialized metabolite production. In tomato (Solanum lycopersicum), SlWRKY73 was found to activate the expression of three monoterpene synthase genes, suggesting that a single WRKY can regulate multiple distinct biosynthetic pathways (Spyropoulou et al., 2014). In Artemisia annua, biosynthesis of the antimalarial drug artemisinin, also produced in trichomes, is regulated by AaWRKY1 (Ma et al., 2009). Recently, the yield of artemisinin in a high-yielding cultivar of A. annua was doubled by overexpressing AaWRKY1 (Han et al., 2014). That work provides compelling evidence that WRKYs can be used successfully to engineer metabolic pathways. Interestingly, in spite of increased artemisinin yield, only one pathway gene was significantly up-regulated in the AaWRKY1 overexpression lines, demonstrating the dramatic outcomes of overcoming a bottleneck in the pathway. Similarly, TcWRKY1 from Taxus chinensis fine-tunes the expression of a rate-limiting gene in the pathway for production of the anticancer drug paclitaxel, commonly called taxol (Li et al., 2013). Together, the studies on AaWRKY1 and TcWRKY1 suggest that WRKY TFs can control critical rate-limiting steps in metabolic pathways.
Rice (Oryza sativa) produces terpenes for defense to pathogens and herbivores. OsWRKY45 is a positive regulator of momilactone, phytocassane, and oryzalexin accumulation by priming biosynthetic gene expression (Akagi et al., 2014). Similarly, rice OsWRKY53 enhances the production of momilactones (Chujo et al., 2014). Overexpression of OsWRKY13 up-regulates genes in the phenylpropanoid pathway while down-regulating those for terpenoid biosynthesis, illustrating the potential of WRKY TFs to differentially modulate discrete classes of metabolites (Qiu et al., 2008a). Both OsWRKY53 and Nicotiana benthamiana WRKY8 (Ishihama et al., 2011) are phosphorylated by the MPK3/MPK6 cascade. Rice OsWRKY76 activates cold stress tolerance but represses terpene synthesis (Yokotani et al., 2013). OsWRKY76 also suppresses production of the phenylpropanoid sakuranetin. As these four WRKYs are responsive to the pathogen Magnaporthe oryzae, these findings suggest the existence of a complex, interconnected signaling network governing within an individual specialized metabolite pathway.
A major engineering problem with plants synthesizing valuable natural products is the long life cycle and resilience to stable transformations. In such species, the correlation of gene expression with metabolite accumulation provides an opportunity to identify important components of the metabolic pathway. The expression of two WRKY TFs has been associated with increased biosynthesis of natural rubber, a polyisoprenoid derived from wounding the bark of the tropical tree Hevea brasiliensis (Zhang et al., 2012; Wang et al., 2013b). Recent characterization of the H. brasiliensis WRKY family identified three WRKYs regulating the expression of the SMALL RUBBER PARTICLE PROTEIN, which binds rubber for storage (Li et al., 2014). An American ginseng (Panax quinquefolius) WRKY TF, PqWRKY1, is associated with increased production of ginsenosides, a group of triterpene compounds (Sun et al., 2013). Heterologous expression of WRKY genes in fast-growing model systems can aid functional characterization. Ectopic expression of PqWRKY1 in Arabidopsis up-regulates triterpene biosynthetic genes, suggesting that WRKYs are capable of regulating metabolic pathways in other species. Collectively, these studies suggest that expression-metabolite association and heterologous gene expression techniques are useful methods to identify WRKY TFs regulating specialized metabolism in long-lived plant species.
