Abstract
Aim:
Interferon-γ inducible protein 16 (IFI16), a DNA sensor for DNA double-strand break (DSB), is expressed in most human hepatocellular carcinoma cell (HCC) lines. In this study we investigated the re-localization of chromatin-bound IFI16 by Nutlin-3, a DNA damage agent, in HCC cells in vitro, and the potential mechanisms.
Methods:
Human HCC SMMC-7721 (wild-type TP53), Huh-7 (mutant TP53), Hep3B (null TP53) and normal fetal liver L02 cell lines were examined. DSB damage in HCC cells was detected via γH2AX expression and foci formation assay. The expression of IFI16 and IFNB mRNA was measured using RT-PCR, and subcellular localization and expression of the IFI16 protein were detected using chromatin fractionation, Western blot analysis, and fluorescence microscopy.
Results:
Treatment of SMMC-7721 cells with Nutlin-3 (10 μmol/L) or etoposide (40 μmol/L) induced significant DSB damage. In SMMC-7721 cells, Nutlin-3 significantly increased the expression levels of IFI16 and IFNB mRNA, and partially redistributed chromatin-bound IFI16 protein to the cytoplasm. These effects were blocked by pretreatment with pifithrin-α, a p53 inhibitor. Furthermore, Nutlin-3 did not induce ectopic expression of IFI16 protein in Huh-7 and Hep3B cells. Moreover, the association of IFI16 with chromatin and Nutlin-3-induced changes in localization were not detected in L02 cells.
Conclusion:
Nutlin-3 regulates the subcellular localization of IFI16 in HCC cells in vitro in a p53-dependent manner.
Keywords: IFI16, DNA double-strand break, Nutlin-3, etoposide, pifithrin-α, p53, human hepatocellular carcinoma, SMMC-7721 cell, Huh-7 cell, Hep3B cell
Introduction
Hepatocellular carcinoma (HCC) is a global health problem usually associated with an inactive form of p531. Murine double minute 2 (MDM2), a p53-selective E3 ligase, negatively controls p53 function by ubiquitination and degradation and ultimately inhibits TP53 transcription and translation2. Restoration of p53 activation by antagonizing MDM2 might offer a new therapeutic strategy.
Nutlin-3, a MDM2 antagonist, disrupts the interaction between p53 and MDM2 and dissociates p53 to bind to other C-terminal modifiers such as interferon-γ inducible protein 16 (IFI16)3. IFI16 belongs to the PYHIN family4, which contains a pyrin domain (PYD) at the N-terminus and two C-terminal HIN200 domains, HIN-A and HIN-B, which can sense double-stranded DNA (dsDNA)3. Meanwhile, the IFI16 HIN-A and HIN-B domains interact with the C-terminus and the core DNA binding region of p53, respectively3. The role of IFI16 is more diverse than that of a traditional interferon-inducible gene5. First, IFI16 regulates cell proliferation6 and cell cycle7 and inhibits cell growth as observed in breast cancer8, head and neck squamous cell carcinoma9, and prostate cancer10. Second, IFI16 contributes to the suppression of viral replication and the promotion of viral clearance to control HBV11 or Herpes viruses12 infection. Third, IFI16, one of the AIM2-like receptors (ALRs), acts as a DNA sensor and triggers innate immune response leading to IFN-β production13 or inflammasome formation14. Additionally, IFI16 is involved in DNA double-strand break (DSB) repair15, autophagy16, cellular senescence17,18, and autoimmune disease such as systemic lupus erythematosus (SLE)19. IFI16 is expressed in most human HCC cell lines and tissues but not in healthy adult liver cells18. IFI16 triggers innate immune responses to suppress HBV/HCV replication and promote viral clearance11,20. Our previous hypothesis showed that IFI16 mis-localization may be a contributing factor to HCC progression21. However, the role of IFI16 subcellular localization is still unclear in HCC chemotherapy.
The present study focused on the relationship between the re-localization of chromatin-bound IFI16 and Nutlin-3 in HCC chemotherapy and the mechanisms underlying the wild-type p53-induced IFI16 re-localization.
Materials and methods
Cell lines and agents
SMMC-7721 (wild-type TP53), Huh-7 (mutant TP53), Hep3B (null TP53) and normal fetal liver L02 cell lines were generous gifts from Prof Cong LIU of the West China Second University Hospital/West China Women's and Children's Hospital.
