Key Points
Significant HLA locus mismatches responsible for transplant-related events were determined in 7898 unrelated marrow donor transplants.
This information provides a rationale for use of an algorithm for unrelated donor selection.
Abstract
We hypothesized that the compatibility of each HLA loci between donor and patient induced divergent transplant-related immunologic responses, which attributed to the individualized manifestation of clinical outcomes. Here, we analyzed 7898 Japanese pairs transplanted with T-cell–replete marrow from an unrelated donor with complete HLA allele typing data. Multivariable competing risk regression analyses were conducted to evaluate the relative risk (RR) of clinical outcomes after transplantation. A significant RR of HLA allele mismatch compared with match was seen with HLA-A, -B, -C, and -DPB1 for grade III-IV acute graft-versus-host disease (GVHD), and HLA-C for chronic GVHD. Of note, only HLA-C and HLA-DPB1 mismatch reduced leukemia relapse, and this graft-versus-leukemia effect of HLA-DPB1 was independent of chronic GVHD. HLA-DRB1 and HLA-DQB1 double (DRB1_DQB1) mismatch was revealed to be a significant RR for acute GVHD and mortality, whereas single mismatch was not. Thus, the number of HLA-A, -B, -C, -DPB1, and DRB1_DQB1 mismatches showed a clear-cut risk difference for acute GVHD, whereas the number of mismatches for HLA-A, -B, -C, and DRB1_DQB1 showed the same for mortality. In conclusion, we determined the biological response to HLA locus mismatch in transplant-related immunologic events, and provide a rationale for use of a personalized algorithm for unrelated donor selection.
Introduction
Allogeneic hematopoietic stem cell transplantation from unrelated donors (UR-HSCT) has been established as a mode of curative therapy for hematologic malignancies and other hematologic or immunologic disorders when an HLA-identical sibling donor is unavailable. Identification of the HLA locus matching at the allele level responsible for immunologic events related to HSCT is important in optimizing HLA matching and minimizing graft-versus-host disease (GVHD) and engraftment failure, as well as in enhancing the graft-versus-leukemia (GVL) effect.1-3
In the late 1990s, the Japan Marrow Donor Program (JMDP) demonstrated for the first time the effect of matching of HLA class I alleles on acute GVHD and the importance of HLA-A and -B allele matching for survival.2 Analysis of a large cohort in the United States also indicated that HLA allele mismatching is a significant risk factor for severe acute GVHD and mortality.3 Subsequent extensive analysis of the JMDP, US National Marrow Donor Program (NMDP), European registries, and the International Histocompatibility Workshop Group (IHWG) revealed considerable evidence that HLA allele compatibility,4-11 HLA haplotype,12,13 and HLA epitope14-16 are significantly associated with clinical outcomes.
We hypothesized that the compatibility of the respective HLA loci between donor and patient accounts for the divergence in transplant-related immunologic responses, and that this effect influences the individualized manifestation of clinical outcomes overall.
Here, to elucidate the biological effects of HLA locus matching on clinical outcomes, we selected pairs transplanted with T-cell–replete marrow for whom precise data for the complete HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles were obtained by retyping.
Methods
Study population
Unrelated donor transplant pairs (7898) from the JMDP database met the following criteria and were included in the analysis: (1) transplantation pairs retyped for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles; (2) T-cell–replete marrow without in vivo use of anti-thymocyte globulin or anti-T-cell monoclonal antibody for GVHD prophylaxis; (3) first transplantation; (4) Japanese ethnicity; and (5) survival for >7 days after transplantation. All pairs were transplanted between January 1993 and December 2010. A total of 12 502 pairs were facilitated through the JMDP during this period. The present 7898 study pairs with retyped HLA data consisted of 74.7% of the 10 575 pairs who matched selection criteria 2 to 5. No significant difference in clinical factors was seen between the HLA retyped and nonretyped pairs (data not shown). Patient diagnosis is listed in Table 1. Standard-risk leukemia was defined as chronic myeloid leukemia (CML) in the first chronic phase or acute lymphoblastic leukemia (ALL) and acute myeloblastic leukemia (AML) in the first complete remission (CR) at the time of transplantation, and diagnosed in 2508 patients, whereas high-risk leukemia was defined as transplantation at a more advanced stage than in standard-risk leukemia, and was diagnosed in 2772 patients. Sex matching between donor and patient was female (donor) to male (patient) in 1494 pairs, male to male in 3253, female to female in 1442, and male to female in 1709. For GVHD prophylaxis, no patient had in vivo use of anti-thymocyte globulin or a monoclonal antibody such as CAMPATH-1H. Tacrolimus-based regimens were used in 4779 patients, in combination with methotrexate in 4529; cyclosporine-based regimens were used in 3078, in combination with methotrexate in 2993; and other regimens were used in 41. The conditioning regimen was classified as myeloablative if it included total body irradiation (TBI) ≥8 Gy, oral busulfan (Bu) ≥9 mg/kg, IV Bu ≥7.2 mg/kg, or melphalan >140 mg/m2; otherwise, it was classified as a reduced-intensity regimen. Transplantation conditioning was done with a myeloablative regimen in 6653 patients and with a reduced-intensity regimen in 1245 patients. Patient and donor characteristics and HLA matching in the GVH direction in total pairs are shown in Table 1, and by HLA locus matching in supplemental Table 1 (see supplemental Data available on the Blood Web site).
Table 1.
Patient and donor characteristics
| Characteristics | Value |
|---|---|
| HLA locus matching match/mismatch, no. (%) | |
| HLA-A | 7048 (89)/850 (11) |
| HLA-B | 7475 (95)/423 (5) |
| HLA-C | 5565 (70)/2333 (30) |
| HLA-DRB1 | 5878 (74)/2020 (26) |
| HLA-DQB1 | 5681 (72)/2217 (28) |
| HLA-DPB1 | 2604 (33)/5294 (67) |
| Patient age, y | |
| Median (range) | 35 (0-77) |
| Donor age, y | |
| Median (range) | 34 (20-56) |
| Disease, no. (%) | |
| Acute lymphoblastic leukemia | 1861 (24) |
| Acute myeloblastic leukemia | 2609 (33) |
| Chronic myeloid leukemia | 983 (12) |
| Myelodysplastic syndrome | 841 (11) |
| Other leukemia | 312 (4) |
| Lymphoid malignancy | 542 (7) |
| Aplastic anemia | 489 (6) |
| Multiple myeloma | 33 (<1) |
| Others | 228 (3) |
| GVHD prophylaxis, no. (%) | |
| Cyclosporine based | 3078 (39) |
| Tacrolimus based | 4779 (61) |
| Others | 41 (<1) |
| Leukemia risk, no. (%) | |
| Standard | 2508 (32) |
| High | 2772 (35) |
| N/A | 2618 (33) |
| Conditioning, no. (%) | |
| Myeloablative | 6653 (84) |
| Reduced intensity | 1245 (16) |
| Sex matching (donor to patient), no. (%) | |
| Female to male | 1494 (19) |
| Male to male | 3253 (41) |
| Female to female | 1442 (18) |
| Male to female | 1709 (22) |
| Transplanted year period, no. (%) | |
| 1993-2000 | 2311 (29) |
| 2001-2005 | 3084 (39) |
| 2006-2010 | 2503 (32) |
Patient and donor characteristics by HLA locus matching are shown in supplemental Table 1.