Orthologs of WRKYs in Specialized Metabolism
Predicted orthologs of several WRKY TFs have been demonstrated to regulate specialized metabolism in multiple plant species (Supplemental Tables S1 and S2). Three WRKYs regulating phenylpropanoids are orthologs of HbWRKY41. HbWRKY33 and TcWRKY1, regulating terpene biosynthesis, have an ortholog that regulates lignin biosynthesis. While AtWRKY1 regulates alkaloid accumulation in California poppy, its ortholog, HbWRKY1, affects rubber accumulation. CrWRKY1 and OsWRKY45 are orthologous proteins regulating TIA and diterpenes in C. roseus and rice, respectively. GaWKRY1 and OsWRKY76 are predicted orthologs but display an antagonistic function in regulating terpene biosynthesis. AaWRKY1 and WRKY109669 are predicted orthologs affecting artemisinin (Ma et al., 2009) and lignin biosynthesis (Naoumkina et al., 2008), respectively. Importantly, six WRKY TFs predicted as orthologs (AtWRKY33, NbWRKY8, OsWRKY53, PsWRKY1, VvWRKY2, and WRKY108715) have independently been found to regulate specialized metabolism (Table I; Supplemental Table S1). Duplicate WRKYs of this group function to regulate the biosynthesis of alkaloids, terpenes, and phenylpropanoids (Naoumkina et al., 2008; Guillaumie et al., 2010; Ishihama et al., 2011; Mao et al., 2011; Mishra et al., 2013; Chujo et al., 2014). Therefore, orthologs of the MPK3/MPK6-AtWRKY33 signaling cascade may be key factors in the regulation of specialized metabolism. Overall, these results reinforce the idea of conserved functions among at least some WRKY TFs. However, other WRKYs involved in specialized metabolism possibly possess species-specific roles (Table I). Collectively, these findings suggest that a core set of WRKYs may be highly conserved in the regulation of specialized metabolism, whereas others may play more restricted roles.
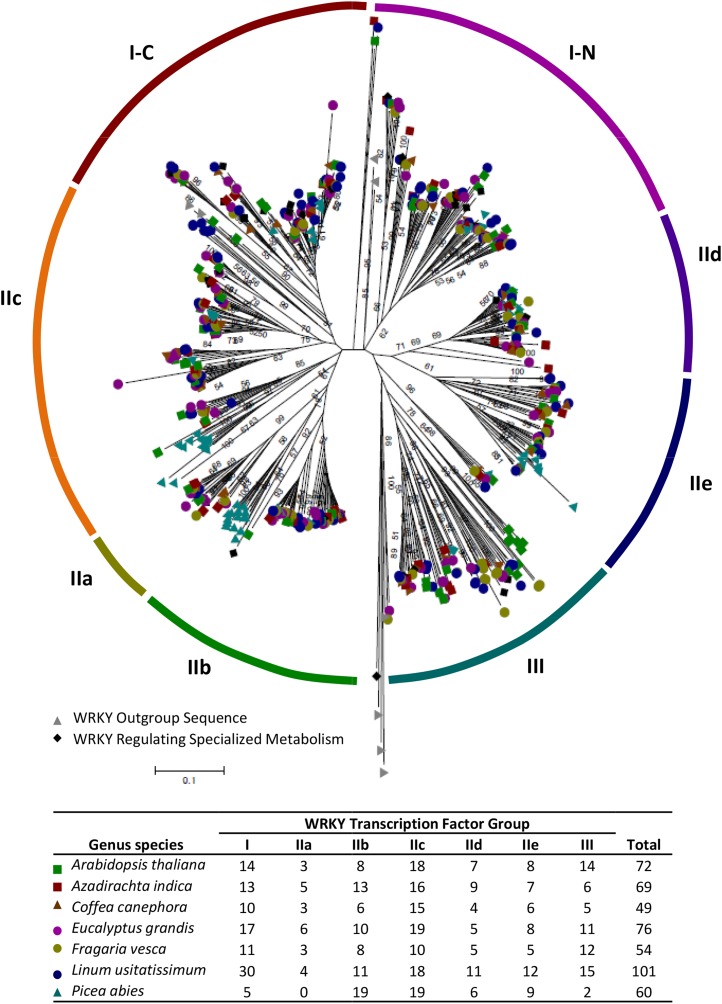
The identification of WRKY TFs regulating specialized metabolism in more species is essential to determine WRKYs with conserved or unique functions. Genomes of several species known for producing valuable compounds have recently been sequenced (Shulaev et al., 2011; Krishnan et al., 2012; Wang et al., 2012; Nystedt et al., 2013; Denoeud et al., 2014; Myburg et al., 2014). The tropical tree neem (Azadirachta indica) produces an array of metabolites with antibiotic, anticancer, and antifeedant properties (Atawodi and Atawodi, 2009) that have applications in pest control (Gahukar, 2014). Flax (Linum usitatissimum) produces podophyllotoxins, precursors used to synthesize the valuable anticancer compounds etoposide and teniposide (Schmidt et al., 2012). Coffee (Coffea canephora and Coffea arabica) synthesizes phenylpropanoids and alkaloids that impact product quality (Franca et al., 2005; Denoeud et al., 2014). Several coffee WRKY TFs have already been identified (Ramiro et al., 2010). Strawberry (Fragaria vesca) is a rich source of anthocyanins and ellagitannins. Eucalyptus (Eucalyptus grandis) and Norway spruce (Picea abies) are desirable timber trees that produce terpene-rich oils with economic importance to the pharmaceutical and aromatherapy industries. We identified a number of WRKY orthologs in these plant species in order to elucidate their roles in regulating specialized metabolism (Supplemental Tables S1 and S2). There are 69, 49, 76, 54, 101, and 60 WRKY TFs in neem, coffee, eucalyptus, strawberry, flax, and spruce, respectively (Fig. 2). Characterization of these orthologs and those from other reported families (Supplemental Table S3) will strengthen our ability to improve natural products from these crops while enhancing our knowledge of WRKY TF function.