Nutlin-3, Etoposide, and Pifithrin-α (PFT-α) were purchased from Sigma-Aldrich Technology Company and stored frozen as a 20 mmol/L stock solution in DMSO (Sigma, USA).
Cell culture and treatment
The cultivation medium contains DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA), 100 U/mL penicillin and 100 μg/mL streptomycin. The cells were cultured at 37 °C under a 5% CO2 atmosphere.
The Nut group of SMMC-7721, Huh-7, Hep3B and L02 cells were cultured with 10 μmol/L Nutlin-3 for 48 h22,23. The PFT group of SMMC-7721 cells was treated with 20 μmol/L PFT-α for 48 h. The PFT+Nut group of SMMC-7721 cells were pretreated with PFT-α (20 μmol/L) for 12 h24 and then exposed to Nutlin-3 (10 μmol/L) for 36 h together with PFT-α. The Eto group of SMMC-7721 cells was cultured with 40 μmol/L Etoposide for 48 h25, which was used as the positive control of DNA DSB damage. The final concentrations of the tested compounds were prepared by diluting the stock solutions in DMEM. DMEM containing 0.1% DMSO was used as a control.
Real time PCR (RT-PCR)
SMMC-7721 cells were seeded in 6-well plates (3×105 cells/well) and treated as described above. Then, total RNA was isolated with TRIzol (Invitrogen) and reverse transcribed using a Prime Script™ Kit (TaKaRa). The RT-PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems, USA). The specific human primers used were as follows (Table 1, forward and reverse).
Table 1. Primer sequences.
|
IFI16 |
5′-GAAGTGCCAGCGTAACTCCTA-3′ |
| 5′-TACCTCAAACACCCCATTCAC-3′ | |
| IFNB1 | 5′-TCCACTACAGCTCTTTCCATGA-3′ |
| 5′-AGTATTCAAGCCTCCCATTCAA-3′ | |
| TP53 | 5′-TAGTGTGGTGGTGCCCTATGAG-3′ |
| 5′-AGTGTGATGATGGTGAGGATGG-3′ | |
| ACTB | 5′-GGCATCCACGAAACTACCTTCA-3′ |
| 5′-GTGATCTCCTTCTGCATCCTGTC-3′ |
The TP53 and ACTB gene were used as the positive and internal control, respectively. The RT-PCR was performed as follows: 40 cycles of 95 °C for 5 s and 60 °C for 30 s. All samples were analyzed in triplicate on the same plate.
Chromatin fractionation and Western blot analysis
SMMC-7721 and L02 cells were seeded in 6-well plates (3×105 cells/well), treated as described above, and fractionated to obtain the chromatin26. Whole-cell extracts were directly prepared in an SDS sample buffer (50 mmol/L Tris-HCl pH 6.8, 1% SDS, 10% glycerol, 5% β-ME, 0.01% bromophenol blue). The primary antibodies were rabbit anti-γH2AX polyclonal antibody (bs-3185R, Bioss, diluted 1:200), mouse anti-IFI16 monoclonal antibody (ab50004, Abcam, diluted 1:1000), rabbit anti-H2b polyclonal antibody (BS1657, Bioworld, diluted 1:500), and β-actin antibody (BA2305, Boster, diluted 1:500). The secondary antibodies, goat anti-mouse IgG-HRP (sc-2005, diluted 1:5000) and goat anti-rabbit IgG-HRP (sc-2004, diluted 1:5000), were purchased from Santa Cruz Biotechnology. H2b and β-actin served as the quality controls for nuclear fraction and cytoplasmic fraction, respectively.
Immunofluorescence (IF) assay
SMMC-7721, Huh-7, Hep3B, and L02 cells were cultured for 48 h on glass coverslips in 24-well plates (2×105 cells/well) with or without Nutlin-3 treatment. The PFT and PFT+Nut group of SMMC-7721 cells were treated as described above. The samples were fixed, permeabilized, blocked, and then incubated for 1 h with rabbit anti-γH2AX polyclonal antibody (bs-3185R, Bioss, diluted 1:200) or mouse anti-IFI16 monoclonal antibody (ab50004, Abcam, diluted 1:1000) at 37 °C and then with goat anti-mouse FITC-conjugated secondary antibody (F0257, Sigma, diluted 1:100) or sheep anti-rabbit Cy3-conjugated secondary antibody (C2306, Sigma, diluted 1:100) at 37 °C for 1 h. Cells were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (10 μg/mL) (Sigma, USA). Images were captured via fluorescence microscopy (Olympus BX51).