N/A, not applicable.
A final clinical survey of patients was completed by September 2012 using the Transplant Registry Unified Management Program.17 Informed consent was obtained from patients and donors in accordance with the Declaration of Helsinki, and approval for the study was obtained from the Institutional Review Board of Aichi Cancer Center and the JMDP.
Outcome definition
Mortality was defined as time from transplantation to death from any cause. Clinical grading of acute GVHD was performed according to established criteria.18,19 Chronic GVHD was defined as limited or extensive chronic GVHD according to the Seattle criteria.20 Neutrophil engraftment was defined as more than 500 cells per cubic millimeter in peripheral blood at 3 consecutive measurements. Relapse was evaluated in patients with AML, ALL, or CML.
HLA typing and matching
All donor-patient pairs were retrospectively genotyped between 2009 and 2011 for all HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles at the field 1 and field 2 level of the 2010 World Health Organization Nomenclature for factors of the HLA system.21 The polymerase chain reaction–sequence specific oligonucleotide method was used for all samples, and the polymerase chain reaction–sequencing based typing method was used to confirm rare alleles and new alleles. HLA alleles were identified with >99.9% accuracy among Japanese. HLA alleles and their number are shown in supplemental Table 2, which also shows HLA loci and their level at confirmatory typing before transplantation.
HLA locus mismatch among the donor-recipient pairs was scored when the recipient’s HLA alleles or antigens were not shared by the donor in the GVH direction for acute GVHD, chronic GVHD, leukemia relapse and survival analysis, and in the HVG direction for neutrophil engraftment. HLA allele match rate in the GVH direction by HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 was 89.2%, 94.6%, 70.5%, 74.4%, 71.9%, and 33.0%, respectively, whereas serological HLA antigen match rate in the GVH direction by HLA-A, -B, -C, and -DR was 99.7%, 99.5%, 72.3%, and 91.8%, respectively.
Statistical analysis
Cumulative incidence of acute GVHD was assessed by a method described elsewhere.22 Overall survival was calculated using the Kaplan-Meier method. Competing events were defined as death without acute GVHD for acute GVHD; death without chronic GVHD for chronic GVHD; death without neutrophil engraftment for neutrophil engraftment; and death without relapse for leukemia relapse. Multivariable competing risk regression analyses23,24 were conducted to evaluate the impact of acute GVHD, chronic GVHD, leukemia relapse and neutrophil engraftment, and a Cox proportional regression model was used to evaluate the impact of mortality. The relative risk (RR) of HLA locus mismatch was compared with HLA locus match in the GVH direction for acute GVHD, chronic GVHD, leukemia relapse and mortality, and in the HVG direction for neutrophil engraftment. Confounders considered were sex (donor-recipient pair), patient age (linear), donor age (linear), disease, risk of leukemia relapse (standard and high), GVHD prophylaxis (cyclosporine-based regimen, tacrolimus-based regimen, and other regimen without cyclosporine and tacrolimus), preconditioning (myeloablative and reduced intensity), and period of transplant year (1992-2000, 2001-2005, 2006-2010). Transplanted cell number and ABO blood type matching were added as confounders in analyses of neutrophil engraftment. Missing data for confounder variables were treated as an unknown group. Acute GVHD, leukemia relapse, neutrophil engraftment, and survival were assessed in patients who survived >7 days, and chronic GVHD at 2 years was assessed in patients who survived 100 or more days after transplantation. Leukemia relapse at 5 years was assessed in patients who survived >7 days after transplantation for leukemia with AML, ALL, and CML. Risk of chronic GVHD on leukemia relapse was assessed by time-dependent covariate analysis in leukemia patients who survived 100 or more days after transplantation. Neutrophil engraftment at 100 days was assessed in all patients. A P value of <.01 was considered significant. All analyses were conducted using STATA version 12 (Stata Corp).
Results
Effect of HLA locus matching on acute GVHD and chronic GVHD
RR of HLA allele mismatch compared with HLA allele match for grade III-IV acute GVHD was highly significant for HLA-A, -B, -C, and -DPB1 (RR 1.29, P = .001; 1.42, P = .001; 1.63, P < .001; and 1.23, P = .001, respectively), but was not significant for HLA-DRB1 or -DQB1 (Table 2). RR of grade II-IV acute GVHD was highly significant for HLA-A, -B, -C, -DRB1, and -DPB1 (RR 1.18, P = .002; 1.28, P = .001; 1.27, P < .001; 1.24, P < .001; and 1.36, P < .001, respectively), but was not significant for HLA-DQB1 (Table 2).
Table 2.
Effect of HLA locus matching on acute GVHD and chronic GVHD in a multivariable competing risk regression model
| HLA | Match or mismatch* | N | Acute GVHD (Grade III-IV)† | Acute GVHD (Grade II-IV)† | N | Chronic GVHD‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR | 95% CI | P | RR | 95% CI | P | RR | 95% CI | P | ||||
| A | Match | 7048 | 1.00 | .001 | 1.00 | .002 | 5892 | 1.00 | .328 | |||
| Mismatch | 850 | 1.29 | 1.10-1.51 | 1.18 | 1.06-1.32 | 636 | 1.06 | 0.94-1.21 | ||||
| B | Match | 7475 | 1.00 | .001 | 1.00 | .001 | 6217 | 1.00 | .235 | |||
| Mismatch | 423 | 1.42 | 1.16-1.73 | 1.28 | 1.11-1.48 | 311 | 1.10 | 0.94-1.30 | ||||
| C | Match | 5565 | 1.00 | <.001 | 1.00 | <.001 | 4716 | 1.00 | <.001 | |||
| Mismatch | 2333 | 1.63 | 1.45-1.83 | 1.27 | 1.17-1.37 | 1812 | 1.24 | 1.13-1.35 | ||||
| DRB1 | Match | 5878 | 1.00 | .022 | 1.00 | <.001 | 4936 | 1.00 | .262 | |||
| Mismatch | 2020 | 1.21 | 1.03-1.43 | 1.24 | 1.11-1.39 | 1592 | 0.93 | 0.82-1.05 | ||||
| DQB1 | Match | 5681 | 1.00 | .336 | 1.00 | .126 | 4758 | 1.00 | .018 | |||
| Mismatch | 2217 | 1.08 | 0.92-1.27 | 1.09 | 0.98-1.22 | 1770 | 1.15 | 1.03-1.30 | ||||
| DPB1 | Match | 2604 | 1.00 | .001 | 1.00 | <.001 | 2223 | 1.00 | .367 | |||
| Mismatch | 5294 | 1.23 | 1.09-1.38 | 1.36 | 1.26-1.47 | 4305 | 1.04 | 0.96-1.12 | ||||
RR of respective HLA locus mismatches at the allele level was compared with HLA match adjusted with other HLA locus matching and clinical factors as listed in Table 1.