Figure 2.
Phylogenetic analysis of Arabidopsis (green squares), coffee (brown triangles), eucalyptus (purple circles), flax (blue circles), neem (red squares), spruce (teal triangles), and strawberry (gold circles) WRKY families alongside those regulating specialized metabolism in other species (top) and the number of TFs present in each group (bottom). Human GCMa and FLYWCH plus G. lamblia, D. discoideum, and C. reinhardtii WRKYs were used to form an outgroup to root the phylogenetic tree (gray triangles). WRKY TFs regulating specialized metabolites are denoted with black diamonds. The neighbor-joining tree was constructed using the proportion of amino acids differing between sequences (i.e. the p-distance method) and 1,000 bootstrap replications using MEGA 6.0. Sequence alignment was performed using the ClustalW algorithm. The phylogenetic tree has been deposited into TreeBase under identification number 16440.
Biological Relevance of WRKY-Regulated Specialized Metabolites
WRKY TFs regulate a diverse array of plant-specialized metabolites that pertain to a broad assortment of biological functions. Biosynthesis of the metabolites listed in Table I is associated with WRKY TFs. In Arabidopsis, camalexin has antifungal properties and aids defense against pathogens including Botrytis cinerea, Alternaria brassicicola, and Sclerotinia sclerotiorum (Thomma et al., 1999; Ferrari et al., 2007; Stotz et al., 2011). Berberine, catharanthine, gossypol, momilactones, phytocassanes, and sanguinarine also have antimicrobial properties and confer tolerance to plant pathogens (Stermitz et al., 2000; Puckhaber et al., 2002; Umemura et al., 2003; Fukuta et al., 2007; Roepke et al., 2010; Obiang-Obounou et al., 2011). In addition to the involvement in auxin signaling (Brown et al., 2001; Buer and Muday, 2004), flavonols are protectants against UV light stress (Li et al., 1993; Bieza and Lois, 2001; Emiliani et al., 2013) and herbivores (Wuyts et al., 2006a; Onkokesung et al., 2014). AtWRKY23 functions in regulating flavonol production (Grunewald et al., 2012) and nematode resistance (Grunewald et al., 2008). Gossypol, another antifeedant compound, deters herbivores, including cotton bollworm (Heliothis armigera) and tobacco budworm (Heliothis virescens; Stipanovic et al., 2008; Kong et al., 2010). OsWRKY13, OsWRKY45, OsWRKY53, and OsWRKY76 regulate the synthesis of the allelopathic diterpene momilactone (Fukuta et al., 2007; Kato-Noguchi et al., 2008). The biopolymer lignin, critical for plant rigidity, also deters herbivory (Wuyts et al., 2006b) and defends against microbes (Xu et al., 2011). Collectively, specialized metabolites regulated by WRKY TFs contribute to overall plant fitness by enhancing tolerance to various abiotic and biotic stresses.