Statistical analysis
The data are presented as the mean±SD. The means were analyzed using one-way ANOVA to compare between the groups. Differences with P<0.05 were considered statistically significant. All statistical tests were performed using SPSS 18.0 (SPSS, Chicago, IL, USA).
Results
Nutlin-3 causes DNA DSB damage in SMMC-7721 cells
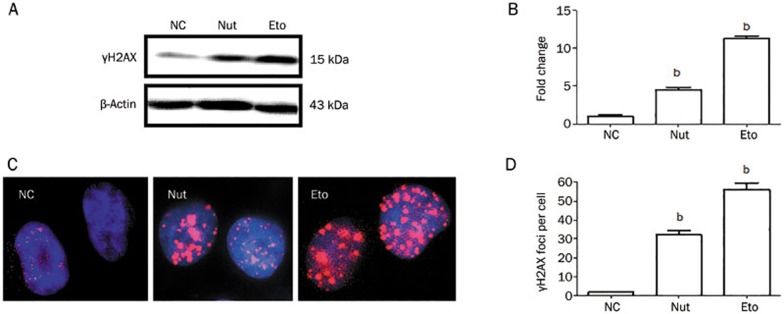
γH2AX, the phosphorylated H2AX (at Ser139), is a sensitive marker of DNA DSB27. To examine the impact of Nutlin-3 on DNA DSB damage, we incubated SMMC-7721 cells with Nutlin-3 (10 μmol/L) or Etoposide (40 μmol/L), an agent capable of inducing DSB, for 48 h and preformed Western blots to assess the expression level of γH2AX. We observed that Nutlin-3 or Etoposide increased the expression level of γH2AX, consistent with Etoposide inducing DSB (Figure 1A). These results suggest that Nutlin-3, like Etoposide, caused DNA DSB damage and triggered H2AX phosphorylation at Ser139 in SMMC-7721 cells.
Figure 1.
Nutlin-3 causes DNA DSB damage in SMMC-7721 cells. (A) Nutlin-3 increased γH2AX expression level. β-Actin served as the loading control. (B) The relative expression levels of γH2AX. The value represents the mean±SD derived from triplicate tests. bP<0.05 vs NC group. (C) Representative images of γH2AX foci formation (1000×). SMMC-7721 cells were treated with 0.1% DMSO, Etoposide (40 μmol/L), or Nutlin-3 (10 μmol/L) for 48 h and analyzed for γH2AX (red). Nuclei were counter-stained with DAPI (blue). The images were merged using Image-Pro plus 6.0. (D) Statistical analysis of the number of γH2AX foci. Data are shown as the mean±SD (n=3). bP<0.05 vs NC group.
γH2AX foci on mitotic chromosomes represent repaired lesions or unrepaired DNA breaks28. We next sought to establish whether the observed Nutlin-3-induced activation of H2AX phosphorylation was indicative of γH2AX foci formation. As expected, treatment of SMMC-7721 cells with Etoposide or Nutlin-3 was observed to induce γH2AX foci formation (Figure 1B). Taken together, Nutlin-3, like Etoposide, caused DNA DSB damage in SMMC-7721 cells, characterized by H2AX phosphorylation (at Ser139) and γH2AX foci formation.