CI, confidence interval.
GVH direction.
Survived 7 or more days.
Survived 100 or more days.
RR of HLA allele mismatch compared with HLA allele match for chronic GVHD was significant for HLA-C (RR 1.24 P < .001), but not significant for HLA-A, -B, -DRB1, -DQB1, or -DPB1 (Table 2).
Effect of HLA locus matching on survival
RR of HLA allele mismatch compared with HLA allele match for mortality was highly significant in the HLA class I locus, namely HLA-A (1.29, P < .001), HLA-B (1.27, P < .001) and HLA-C (1.21, P < .001), but was not significant in the HLA class II locus, namely HLA-DRB1, -DQB1, and -DPB1 (Table 3).
Table 3.
Effect of HLA locus matching on leukemia relapse, engraftment, and mortality
| HLA | Match or mismatch* | Leukemia relapse† | Engraftment‡ | Mortality | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | RR | 95% CI | P | N | RR | 95% CI | P | N | RR | 95% CI | P | ||
| A | Match | 4847 | 1.00 | .381 | 6898 | 1.00 | .035 | 7048 | 1.00 | <.001 | |||
| Mismatch | 606 | 0.92 | 0.76-1.11 | 851 | 0.93 | 0.87-0.99 | 850 | 1.29 | 1.17-1.42 | ||||
| B | Match | 5163 | 1.00 | .493 | 7320 | 1.00 | .146 | 7475 | 1.00 | <.001 | |||
| Mismatch | 290 | 0.91 | 0.69-1.20 | 429 | 0.93 | 0.84-1.03 | 423 | 1.27 | 1.11-1.45 | ||||
| C | Match | 3865 | 1.00 | <.001 | 5511 | 1.00 | .049 | 5565 | 1.00 | <.001 | |||
| Mismatch | 1588 | 0.70 | 0.61-0.80 | 2238 | 0.95 | 0.90-1.00 | 2333 | 1.21 | 1.13-1.30 | ||||
| DRB1 | Match | 4045 | 1.00 | .468 | 5763 | 1.00 | .212 | 5878 | 1.00 | .125 | |||
| Mismatch | 1408 | 0.93 | 0.76-1.14 | 1986 | 0.95 | 0.89-1.03 | 2020 | 1.09 | 0.98-1.21 | ||||
| DQB1 | Match | 3924 | 1.00 | .974 | 5583 | 1.00 | .014 | 5681 | 1.00 | .145 | |||
| Mismatch | 1529 | 1.00 | 0.83-1.22 | 2166 | 0.91 | 0.85-0.98 | 2217 | 1.08 | 0.97-1.19 | ||||
| DPB1 | Match | 1792 | 1.00 | <.001 | 2531 | 1.00 | .126 | 2604 | 1.00 | .349 | |||
| Mismatch | 3661 | 0.69 | 0.61-0.77 | 5218 | 0.97 | 0.92-1.01 | 5294 | 1.03 | 0.96-1.11 | ||||
Multivariable competing risk regression analyses were conducted to evaluate the impact of leukemia relapse and neutrophil engraftment, and a Cox proportional regression model was conducted for mortality. RR of respective HLA locus mismatches at the allele level was compared with HLA match adjusted with other HLA locus matching and the clinical factors listed in Table 1 for leukemia relapse and mortality. Transplanted cell number and ABO blood type matching were added for neutrophil engraftment.
GVH direction for leukemia relapse and mortality; HVG direction for engraftment.
At 5 years after transplantation.
Neutrophil recovery to successive >500 per microliter measurement at 3 time points in 100 days.
Positive interaction of HLA-DRB1 mismatch and HLA-DQB1 mismatch in the risk of acute GVHD and survival
As HLA-DRB1 and HLA-DQB1 matching are closely linked in the HLA region and matching probability for HLA-DRB1 and HLA-DQB1 was 89%, stratified analysis of HLA-DRB1 matching and HLA-DQB1 matching was performed (Table 4). Pairs with HLA-DRB1 and HLA-DQB1 double (DRB1_DQB1) mismatch showed a significant risk of acute GVHD compared with pairs with both DRB1_DQB1 match (RR of grade III-IV, 1.32, P < .001; and RR of grade II-IV, 1.34, P < .001). HLA-DRB1 mismatch alone or HLA-DQB1 mismatch alone showed no significant difference in either grade III-IV or grade II-IV acute GVHD from DRB1_DQB1 match, respectively. Thus, DRB1_DQB1 mismatch induced a greater effect on acute GVHD than would be expected from the independent effect of either HLA-DRB1 or HLA-DQB1 mismatch alone.
Table 4.
Stratified analysis of HLA-DRB1 and HLA-DQB1 matching on acute GVHD and survival
| HLA matching* | N | Acute GVHD (Grade III-IV)† | Acute GVHD (Grade II-IV)† | Mortality† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RR | 95% CI | P | RR | 95% CI | P | RR | 95% CI | P | ||
| DRB1 match and DQB1 match | 5356 | 1.00 | 1.00 | 1.00 | ||||||
| DRB1 mismatch and DQB1 match | 325 | 0.98 | 0.74-1.28 | .866 | 1.19 | 1.00-1.42 | .046 | 1.04 | 0.88-1.22 | .662 |
| DRB1 match and DQB1 mismatch | 522 | 0.92 | 0.73-1.16 | .482 | 1.05 | 0.91-1.21 | .517 | 1.04 | 0.92-1.19 | .532 |
| DRB1 mismatch and DQB1 mismatch | 1695 | 1.32 | 1.16-1.50 | <.001 | 1.34 | 1.23-1.46 | <.001 | 1.17 | 1.08-1.27 | <.001 |
Multivariable competing risk regression analyses were conducted to evaluate the impact of acute GVHD and Cox proportional regression model for mortality. RR of the combination of HLA-DRB1 and/or -DQB1 mismatch was compared with HLA-DRB1 and -DQB1 match. Adjusted confounders were HLA-A, -B, -C, and -DPB1 locus matching and the clinical factors listed in Table 1.
GVH direction.
Survived 7 or more days.
As with acute GVHD, stratified analysis of both HLA locus matching showed that pairs with DRB1_DQB1 mismatch were at significantly higher risk of mortality than pairs with DRB1_ DQB1 match (RR 1.17, P < .001) (Table 4). In contrast, risk with HLA-DRB1 mismatch alone or HLA-DQB1 mismatch alone was not significantly different from that with DRB1_DQB1 match (RR 1.04, P = .662 and RR 1.04, P = .532, respectively).
The risk of double HLA locus mismatch combinations other than DRB1_DQB1 for grade III to IV acute GVHD and mortality were analyzed. As shown in supplemental Table 3, none of these double mismatch combinations revealed an epistatic effect of double HLA locus mismatch.