THE FUTURE: WHERE TO GO FROM HERE?
Significant progress has been made in understanding the role of WRKY TFs over the last two decades; however, much more waits to be explored. Clearly, the regulation of specialized metabolism by WRKY TFs is highly integrated into plant defense. Understanding WRKY regulation will unravel the little-known but highly intricate regulatory network that governs plant health and metabolic diversity. In the future, we envision several major areas of research that will contribute significantly to understanding WRKY involvement in specialized metabolism.
(1) No study has specifically focused on determining the number of TFs involved in specialized metabolism. Evidence from the more thoroughly characterized R2R3-MYB and bHLH families suggests that approximately 23% and 12% of the subgroups, respectively, regulate specialized metabolism (Feller et al., 2011). By contrast, we determined up to 43% of the WRKY ortholog clusters involved in the biosynthesis of specialized metabolites (Supplemental Table S1). Future efforts should focus on determining how many different WRKY TFs within a given species regulate specialized metabolite biosynthesis, although this could be complicated by the type and size of the pathway of interest. Many plants, such as Catharanthus spp., possess complex specialized metabolic pathways that produce structurally similar compounds. In such plants, it will be interesting to explore whether expressing WRKY genes from foreign species can differentially modulate biosynthetic genes, leading to the synthesis of novel specialized metabolites with pharmaceutical potential. Identification of all the WRKYs involved in specialized metabolism is essential to unraveling the interwoven networks connecting the syntheses of natural products.
(2) Understanding how the numerous WRKYs of a family differentially regulate unique sets of target genes has long been a major enigma and is just now starting to be deciphered. Previously, only the C-terminal domain of group I WRKY TFs was known to bind DNA; however, recently, this function was extended to the N-terminal WRKY domain (Brand et al., 2013). How dual WRKY domains affect DNA binding and what role each plays in gene regulation necessitate further study. Furthermore, over the last several years, WRKYs have been found to recognize less stringent cis-elements than were suspected previously (Brand et al., 2013). WRKY interactions with VQ proteins, calmodulin, histone deacetylases, and phosphorylation (Kim et al., 2008; Mao et al., 2011; Chi et al., 2013) indicate posttranslational modifications in regulating WRKY TF activity. Many other proteins are anticipated to be involved in the interactions and regulation of WRKY TFs. Identification of these factors and their functions will help further elucidate where WRKYs function in the grand signaling hierarchy of plant-specialized metabolism.
(3) In the future, the ability to use phylogenetic information to predict the functions of WRKYs, especially those in nonmodel plants, and to identify those with novel regulatory roles will be beneficial to crop genetic manipulation for the production of specialized metabolites. With ease of access to sequenced plant genomes, WRKY families have been identified in many plants (Supplemental Table S3). To date, studies involving the complementation of WRKY function between species have been mainly restricted to model plants. The conserved function of WRKY genes and the activation of similar target genes between species are known (Proietti et al., 2011) but only beginning to be understood. Similarly, although functional redundancy is well known in the WRKY family (Jiang and Deyholos, 2009), predicting which TFs perform overlapping roles is still primarily conjecture. Identifying potentially functionally redundant WRKYs may be improved by utilizing programs that categorize groups of orthologous sequences (e.g. OrthoMCL). Understanding and accurately predicting each WRKY’s function will aid the identification of orthologous factors that will improve the engineering of less characterized medicinal species.