Nutlin-3 induces the chromatin-bound protein IFI16 to partially localize in the cytoplasm of SMMC-7721 cells
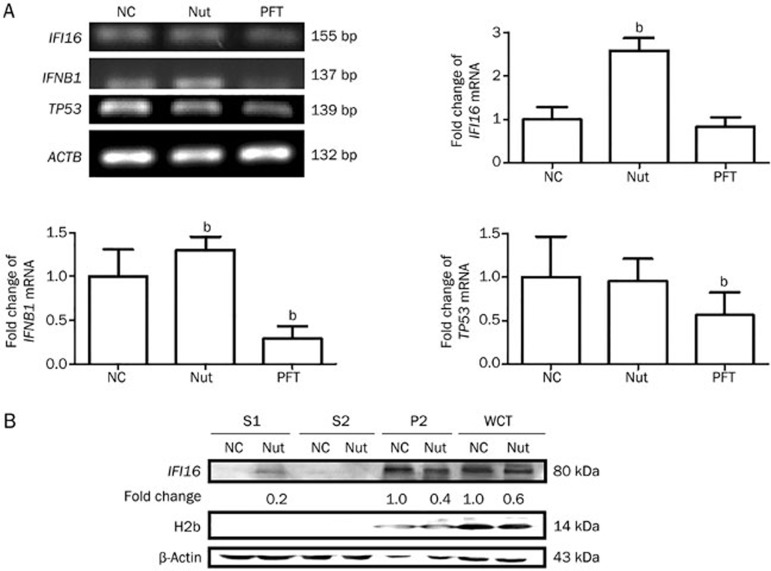
IFI16 is a member of the DNA sensors involved in DNA DSB repair15. First, because IFI16 is regulated at the transcriptional and post-transcriptional level29, we preformed RT-PCR to determine the expression level of IFI16 mRNA. We treated SMMC-7721 cells with PFT-α, a p53 transcriptional inhibitor30, for 48 h to test TP53 mRNA levels as a positive control. These data showed that Nutlin-3 significantly increased the expression level of IFI16 mRNA (2.58 fold, P<0.05) (Figure 2A). These results suggest that Nutlin-3 promoted IFI16 expression at the transcriptional level.
Figure 2.
Nutlin-3 induces the chromatin-bound protein IFI16 to partially localize to the cytoplasm of SMMC-7721 cells and increases the expression level of IFNB1 mRNA. (A) Nutlin-3 increased the expression level of IFI16 mRNA. The left panel shows representative gels of IFI16 and IFNB1. The TP53 and ACTB gene were used as the positive and internal control, respectively. The right graph shows the relative expression levels of IFI16 and IFNB1 mRNA. The value represents the mean±SD (n=3). bP<0.05 vs NC group. (B) Chromatin fractions were analyzed via Western blot in SMMC-7721 cells. H2b and β-actin served as the quality control for the nuclear fraction and the cytoplasmic fraction, respectively. S1, cytoplasmic proteins; S2, soluble nuclear proteins; P2, chromatin-enriched sediment; WCE, whole-cell extracts.
As the IFI16-HIN200 domain contains a DNA binding region at the C-terminus, we then extracted the chromatin fractions26 and used Western blots to investigate the association of IFI16 with chromatin and the expression level of the IFI16 protein. However, we detected that Nutlin-3 down-regulated the expression level of the IFI16 protein in SMMC-7721 cells (Figure 2B).
Next, we sought to establish whether the observed decrease in IFI16 levels was due to its subcellular localization. Interestingly, IFI16 was detected in only the chromatin-binding fraction of control cells, suggesting that it is a chromatin-bound protein (Figure 2B). We have previously confirmed that IFI16 is mainly localized in the nucleus of SMMC-7721 cells31. However, IFI16 was partially detected in the cytoplasm of Nutlin-3-treated cells (Figure 2B). Nuclear IFI16 is induced in the cytoplasm of stratified squamous epithelia in response to UVB exposure and acts as a mechanism of auto-antigen processing in SLE19. Meanwhile, endogenous IFI16 released by apoptotic cells acts as a novel alarmin, binding to neighbor cells and propagating the damaged-signal32. In addition, nuclear IFI16 is relocalized to the cytoplasm leading to proteasomal degradation by infection with HSV-133. According to the results that Nutlin-3 up-regulates IFI16 mRNA and down-regulates IFI16 protein levels, we proposed that IFI16 might be partially degraded in the cytoplasm or released into the extracellular milieu. These results indicate that IFI16 distribution is dynamic in response to Nutlin-3 treatment in SMMC-7721 cells.