The same results were obtained using the same stratified analysis of HLA-DRB1 and -DQB1 with serological HLA-A, -B, and -DR match pairs (supplemental Table 4).
Effect of HLA locus matching on leukemia relapse
The occurrence of leukemia relapse within 5 years after transplantation was analyzed in patients with AML, ALL, and CML. RR of HLA allele mismatch compared with HLA allele match for leukemia relapse was low with high significance in HLA-C (RR 0.70, P < .001) and -DPB1 (RR 0.69, P < .001), but was not significant in HLA-A, -B, -DRB1, or -DQB1 (Table 3).
Independence of GVL effect of HLA-DPB1 mismatch from chronic GVHD
As described in the previous paragraph, HLA-DPB1 mismatch induced the GVL effect, but did not induce chronic GVHD. Chronic GVHD also induced the GVL effect. Therefore, the GVL effect of HLA-DPB1 matching in relation to chronic GVHD was analyzed in 2129 leukemia patients with HLA-A, -B, -C, -DRB1, and -DQB1 allele complete match donors who survived 100 or more days after transplantation. Multivariate competing risk regression analysis, including HLA-DPB1 matching and chronic GVHD, were performed with chronic GVHD treated as a time-dependent covariate (Table 5). Both limited-type chronic GVHD and extensive-type chronic GVHD were associated with a significantly lower leukemia relapse risk than no chronic GVHD. Furthermore, 1 and 2 DPB1 allele mismatch was associated with a significantly lower leukemia relapse risk than HLA-DPB1 match. Interaction analysis between HLA-DPB1 matching and chronic GVHD was not significant (RR 1.26, 95% CI 0.85-1.88, P = .255), indicating the lack of any effect modification between HLA-DPB1 matching and chronic GVHD.
Table 5.
Effect of chronic GVHD and HLA-DPB1 matching on leukemia relapse
| N | RR | 95% CI | P | |
|---|---|---|---|---|
| HLA-DPB1 | ||||
| Match* | 804 | 1.00 | ||
| 1-allele mismatch* | 971 | 0.70 | 0.58-0.84 | <.001 |
| 2-allele mismatch* | 354 | 0.54 | 0.41-0.72 | <.001 |
| Chronic GVHD | ||||
| No | 1232 | 1.00 | ||
| Limited type | 345 | 0.56 | 0.42-0.74 | <.001 |
| Extensive type | 552 | 0.46 | 0.36-0.58 | <.001 |
Multivariate competing risk regression analysis including HLA-DPB1 matching and chronic GVHD was performed by treating chronic GVHD as a time-dependent covariate adjusted for the clinical confounders listed in Table 1.
GVH direction.
When acute GVHD was added to this analysis, RR of grade III-IV acute GVHD and grade II-IV acute GVHD was 0.77 (95% CI 0.57-1.04, P = .091) and 0.82 (95% CI 0.68-0.99, P = .038), respectively. Thus, the effect of acute GVHD on leukemia relapse was not significant in patients who survived more than 100 days after transplantation.
Effect of HLA locus matching on neutrophil engraftment
Engraftment risk of neutrophils at 100 days after transplantation was assessed in all patients. Although RR of engraftment by HLA locus mismatch in the HVG direction showed the relatively lower risk range of 0.91 to 0.97 compared with HLA locus match in all 6 HLA loci, there was no significant HLA locus matching for neutrophil engraftment (Table 4).
Effect of multiple HLA locus mismatch on acute GVHD and survival
As the above HLA locus matching analysis indicated that multiple HLA locus mismatch was associated with a higher risk of adverse clinical outcomes of acute GVHD and survival, we next explored the appropriate HLA mismatch locus combination which revealed the effect of the number of HLA mismatch loci for acute GVHD and survival. The number of HLA 1-allele mismatches was summed after exclusion of 2-allele mismatches in each HLA locus. The combination of HLA-DRB1 1-allele mismatch and HLA-DQB1 1-allele mismatch (DRB1_DQB1 mismatch) was adopted and treated as 1 HLA locus mismatch.
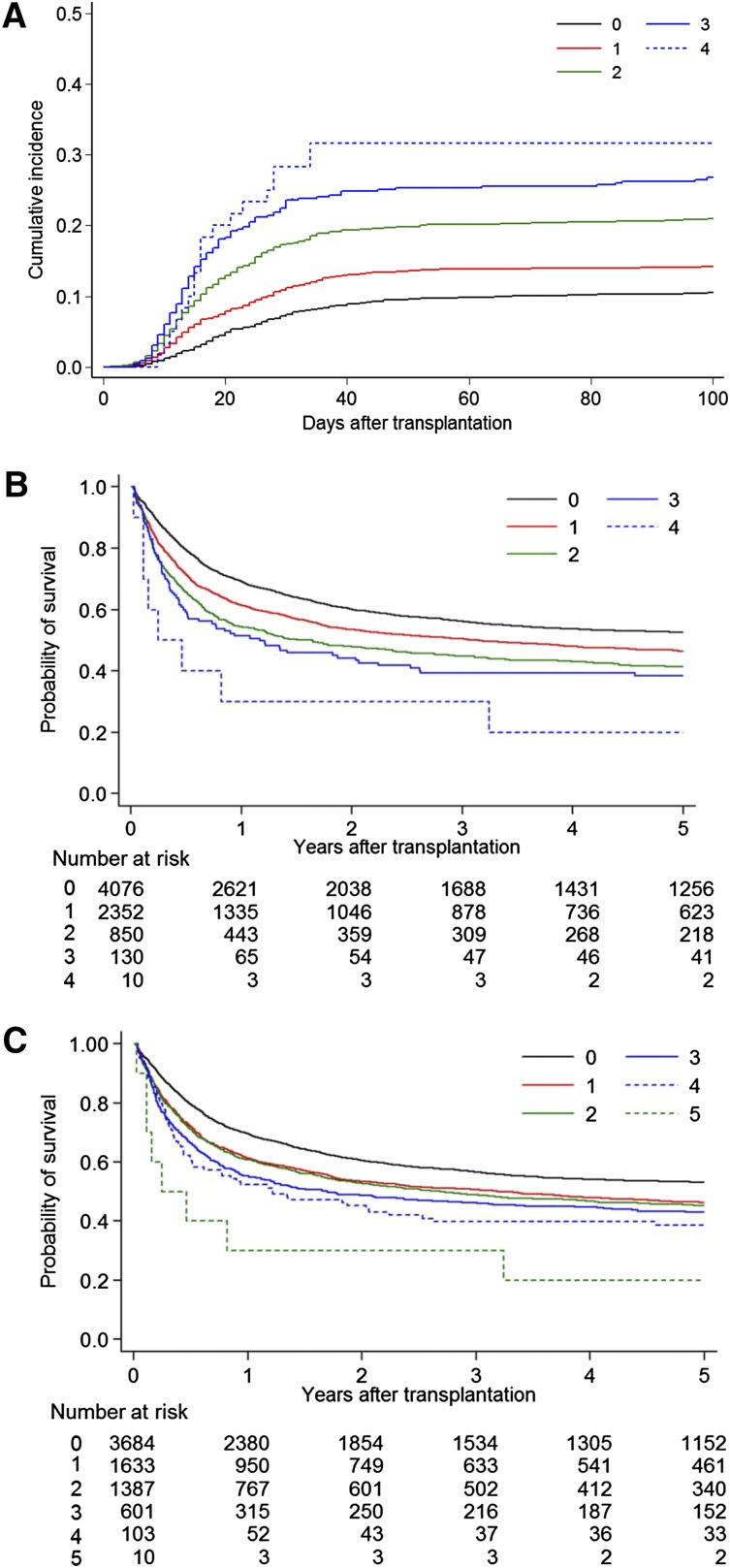
The cumulative incidence curve of grade III-IV acute GVHD by the number of HLA-A, -B, -C, -DPB1 locus mismatches and DRB1_DQB1 mismatch showed a clear-cut risk difference which discriminated 0, 1, 2, 3, and 4 HLA locus mismatches (Figure 1A). Specifically, compared with 0 mismatches (n = 1476), RRs for grade III-IV acute GVHD were 1.37 with 1 mismatch (n = 2549), 2.19 with 2 mismatches (n = 1377), 2.82 with 3 mismatches (n = 415), and 3.25 with 4 mismatches (n = 60) (P < .001).