(4) Many key features of the evolution of the WRKY family still need to be resolved. For example, why do certain species undergo increased expansion of one specific subgroup compared with other WRKY groups? While WRKY TFs have been shown to arise from tandem and segmental gene duplication events in rice (Wu et al., 2005), differential expression, not sequence divergence, probably led to most WRKY family diversification (Babu et al., 2006). WRKYs have been proposed to play a role in preventing stress as plants emerged from aquatic environments (Wen et al., 2014), but more studies are hastily needed to determine which WRKY members were involved and what functions they possess. Equally important is to identify which core set of these genes were present in ancestral plants and the function of WRKYs in algae. The sole group I WRKY identified from C. reinhardtii is orthologous to Arabidopsis WRKY33, which functions in diverse hormone, stress, and metabolic pathways (Zheng et al., 2006; Jiang and Deyholos, 2009; Li et al., 2011, 2012; Birkenbihl et al., 2012; Logemann et al., 2013; Wang et al., 2013a; Schluttenhofer et al., 2014). C. reinhardtii WRKY1, therefore, may be an early WRKY mediating cross-talk between pathways in the alga. Further supporting the hypothesis that WRKYs played an ancient role in modulating metabolism, a group I-like WRKY from G. lamblia functions downstream of a MAPK cascade to regulate cyst cell wall formation (Pan et al., 2009). Understanding WRKY family evolution will reveal the extent, when, and how WRKYs specifically contributed to the rise of plant-specialized metabolism.
(5) While the number of WRKY TFs known to regulate specialized metabolism has increased appreciably over the past several years, much remains to be learned about how they function with other TFs in regulatory networks. Currently, minimal information is available on where WRKYs fall in the regulatory hierarchy in specialized metabolism. CrWRKY1 regulates the expression of several key TFs involved in TIA biosynthesis (Suttipanta et al., 2011). Additionally, AtWRKY33, NbWRKY8, and OsWRKY45 are targets of MAPK cascades (Andreasson et al., 2005; Qiu et al., 2008b; Mao et al., 2011; Ueno et al., 2013), further suggesting that some WRKYs may function as top-level regulators of specialized metabolism. AtWRKY23 functions downstream of auxin-response factors to synthesize flavonols that are induced to abate auxin signaling (Grunewald et al., 2012). Furthermore, OsWRKY13 regulates over 500 genes, including 39 TFs, and binds the promoters of other WRKY TFs (Qiu et al., 2009). While exceptions probably exist, WRKY TFs, thus far, appear to be key top-tier factors directly regulating the biosynthesis of specialized metabolism.
CONCLUSION
The year 2014 marks two decades since the discovery of the first WRKY, a major milestone in TF research. Over the last score of years, various research groups have identified many WRKY TF families (Supplemental Table S3), and we have learned a considerable amount about how WRKYs function. In the past, most studies have been focused on WRKY TFs in plant defense against abiotic and biotic stresses; however, a neglected role of this family is in regulating specialized metabolism. Future work will entail defining which WRKYs do what task as well as elucidating the mechanisms that govern each TF, which will likely vary between species. As WRKY TFs are essential to plant-specialized metabolism, understanding their functions will provide ways to successfully reengineer medicinal species to provide continuous and higher yielding materials for drug synthesis. Elucidating how WRKYs regulate metabolism thus is critical not only for improving crop stress tolerance but also for increasing the production of valuable plant-derived natural products.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Orthologs of WRKY TFs involved in specialized metabolism.
Supplemental Table S2. WRKY TF sequences from species producing valuable metabolites.
Supplemental Table S3. List of reported WRKY TF families.