Nutlin-3 increases the expression level of IFNB1 mRNA
In our previous study, we have shown that Nutlin-3 causes apoptosis in HCC cells with different TP53 genotypes23. It has been reported that IFI16 cytoplasmic accumulation can maximize innate immune system sensitivity in cancer chemotherapies34. To determine whether the ectopic expression of IFI16 triggered IFN-β production in the cytoplasm of Nutlin-3-treated cells, we investigated the level of IFNB1 mRNA using RT-PCR analysis. The TP53 mRNA in PFT-treated cells was used as the positive control (Figure 2A). Compared with the NC group, the expression level of IFNB1 mRNA was increased in Nutlin-3-treated SMMC-7721 cells (1.30 fold, P<0.05) (Figure 2A). These data suggest that the ectopic expression of IFI16 might trigger the expression of IFNB1 at the transcriptional level in SMMC-7721 cells.
Nutlin-3 causes the ectopic expression of IFI16 in HCC cells in a p53-dependent manner
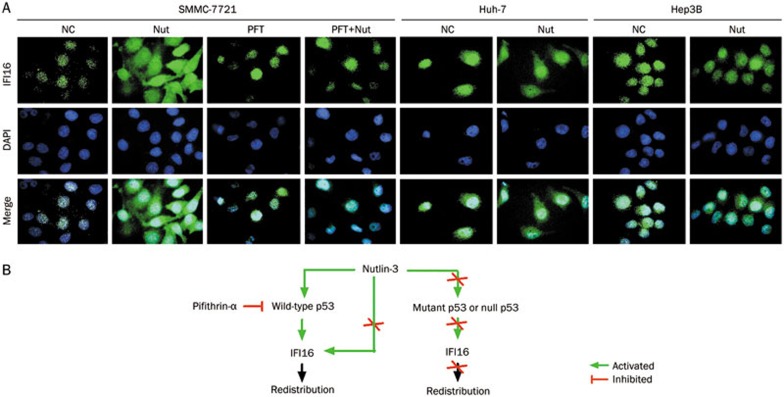
During DNA DSB damage, p300 acetylates p5335 and IFI16-NLS leading to IFI16 cytoplasmic localization34. To clarify whether IFI16 re-localization is a p53-dependent response by Nutlin-3, we used PFT-α to pre-treat SMMC-7721 cells (wild-type TP53) for 12 h to inhibit p53 activity and then exposed the cells to Nutlin-3 together with PFT-α for 36 h at the same conditions. After treatment, we performed IF analysis with anti-IFI16. Consistent with the Western blot results, IFI16 was distributed in only the nucleus of SMMC-7721 cells, but in the nucleus and cytoplasm of Nutlin-3-treated cells (Figure 3A). As expected, we detected IFI16 in only the nucleus of PFT and PFT+Nut group cells (Figure 3A), suggesting that IFI16 re-localization was not detected in p53-inhibited SMMC-7721 cells.
Figure 3.
Nutlin-3 causes IFI16 ectopic expression in a p53-dependent manner. (A) IFI16 subcellular localization was detected in HCC cells with different TP53 genotypes using fluorescence microscopy (1000×). Cells were treated as described above and stained for IFI16 (green). Nuclei were counter-stained with DAPI (blue). Images were merged using Image-Pro plus 6.0. (B) Proposed model for IFI16 subcellular localization regulated by Nutlin-3 in a p53-dependent manner in HCC cells.
IFI16 is also a chromatin-bound protein in Huh-7 (mutant TP53) and Hep3B (null TP53) cells21. To clarify whether the change in IFI16 localization is associated with p53 status in HCC cells, we used Huh-7 and Hep3B cells to perform IF analysis with the anti-IFI16. We detected IFI16 in only the nucleus of Huh-7 and Hep3B cells (Figure 3A). Interestingly, IFI16 re-localization was not detected in Huh-7 or Hep3B cells treated with Nutlin-3 (Figure 3A). Taken together, these results show that Nutlin-3-induced IFI16 redistribution occurs in a p53-dependent manner (Figure 3B).