Figure 1.
Acute GVHD and survival curve by the number of multiple HLA locus mismatches. The number of HLA 1-allele mismatches in the GVH direction, with exclusion of 2-allele mismatches, in each HLA locus was summed. (A) Cumulative incidence of grade III-IV acute GVHD by the mismatch number of HLA-A, -B, -C, -DRB1_DQB1, and -DPB1 at the allele level in the GVH direction. DRB1_DQB1: both HLA-DRB1 mismatch and HLA-DQB1 mismatch treated as 1 mismatch. 0: no mismatch (n = 1476); 1: 1 mismatch (n = 2549); 2: 2 mismatches (n = 1379); 3: 3 mismatches (n = 415); 4: 4 mismatches (n = 60). Cumulative incidence at 100 days was 0, 11% (95% CI, 9%-12%); 1, 14% (13%-16%); 2, 21% (19%-23%); 3, 27% (23%-31%); and 4, 32% (20%-44%). (B) Kaplan-Meier curve of survival by the mismatch number of HLA-A, -B, -C, and -DRB1_DQB1 at the allele level. Survival rate at 5 years was 0, 53% (95% CI, 51%-54%); 1, 46% (44%-49%); 2, 41% (38%-45%); 3, 38% (30%-47%); and 4, 20% (3%-47%). (C) Kaplan-Meier curve of survival by the mismatch number of HLA-A, -B, -C, -DRB1, and -DQB1 at the allele level.
To clarify the risk of a 2 HLA loci single-mismatch combination, each 2 mismatch combination was compared with the combination of HLA-A and -C mismatch for grade III-IV GVHD. As shown in supplemental Table 5, the risk of double mismatch combination pairs showed no significant differences, except DRB1_DQB1 mismatch and -DPB1 mismatch combination, albeit that the number of some of these combinations was too small for any precise evaluation of risk.
The most clear-cut risk difference discriminating 0, 1, 2, 3, and 4 HLA locus mismatches is seen in the Kaplan-Meier curve for survival by the number of HLA locus mismatches of HLA-A, -B, -C, and DRB1_DQB1 (Figure 1B). Compared with 0 mismatches (n = 4076), the RR for mortality was 1.28 with 1 mismatch (n = 2352), 1.57 with 2 mismatches (n = 850), and 1.73 with 3 mismatches (n = 130) (P < .001). To clarify the risk of a 2 HLA loci single-mismatch combination, each 2 mismatch combination was compared with the combination of HLA-A and -C mismatch for mortality. As shown in supplemental Table 5, there were no significant differences between each double mismatch combination.
When HLA-DRB1 mismatch and HLA-DQB1 mismatch were added separately to this analysis, the survival curves of 1, 2, 3, 4, and 5 mismatches showed less clear-cut differences (Figure 1C).
Significant clinical factors other than HLA matching which affected transplant-related clinical outcomes
Significant variables (P < .01) other than HLA locus matching for acute GVHD, chronic GVHD, leukemia relapse, neutrophil engraftment, and mortality are listed in Table 6. Patient age affected acute GVHD, chronic GVHD and mortality, and donor age affected chronic GVHD and mortality. Compared with ALL, CML showed a lower risk of chronic GVHD, leukemia relapse and mortality, and a higher risk of neutrophil engraftment. AML showed a lower risk of mortality, and aplastic anemia showed a lower risk of acute GVHD, chronic GVHD and mortality. A reduced conditioning regimen showed a higher risk of acute GVHD (grade III-IV) compared with a myeloablative regimen. Tacrolimus-based GVHD prophylaxis showed a higher rate of neutrophil engraftment compared with cyclosporine-based GVHD prophylaxis, but no increase for acute GVHD and chronic GVHD. Sex matching conversely affected acute GVHD and neutrophil engraftment. ABO blood type matching and transplanted cell number affected neutrophil engraftment. The passage of time, reflecting an improvement in clinical selection for variables, was associated with a lower risk of mortality as a whole. RR of all variables for each factor are shown in supplemental Table 6.
Table 6.