Supplementary Material
Glossary
- TF
transcription factor
- bHLH
basic helix-loop-helix
- TIA
terpene indole alkaloid
- BIA
benzylisoquinoline alkaloid
References
- Aerts RJ, Gisi D, De Carolis E, De Luca V, Baumann TW (1994) Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. Plant J 5: 635–643 [Google Scholar]
- Akagi A, Fukushima S, Okada K, Jiang CJ, Yoshida R, Nakayama A, Shimono M, Sugano S, Yamane H, Takatsuji H (2014) WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol Biol 86: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NHT, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Park JH, Zhang L, Ahyow M, Davidow P, Van Fleet J, Rarang JC, Hippley M, Johnson TW, Yoo HD, et al. (2008) Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol J 6: 160–175 [DOI] [PubMed] [Google Scholar]
- Atawodi S, Atawodi J (2009) Azadirachta indica (neem): a plant of multiple biological and pharmacological activities. Phytochem Rev 8: 601–620 [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 34: 6505–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi M, Oelmüller R (2014) WRKY transcription factors: jack of many trades in plants. Plant Signal Behav 9: e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieza K, Lois R (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LH, Fischer NM, Harter K, Kohlbacher O, Wanke D (2013) Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res 41: 9764–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Niu E, Du H, Zhao L, Feng Y, Guo W (2014) Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop Journal 2: 87–101 [Google Scholar]
- Chabner BA. (2011) Drug shortages: a critical challenge for the generic-drug market. N Engl J Med 365: 2147–2149 [DOI] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D (2013) WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 110: E1963–E1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhou Y, Yang Y, Chi YJ, Zhou J, Chen JY, Wang F, Fan B, Shi K, Zhou YH, et al. (2012) Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol 159: 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6: 287–300 [DOI] [PubMed] [Google Scholar]
- Chujo T, Miyamoto K, Ogawa S, Masuda Y, Shimizu T, Kishi-Kaboshi M, Takahashi A, Nishizawa Y, Minami E, Nojiri H, et al. (2014) Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 9: e98737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345: 1181–1184 [DOI] [PubMed] [Google Scholar]
- Duan MR, Nan J, Liang YH, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su XD (2007) DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res 35: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani J, Grotewold E, Falcone Ferreyra ML, Casati P (2013) Flavonols protect Arabidopsis plants against UV-B deleterious effects. Mol Plant 6: 1376–1379 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca AS, Mendonça JCF, Oliveira SD (2005) Composition of green and roasted coffees of different cup qualities. Food Sci Technol (Campinas) 38: 709–715 [Google Scholar]
- Fukuta M, Dang Xuan T, Deba F, Tawata S, Dang Khanh T, Min Chung I (2007) Comparative efficacies in vitro of antibacterial, fungicidal, antioxidant, and herbicidal activities of momilactones A and B. J Plant Interact 2: 245–251 [Google Scholar]
- Gahukar RT. (2014) Factors affecting content and bioefficacy of neem (Azadirachta indica A. Juss.) phytochemicals used in agricultural pest control: a review. Crop Prot 62: 93–99 [Google Scholar]
- Grunewald W, De Smet I, Lewis DR, Löfke C, Jansen L, Goeminne G, Vanden Bossche R, Karimi M, De Rybel B, Vanholme B, et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109: 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inzé D, Beeckman T, Gheysen G (2008) A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol 148: 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10: e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie S, Mzid R, Méchin V, Léon C, Hichri I, Destrac-Irvine A, Trossat-Magnin C, Delrot S, Lauvergeat V (2010) The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol 72: 215–234 [DOI] [PubMed] [Google Scholar]
- Guo C, Guo R, Xu X, Gao M, Li X, Song J, Zheng Y, Wang X (2014) Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot 65: 1513–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Wang H, Lundgren A, Brodelius PE (2014) Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102: 89–96 [DOI] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, et al. (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet 244: 563–571 [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yamada R, Yoshioka M, Katou S, Yoshioka H (2011) Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23: 1153–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama N, Yoshioka H (2012) Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol 15: 431–437 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D (2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Dubouzet E, Kokabu Y, Yoshida S, Taniguchi Y, Dubouzet JG, Yazaki K, Sato F (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol 48: 8–18 [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H, Ino T, Ota K (2008) Secretion of momilactone A from rice roots to the rhizosphere. J Plant Physiol 165: 691–696 [DOI] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong G, Daud MK, Zhu S (2010) Effects of pigment glands and gossypol on growth, development and insecticide-resistance of cotton bollworm (Heliothis armigera (Hübner)). Crop Prot 29: 813–819 [Google Scholar]