IFI16 re-localization is a unique response in SMMC-7721 cells
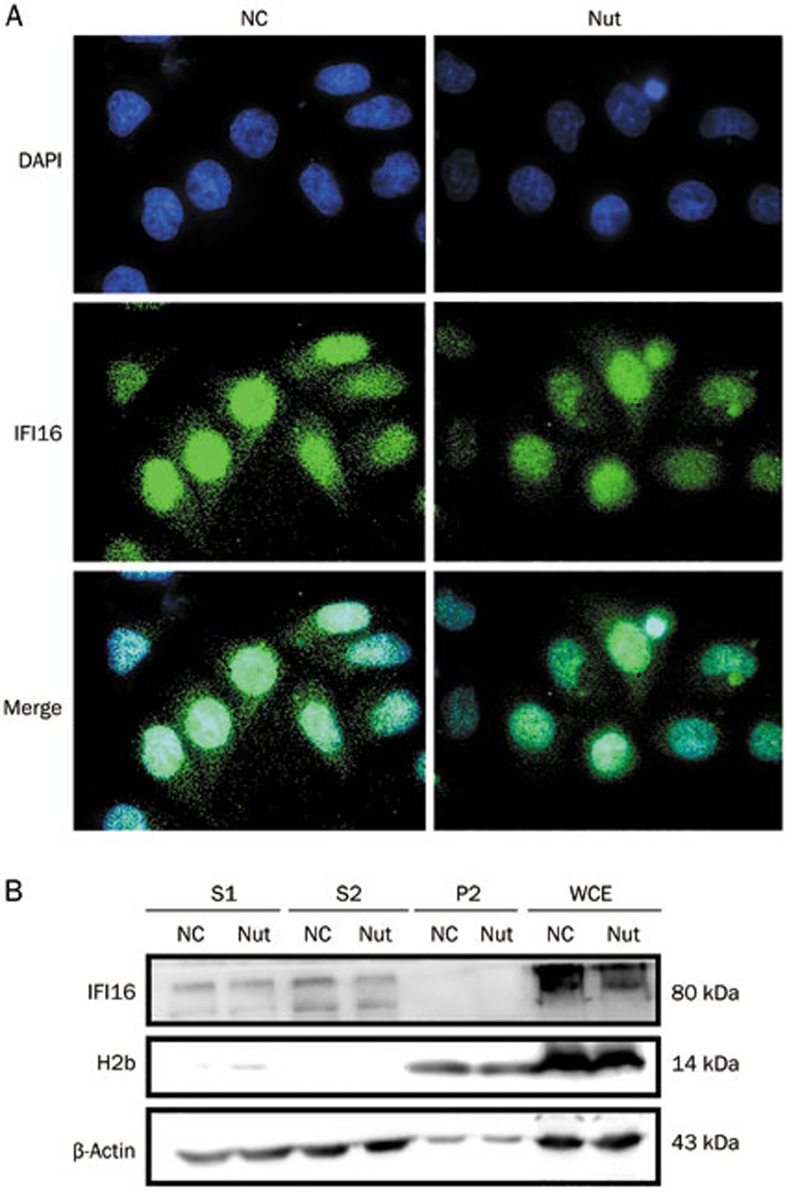
To clarify whether Nutlin-3 causes IFI16 re-localization in normal liver cells, we used the L02 cells to perform IF analysis with an IFI16 antibody. Surprisingly, IFI16 re-localization was not detected in Nutlin-3-treated L02 cells (Figure 4A). This result suggests that Nutlin-3 does not change IFI16 subcellular localization in L02 cells with wild-type TP53.
Figure 4.
IFI16 subcellular localization regulated by Nutlin-3 in L02 cells. (A) IFI16 subcellular localization was detected in L02 cells using fluorescence microscopy (1000×). Cells were treated as described above and stained for IFI16 (green). Nuclei were counter-stained with DAPI (blue). Images were merged using Image-Pro plus 6.0. (B) Chromatin fractions were analyzed via Western blot in L02 cells. H2b and β-actin served as the quality control for the nuclear fraction and the cytoplasmic fraction, respectively. S1, cytoplasmic proteins; S2, soluble nuclear proteins; P2, chromatin-enriched sediment; WCE, whole-cell extracts.
Next, we investigated the association of IFI16 with chromatin using Western blot analysis. Interestingly, IFI16 was found in the cytoplasm and nucleoplasm, revealing that it is not a chromatin-bound protein in L02 cells. Consistent with the IF analysis (Figure 4A), Nutlin-3 did not change IFI16 localization (Figure 4B). Together, these results show that the re-localization of IFI16, a chromatin-bound protein, is a unique response in Nutlin-3-treated SMMC-7721 cells.