Significant factors other than HLA locus matching for clinical outcomes
| Outcomes, Significant factor (P < .01) | N | RR | 95% CI | P |
|---|---|---|---|---|
| Acute GVHD (grade III-IV) | ||||
| Patient age, year linear | 7898 | 0.99 | 0.99-1.00 | <.001 |
| Disease | ||||
| ALL (Ref.) | 1861 | 1.00 | ||
| Aplastic anemia | 489 | 0.41 | 0.26-0.64 | <.001 |
| Conditioning | ||||
| Myeloablative (Ref.) | 6653 | 1.00 | ||
| Reduced intensity | 1245 | 1.26 | 1.07-1.50 | .007 |
| Sex matching | ||||
| Female to male (Ref.) | 1494 | 1.00 | ||
| Female to female | 1442 | 0.77 | 0.64-0.92 | .005 |
| Chronic GVHD | ||||
| Patient age, year linear | 6528 | 1.01 | 1.00-1.01 | <.001 |
| Donor age, year linear | 6528 | 1.00 | 1.00-1.00 | <.001 |
| Disease | ||||
| ALL (Ref.) | 1568 | 1.00 | ||
| CML | 813 | 1.28 | 1.13-1.46 | <.001 |
| Aplastic anemia | 425 | 0.64 | 0.46-0.89 | .008 |
| Transplanted year | ||||
| 1993-2000 (Ref.) | 1865 | 1.00 | ||
| 2006-2010 | 2117 | 0.74 | 0.65-0.83 | <.001 |
| Leukemia relapse | ||||
| Disease | ||||
| ALL (Ref.) | 1861 | 1.00 | ||
| CML | 983 | 0.49 | 0.39-0.60 | <.001 |
| Leukemia risk | ||||
| Standard (Ref.) | 2508 | 1.00 | ||
| High | 2772 | 2.62 | 2.31-2.98 | <.001 |
| Transplanted year | ||||
| 1993-2000 (Ref.) | 1815 | 1.00 | ||
| 2001-2005 | 2079 | 1.34 | 1.14-1.56 | <.001 |
| 2006-2010 | 1559 | 1.31 | 1.09-1.57 | .004 |
| Neutrophil engraftment | ||||
| Disease | ||||
| ALL (Ref.) | 1831 | 1.00 | ||
| CML | 959 | 0.90 | 0.84-0.97 | .005 |
| GVHD prophylaxis | ||||
| Cyclosporin based (Ref.) | 2998 | 1.00 | ||
| Tacrolimus based | 4716 | 1.12 | 1.07-1.18 | <.001 |
| Leukemia risk | ||||
| Standard (Ref.) | 2486 | 1.00 | ||
| High | 2703 | 0.81 | 0.77-0.85 | <.001 |
| Sex matching | ||||
| Female to male (Ref.) | 1462 | 1.00 | ||
| Male to male | 3182 | 1.10 | 1.03-1.16 | .002 |
| Male to female | 1686 | 1.12 | 1.05-1.20 | .001 |
| ABO blood type matching | ||||
| Match (Ref.) | 3455 | 1.00 | ||
| Major mismatch | 1452 | 0.88 | 0.83-0.94 | <.001 |
| Transfused nuclear cell no./weight, kg, ×10E8 | ||||
| <2.0 (Ref.) | 1038 | 1.00 | ||
| 2.0-4.0 | 4999 | 1.34 | 1.26-1.42 | <.001 |
| ≤4.0 | 1068 | 1.42 | 1.31-1.55 | <.001 |
| Mortality | ||||
| Patient age, year linear | 7898 | 1.02 | 1.02-1.02 | <.001 |
| Donor age, year linear | 7898 | 1.01 | 1.01-1.02 | <.001 |
| Disease | ||||
| ALL (Ref.) | 1861 | 1.00 | ||
| AML | 2609 | 0.81 | 0.74-0.89 | <.001 |
| CML | 983 | 0.72 | 0.63-0.81 | <.001 |
| MDS | 841 | 0.50 | 0.40-0.64 | <.001 |
| Other leukemia | 312 | 0.68 | 0.52-0.89 | .005 |
| Lymphoid malignancy | 542 | 0.54 | 0.42-0.70 | <.001 |
| Aplastic anemia | 489 | 0.30 | 0.23-0.40 | <.001 |
| Leukemia risk | ||||
| Standard (Ref.) | 2508 | 1.00 | ||
| High | 2772 | 2.19 | 2.01-2.39 | <.001 |
| Sex matching | ||||
| Female to male (Ref.) | 1494 | 1.00 | ||
| Female to female | 1442 | 0.81 | 0.72-0.90 | <.001 |
| Transplanted year | ||||
| 1993-2000 (Ref.) | 2311 | 1.00 | ||
| 2001-2005 | 3084 | 0.81 | 0.74-0.89 | <.001 |
| 2006-2010 | 2503 | 0.67 | 0.60-0.75 | <.001 |
Multivariable competing risk regression analyses were conducted to evaluate the impact of acute GVHD, chronic GVHD, leukemia relapse and neutrophil engraftment, and a Cox proportional regression model for mortality. RR of respective factors was compared with the reference factor adjusted by HLA locus matching and clinical factors. Factors with significance (P < .01) were listed. RR of all variables is shown in supplemental Table 6.
Ref., reference factor.
Discussion
In this study, the accumulation of UR-HSCT clinical data and HLA retyping data through the JMDP allowed us to analyze biological immune responses of transplant-related events by HLA locus matching at the allele level. As data for some of the previously identified HLA alleles were no longer up to date, precise assessment of HLA matching required that we renew HLA allele types to meet the recent HLA nomenclature. We performed HLA allele typing for all HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1. In addition, to elucidate the biological immune responses, we strictly restricted pairs to non-T-cell–depleted bone marrow as stem cell source and to Japanese pairs as ethnic background.
Significant RRs of HLA allele mismatch compared with match were HLA-A, -B, -C and -DPB1 for grade III-IV acute GVHD; HLA-C for chronic GVHD; HLA-C and HLA-DPB1 for leukemia relapse; and HLA-A, -B, -C for mortality. Furthermore, stratified analysis of HLA-DRB1 and -DQB1 revealed that HLA-DRB1_DQB1 double mismatch was a significant RR for severe acute GVHD and mortality. These findings supersede previous JMDP studies2,4,5 and provide a rationale for the development of an algorithm for unrelated donor selection.
HLA-A and/or -B locus mismatch induced significant severe acute GVHD but not the GVL effect, and resulted in a lower survival rate than in HLA match pairs. Since the first report from the JMDP showing the risk of HLA-A and/or -B for acute GVHD and survival, both the selection of HLA-A and/or -B mismatch donors and the impact of this mismatch have dramatically decreased. In spite of this information bias, HLA-A and/or -B allele mismatch should be considered in donor selection and GVHD prophylaxis as a high-risk HLA locus of severe acute GVHD and mortality. The NMDP6,7 and IHWG reports10 also indicated the risk of HLA-A and/or -B mismatch.
HLA-C mismatch induces not only a high risk of acute GVHD but also a high risk of chronic GVHD and low risk of leukemia relapse. When an HLA-C mismatch donor is considered for the induction of GVL effect in general practice, the risk of acute GVHD and chronic GVHD should be kept in mind. This effect of HLA-C mismatch on leukemia relapse and survival confirms findings of previous JMDP5,25 and NMDP reports.6 In addition to T-cell recognition of the mismatched amino acid difference in HLA-C molecules,14 NK-cell receptor KIR2DL ligand mismatch should also be considered, as described elsewhere.5,26 The effect of KIR ligand mismatch remains controversial worldwide. Further analysis of HLA-C allele mismatch combination in conjunction with KIR receptor using JMDP pairs and comparison with non-JMDP pairs will help to elucidate the mechanism of HLA-C and KIR-related immunologic reaction and solve these discrepancies.
Our stratified analysis showed that the concurrent presence of HLA-DRB1 mismatch and HLA-DQB1 mismatch was associated with a high risk of severe acute GVHD and mortality, whereas the presence of HLA-DRB1 mismatch or HLA-DQB1 mismatch only did not induce a significantly higher risk of severe acute GVHD or survival. This epistasis of 2 HLA loci mismatch needs to be interpreted with care. In particular, the relatively small number of DRB1 alone mismatch pairs (n = 325) might have limited the statistical power. An additional consideration is that no other HLA 2 locus mismatch combination showed such an epistatic effect of DRB1 and DQB1 on the risk of severe acute GVHD and mortality (supplemental Table 3). Interaction of the HLA-DQB1 molecule with that of HLA-DR groups might evoke unique immune reactions related to allogeneic transplantation for severe acute GVHD. As reported by Fernández-Viña et al,27 the effect of the low expression of HLA loci, not only of DP, DQ but also the DRB3/4/5 locus, needs to be explored.