- Krishnan NM, Pattnaik S, Jain P, Gaur P, Choudhary R, Vaidyanathan S, Deepak S, Hariharan AK, Krishna PB, Nair J, et al. (2012) A draft of the genome and four transcriptomes of a medicinal and pesticidal angiosperm Azadirachta indica. BMC Genomics 13: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka SK, Boutaoui N, Paulose B, Vongpaseuth K, Normanly J, Roberts SC, Walker EL (2012) Identification and expression analysis of methyl jasmonate responsive ESTs in paclitaxel producing Taxus cuspidata suspension culture cells. BMC Genomics 13: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Guo D, Yang ZP, Tang X, Peng SQ (2014) Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 104: 14–23 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233: 1237–1252 [DOI] [PubMed] [Google Scholar]
- Li S, Zhang P, Zhang M, Fu C, Yu L (2013) Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol (Stuttg) 15: 19–26 [DOI] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Rawat V, Schneeberger K, Schmelzer E, Somssich IE (2013) Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol 198: 1165–1177 [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Pu G, Lei C, Ma L, Wang H, Guo Y, Chen J, Du Z, Wang H, Li G, et al. (2009) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol 50: 2146–2161 [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H (2012) The nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 73: 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Triptahi V, Singh S, Phukan UJ, Gupta MM, Shanker K, Shukla RK (2013) Wound induced transcriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS ONE 8: e52784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, et al. (2014) The genome of Eucalyptus grandis. Nature 510: 356–362 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, He X, Dixon RA (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Obiang-Obounou BW, Kang OH, Choi JG, Keum JH, Kim SB, Mun SH, Shin DW, Kim KW, Park CB, Kim YG, et al. (2011) The mechanism of action of sanguinarine against methicillin-resistant Staphylococcus aureus. J Toxicol Sci 36: 277–283 [DOI] [PubMed] [Google Scholar]
- Onkokesung N, Reichelt M, van Doorn A, Schuurink RC, van Loon JJA, Dicke M (2014) Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J Exp Bot 65: 2203–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YJ, Cho CC, Kao YY, Sun CH (2009) A novel WRKY-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J Biol Chem 284: 17975–17988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L (2013) Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim Biophys Acta 1829: 1236–1247 [DOI] [PubMed] [Google Scholar]
- Pecher P, Eschen-Lippold L, Herklotz S, Kuhle K, Naumann K, Bethke G, Uhrig J, Weyhe M, Scheel D, Lee J (2014) The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytol 203: 592–606 [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Qiu JL, Lütje J, Fiil BK, Hansen S, Mundy J, Petersen M (2010) Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS ONE 5: e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti S, Bertini L, Van der Ent S, Leon-Reyes A, Pieterse CMJ, Tucci M, Caporale C, Caruso C (2011) Cross activity of orthologous WRKY transcription factors in wheat and Arabidopsis. J Exp Bot 62: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckhaber LS, Dowd MK, Stipanovic RD, Howell CR (2002) Toxicity of (+)- and (−)-gossypol to the plant pathogen, Rhizoctonia solani. J Agric Food Chem 50: 7017–7021 [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD- repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S (2009) Exploring transcriptional signalling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biol 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S (2008a) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant 1: 538–551 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. (2008b) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina SK, Wankhede DP, Jaggi M, Singh P, Jalmi SK, Raghuram B, Sheikh AH, Sinha AK (2012) CrMPK3, a mitogen activated protein kinase from Catharanthus roseus and its possible role in stress induced biosynthesis of monoterpenoid indole alkaloids. BMC Plant Biol 12: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro D, Jalloul A, Petitot AS, Grossi De Sá M, Maluf M, Fernandez D (2010) Identification of coffee WRKY transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genet Genomes 6: 767–781 [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke J, Salim V, Wu M, Thamm AMK, Murata J, Ploss K, Boland W, De Luca V (2010) Vinca drug components accumulate exclusively in leaf exudates of Madagascar periwinkle. Proc Natl Acad Sci USA 107: 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, et al. (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J 10: 2–11 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer C, Pattanaik S, Patra B, Yuan L (2014) Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics 15: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TJ, Klaes M, Sendker J (2012) Lignans in seeds of Linum species. Phytochemistry 82: 89–99 [DOI] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P (2013) Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears MT, Zhang H, Rushton PJ, Wu M, Han S, Spano AJ, Timko MP (2014) NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol Biol 84: 49–66 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2011) Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol 52: 1117–1130 [DOI] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22: 3390–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Gavrieli J, Oakey JS, Curtis WR (1998) Interaction of methyl jasmonate, wounding and fungal elicitation during sesquiterpene induction in Hyoscyamus muticus in root cultures. Plant Cell Rep 17: 391–395 [DOI] [PubMed] [Google Scholar]
- Spyropoulou EA, Haring MA, Schuurink RC (2014) RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics 15: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K (2000) Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci USA 97: 1433–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanovic RD, López JD Jr, Dowd MK, Puckhaber LS, Duke SE (2008) Effect of racemic, (+)- and (−)-gossypol on survival and development of Heliothis virescens larvae. Environ Entomol 37: 1081–1085 [DOI] [PubMed] [Google Scholar]
- Stotz HU, Sawada Y, Shimada Y, Hirai MY, Sasaki E, Krischke M, Brown PD, Saito K, Kamiya Y (2011) Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J 67: 81–93 [DOI] [PubMed] [Google Scholar]
- Sun Y, Niu Y, Xu J, Li Y, Luo H, Zhu Y, Liu M, Wu Q, Song J, Sun C, et al. (2013) Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tissue Organ Cult 114: 269–277 [Google Scholar]
- Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L (2011) The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol 157: 2081–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J 19: 163–171 [DOI] [PubMed] [Google Scholar]
- Tripathi P, Rabara R, Rushton P (2014) A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239: 255–266 [DOI] [PubMed] [Google Scholar]
- Ueno Y, Yoshida R, Kishi-Kaboshi M, Matsushita A, Jiang CJ, Goto S, Takahashi A, Hirochika H, Takatsuji H (2013) MAP kinases phosphorylate rice WRKY45. Plant Signal Behav 8: e24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura K, Ogawa N, Shimura M, Koga J, Usami H, Kono T (2003) Possible role of phytocassane, rice phytoalexin, in disease resistance of rice against the blast fungus Magnaporthe grisea. Biosci Biotechnol Biochem 67: 899–902 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA 107: 22338–22343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65: 799–815 [DOI] [PubMed] [Google Scholar]
- Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF (2014) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol 164: 2020–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Du B, Liu M, Sun N, Qi X (2013a) Arabidopsis transcription factor WRKY33 is involved in drought by directly regulating the expression of CesA8. Am J Plant Sci 4: 21 [Google Scholar]
- Wang Y, Guo D, Li HL, Peng SQ (2013b) Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol Biochem 71: 283–289 [DOI] [PubMed] [Google Scholar]
- Wang Z, Hobson N, Galindo L, Zhu S, Shi D, McDill J, Yang L, Hawkins S, Neutelings G, Datla R, et al. (2012) The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J 72: 461–473 [DOI] [PubMed] [Google Scholar]
- Wen F, Zhu H, Li P, Jiang M, Mao W, Ong C, Chu Z (2014) Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res 21: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12: 9–26 [DOI] [PubMed] [Google Scholar]
- Wuyts N, Lognay G, Swennen R, De Waele D (2006a) Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J Exp Bot 57: 2825–2835 [DOI] [PubMed] [Google Scholar]
- Wuyts N, Swennen R, De Waele D (2006b) Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology 8: 89–101 [Google Scholar]
- Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62: 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Wang JW, Wang S, Wang JY, Chen XY (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol 135: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, et al. (2005) Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17: 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol Biochem 46: 394–401 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Watanabe S, Inoue M, Yamasaki T, Seki M, Shinozaki K, Yokoyama S (2012) Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J Biol Chem 287: 7683–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Patra B, Li R, Pattanaik S, Yuan L (2013) Promoter analysis reveals cis-regulatory motifs associated with the expression of the WRKY transcription factor CrWRKY1 in Catharanthus roseus. Planta 238: 1039–1049 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, et al. (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64: 5085–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Xu W, Wang J, Wang L, Yao W, Yang Y, Xu Y, Ma F, Du Y, Wang Y (2013) The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol 200: 834–846 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J (2011) The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J 67: 61–71 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhu J, Ni Y, Cai Y, Zhang Z (2012) Expression profiling of HbWRKY1, an ethephon-induced WRKY gene in latex from Hevea brasiliensis in responding to wounding and drought. Trees (Berl) 26: 587–595 [Google Scholar]
- Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.