As also reported by Shaw et al,8 the present study found that HLA-DPB1 mismatch induced acute GVHD and the GVL effect, but did not affect survival. HLA-DP antigen was originally typed using the in vitro–primed lymphocyte test. From this, HLA-DPB1 and its matching are known to play a distinct biological role in immunologic reactions. Indeed, the GVL effect in HLA-DPB1 mismatch combination in our previous analysis provided a rationale to explain the induction of the GVL effect and less acute GVHD.25 In addition, our present results show for the first time that HLA-DPB1 mismatch and the occurrence of chronic GVHD affect the GVL effect independently of each other. The mechanism of the GVL effect induced by T-cell recognition of the HLA-DPB1 allele mismatch might differ from that induced by chronic GVHD. Potential candidates for the molecular implications of acute GVHD and the GVL effect include the high-risk HLA-DPB1 mismatch combinations for severe acute GVHD reported from the JMDP14,25 and the effect of T-cell-epitope matching at HLA-DPB1 reported by Fleischhauer et al.16
When the impacts of the respective HLA locus matching described above are taken together, RR of mismatch of HLA class I loci is heightened, with a range of RR 1.29 to 1.63 for severe acute GVHD and RR 1.21 to 1.27 for mortality. For HLA class II loci, mismatch of double HLA-DRB1 and -DQB1 should be considered, with RR 1.32 for severe acute GVHD and 1.14 for mortality. Thus, appropriate combinations of HLA loci need to be selected according to the risk of each HLA locus and the interaction of HLA-DRB1 and -DQB1 for donor selection.
The number of multiple mismatches of HLA-A, -B, -C, -DRB1_DQB1 and -DPB1 showed good predictive value for the risk of severe acute GVHD. Furthermore, prediction of the risk of mortality after transplantation should consider the number of multiple mismatches of HLA-A, -B, -C, and -DRB1_DQB1 locus, and not of HLA-A, -B, -C, -DRB1, and -DQB1. This mismatch score is in agreement with reports from the NMDP6,7,11 and Loiseau et al28 showing that mismatch of HLA-DQB1 demonstrated an additive adverse effect in outcomes. Our analysis using the present data set is consistent with findings from a recent report29 which showed a significant risk with single HLA-DRB1 mismatch using the Japanese HSCT dataset in leukemia patients with HLA-A, -B, -C and -DRB1 allele data.
Our analysis also provides further information for personalized unrelated donor selection. In cases where the transplant team is particularly concerned about the prevention of severe acute GVHD, leukemia relapse or early mortality, the specific HLA locus mismatches and number of mismatched locus should be considered with regard to the patient’s disease, disease status, and clinical condition. The benefit of HLA-C mismatch and HLA-DPB1 mismatch for a specific GVL effect in leukemia patients is noted.
A number of other important factors will also impact clinical outcomes and change the magnitude of the HLA barrier. In the present study, clinical risk factors other than HLA matching are shown in Table 6. The magnitude of risks for HLA locus mismatch is compatible with that for clinical factors as a whole.
Candidates range widely, from ethnicity of the donor and patient30 to HLA haplotype12,13 and other genetic polymorphisms both inside and outside the HLA region.31-33 Clinical risk factors in the present study agree with those reported previously, including procedures for GVHD prophylaxis, intensity of the conditioning regimen,34 disease,35,36 leukemia relapse risk, and stem cell source.37 It will be interesting to determine whether these candidates shift the HLA barrier quantitatively and maintain the same divergent effect of each HLA locus, or qualitatively alter the HLA locus-specific barrier. As unrelated peripheral blood stem cell transplantation was not facilitated by the JMDP during the period of this study, we were unable to analyze the data for unrelated PBSCT. PBSCT might heighten the threshold of the HLA barrier, as reported by the NMDP.37 Analysis for unrelated cord blood transplantation compared with unrelated donor transplantation38,39 might shed light on the latter possibility and help elucidate the altered immune mechanisms which cause transplant-related events.
Our homogeneous cohort was restricted to Japanese pairs, which allowed us to elucidate biological responses based on this particular genetic background. However, individual ethnic groups present distinct HLA allele and HLA haplotypes, and these differences in the ethnic background of patient and donor might impact transplant-related clinical outcomes.40 Our findings need to be validated using unrelated donor transplantation data for other ethnic groups.
In conclusion, we clearly determined the HLA locus mismatches responsible for diverse transplant-related immunologic events. Furthermore, we provide a rationale for the development of an algorithm for unrelated donor selection.
Acknowledgments
The authors thank the staff members of the transplantation centers, donor centers, and the Japan Marrow Donor Program office for their generous cooperation.
This work was supported by grants from the Japanese Ministries of Health, Labor and Welfare (H23-Immunology-010 and H26-Immunology-106) and Education, Culture, Sports, Science and Technology (MEXT KAKENHI grant no. 22133011).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.M., K. Kashiwase, K. Matsuo, M.M., T.I., H. Saji, S.K., Y.K., and T.S. participated in the design of the study; K. Kashiwase, F.A., and T.Y. performed the histocompatibility analysis; M.O., N.D., T.E., Y.M., K. Miyamura, T.M., H. Sao, Y.A., and K. Kawa organized and collected the clinical data and samples for transplantation; Y.M., S.M., and K. Matsuo performed statistical data analysis; Y.M., S.M., and K. Kashiwase performed the analysis and wrote the paper; and all authors checked the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yasuo Morishima, Division of Epidemiology and Prevention, Aichi Cancer Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan; e-mail: ymorisim@aichi-cc.jp.
References
- 1.Kernan NA, Bartsch G, Ash RC, et al. Analysis of 462 transplantations from unrelated donors facilitated by the National Marrow Donor Program. N Engl J Med. 1993;328(9):593–602. doi: 10.1056/NEJM199303043280901. [DOI] [PubMed] [Google Scholar]
- 2.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339(17):1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 3.Petersdorf EW, Gooley TA, Anasetti C, et al. Optimizing outcome after unrelated marrow transplantation by comprehensive matching of HLA class I and II alleles in the donor and recipient. Blood. 1998;92(10):3515–3520. [PubMed] [Google Scholar]
- 4.Morishima Y, Sasazuki T, Inoko H, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002;99(11):4200–4206. doi: 10.1182/blood.v99.11.4200. [DOI] [PubMed] [Google Scholar]
- 5.Morishima Y, Yabe T, Matsuo K, et al. Japan Marrow Donor Program. Effects of HLA allele and killer immunoglobulin-like receptor ligand matching on clinical outcome in leukemia patients undergoing transplantation with T-cell-replete marrow from an unrelated donor. Biol Blood Marrow Transplant. 2007;13(3):315–328. doi: 10.1016/j.bbmt.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 7.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 8.Shaw BE, Gooley TA, Malkki M, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110(13):4560–4566. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 9.Fürst D, Müller C, Vucinic V, et al. High-resolution HLA matching in hematopoietic stem cell transplantation: a retrospective collaborative analysis. Blood. 2013;122(18):3220–3229. doi: 10.1182/blood-2013-02-482547. [DOI] [PubMed] [Google Scholar]
- 10.Petersdorf EW, Malkki M, Hsu K, et al. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. 16th IHIW: International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Int J Immunogenet. 2013;40(1):2–10. doi: 10.1111/iji.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidala J, Lee SJ, Ahn KW, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124(16):2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4(1):e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morishima S, Ogawa S, Matsubara A, et al. Japan Marrow Donor Program. Impact of highly conserved HLA haplotype on acute graft-versus-host disease. Blood. 2010;115(23):4664–4670. doi: 10.1182/blood-2009-10-251157. [DOI] [PubMed] [Google Scholar]
- 14.Kawase T, Morishima Y, Matsuo K, et al. Japan Marrow Donor Program. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110(7):2235–2241. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 15.Pidala J, Wang T, Haagenson M, et al. Amino acid substitution at peptide-binding pockets of HLA class I molecules increases risk of severe acute GVHD and mortality. Blood. 2013;122(22):3651–3658. doi: 10.1182/blood-2013-05-501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischhauer K, Shaw BE, Gooley T, et al. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atsuta Y, Suzuki R, Yoshimi A, et al. Unification of hematopoietic stem cell transplantation registries in Japan and establishment of the TRUMP System. Int J Hematol. 2007;86(3):269–274. doi: 10.1532/IJH97.06239. [DOI] [PubMed] [Google Scholar]
- 18.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18(4):295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Przepiorka D, Weisdorf D, Martin P, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15(6):825–828. [PubMed] [Google Scholar]
- 20.Shulman HM, Sullivan KM, Weiden PL, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med. 1980;69(2):204–217. doi: 10.1016/0002-9343(80)90380-0. [DOI] [PubMed] [Google Scholar]
- 21.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75(4):291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gooley TA, Leisenring W, Crowley J, Storer BE. Estimation of failure probabilities in the presence of competing risks: new representations of old estimators. Stat Med. 1999;18(6):695–706. doi: 10.1002/(sici)1097-0258(19990330)18:6<695::aid-sim60>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 23.Cox DR. Regression models and life-tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 24.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 25.Kawase T, Matsuo K, Kashiwase K, et al. Japan Marrow Donor Program. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009;113(12):2851–2858. doi: 10.1182/blood-2008-08-171934. [DOI] [PubMed] [Google Scholar]
- 26.Yabe T, Matsuo K, Hirayasu K, et al. Japan Marrow Donor Program. Donor killer immunoglobulin-like receptor (KIR) genotype-patient cognate KIR ligand combination and antithymocyte globulin preadministration are critical factors in outcome of HLA-C-KIR ligand-mismatched T cell-replete unrelated bone marrow transplantation. Biol Blood Marrow Transplant. 2008;14(1):75–87. doi: 10.1016/j.bbmt.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Viña MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121(22):4603–4610. doi: 10.1182/blood-2013-02-481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loiseau P, Busson M, Balere ML, et al. HLA Association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant. 2007;13(8):965–974. doi: 10.1016/j.bbmt.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Kanda Y, Kanda J, Atsuta Y, et al. Impact of a single human leucocyte antigen (HLA) allele mismatch on the outcome of unrelated bone marrow transplantation over two time periods. A retrospective analysis of 3003 patients from the HLA Working Group of the Japan Society for Blood and Marrow Transplantation. Br J Haematol. 2013;161(4):566–577. doi: 10.1111/bjh.12279. [DOI] [PubMed] [Google Scholar]
- 30.Morishima Y, Kawase T, Malkki M, et al. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Significance of ethnicity in the risk of acute graft-versus-host disease and leukemia relapse after unrelated donor hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2013;19(8):1197–1203. doi: 10.1016/j.bbmt.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersdorf EW, Malkki M, Gooley TA, et al. MHC-resident variation affects risks after unrelated donor hematopoietic cell transplantation. Sci Transl Med. 2012;4(144):144ra101. [DOI] [PMC free article] [PubMed]
- 32.Petersdorf EW, Malkki M, Horowitz MM, Spellman SR, Haagenson MD, Wang T. Mapping MHC haplotype effects in unrelated donor hematopoietic cell transplantation. Blood. 2013;121(10):1896–1905. doi: 10.1182/blood-2012-11-465161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harkensee C, Oka A, Onizuka M, et al. Japan Marrow Donor Program. Single nucleotide polymorphisms and outcome risk in unrelated mismatched hematopoietic stem cell transplantation: an exploration study. Blood. 2012;119(26):6365–6372. doi: 10.1182/blood-2012-01-406785. [DOI] [PubMed] [Google Scholar]
- 34.Weisdorf D, Zhang MJ, Arora M, Horowitz MM, Rizzo JD, Eapen M. Graft-versus-host disease induced graft-versus-leukemia effect: greater impact on relapse and disease-free survival after reduced intensity conditioning. Biol Blood Marrow Transplant. 2012;18(11):1727–1733. doi: 10.1016/j.bbmt.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horan J, Wang T, Haagenson M, et al. Evaluation of HLA matching in unrelated hematopoietic stem cell transplantation for nonmalignant disorders. Blood. 2012;120(14):2918–2924. doi: 10.1182/blood-2012-03-417758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yagasaki H, Kojima S, Yabe H, et al. Japan Marrow Donor Program. Acceptable HLA-mismatching in unrelated donor bone marrow transplantation for patients with acquired severe aplastic anemia. Blood. 2011;118(11):3186–3190. doi: 10.1182/blood-2011-04-349316. [DOI] [PubMed] [Google Scholar]
- 37.Anasetti C, Logan BR, Lee SJ, et al. Blood and Marrow Transplant Clinical Trials Network. Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med. 2012;367(16):1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atsuta Y, Morishima Y, Suzuki R, et al. Japan Marrow Donor Program and Japan Cord Blood Bank Network. Comparison of unrelated cord blood transplantation and HLA-mismatched unrelated bone marrow transplantation for adults with leukemia. Biol Blood Marrow Transplant. 2012;18(5):780–787. doi: 10.1016/j.bbmt.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Spellman SR, Eapen M, Logan BR, et al. National Marrow Donor Program; Center for International Blood and Marrow Transplant Research. A perspective on the selection of unrelated donors and cord blood units for transplantation. Blood. 2012;120(2):259–265. doi: 10.1182/blood-2012-03-379032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morishima Y, Kawase T, Malkki M, Petersdorf EW International Histocompatibility Working Group in Hematopoietic Cell Transplantation Component. Effect of HLA-A2 allele disparity on clinical outcome in hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007;69(suppl 1):31–35. doi: 10.1111/j.1399-0039.2006.759_3.x. [DOI] [PubMed] [Google Scholar]