Discussion
The effect of Nutlin-3 on HCC with mainly wild-type p53 are already known22,23. In this article, we demonstrated a link between Nutlin-3, a DNA damage agent36, and IFI16, a DNA sensor, through wild-type p53 activation in HCC cells. Nutlin-3 treatment increased the expression level of IFI16 and IFNB1 mRNA, and partially triggered chromatin-bound IFI16 redistribution to the cytoplasm of SMMC-7721 cells in a p53-dependent manner but not in L02 cells. This study provides new insight into the relationship between Nutlin-3 and the re-localization of chromatin-bound IFI16 in HCC therapy.
IFI16 is involved in liver cancer progression. IFI16 is up-regulated in chronic HCV and/or HBV liver tissue, suggesting that elevated IFI16 blocks viral replication and promotes viral clearance11,37. In the present study, we also showed that Nutlin-3 triggers IFI16 mRNA expression in SMMC-7721 cells. Therefore, we proposed that increased IFI16 expression in HCC cells by Nutlin-3 is similar to acute liver graft rejection38 and virus-infected liver cells39, which might reflect hepatic reversion to fetal status after liver injury.
IFI16 was predominantly in the nucleus34. We have confirmed that IFI16 is a chromatin-bound protein in HCC cells but not in L02 cells21. Recently, a study clearly demonstrated that the IFI16-HIN domains recognize the dsDNA sugar-phosphate backbone in non-sequence-specific manner and that the PYD domain inhibits the HIN:DNA interaction, even though it has no DNA-binding capacity40. For example, the PYD and HIN domains of AIM2, the other ALRs, form an intra-molecular complex in an auto-inhibited 'resting' state in HEK293T cells40. In the present study, we found that the subcellular localization of IFI16 was partly changed in response to Nutlin-3 treatment in SMMC-7721 cells. Consistently, some studies reported that IFI16 localization is involved in the DNA damage pathway by DNA-damaging agents such as IR15 and UVB19 in some cancer cells. The strong preference of IFI16 protein binding to supercoiled DNA and cruciform structures suggests that it may play an important role in DNA damage recognition during the repair response41. Further studies need to be conducted to address the mechanism of Nutlin-3 and re-localization of IFI16 in normal fetal liver cells.
IFI16 localization is important for its biological activities. IFI16 sensing of pathogenic or host-damaged DNA42 depends on the localization of the sensor and the DNA target, which is consistent with a two-signal model of innate immunity43. Therefore, re-localization of IFI16, a scaffold protein, may be a means for its activation. For example, IFI16 acts as a DNA sensor in the nucleus and redistributes to cytoplasm and forms inflammasomes upon KSHV14, EBV44, and HSV-133 infection. IFI16 cytoplasmic accumulation can also benefit innate immunity in cancer chemotherapies34. In the present study, we detected the increased expression level of IFNB1 mRNA in Nutlin-3-treated SMMC-7721 cells, which promotes the anti-inflammatory activation of IFI1645.
The function of the injury-activated innate immune system is similar to the two sides of a coin46. According to the danger hypothesis47, over-expressed IFI16 protein released in the extracellular niche acts as a novel alarmin propagating the damaged-signal32. For example, nuclear IFI16 delocalizes to the cytoplasm and then exists in the extracellular environment circulating as a new danger signal in response to UV or infected DNA, leading to impaired endothelial cells through high-affinity membrane binding in autoimmune diseases32.
Mechanically, we found that the IFI16 re-localization induced by Nutlin-3 treatment was involved in the p53 pathway but not in L02 cells. Consistent with our results, a study reported that IFI16-NLS acetylation by p300, a histone acetyl-transferase, promotes IFI16 cytoplasmic localization34. p300 binds to and activates p53 in response to DNA-damaged agents48. Novel anticancer agents target p53 activation, a critical cellular pathway. Loss of IFI16 results in deregulation of p53-mediated apoptosis in breast cancer8. Consistently, over-expressed IFI16 has no effect on cell growth in p53-inactived oral squamous cell carcinoma49. Our previous hypothesis also proposed that IFI16 mis-localization may be a contributing factor to HCC progression21. If we are able to redistribute chromatin-bounding IFI16 into the cytoplasm with activated-p53 restoration, we may offer an alternative for HCC therapy.
In recent years, some studies have focused on the role of IFI16 and targeted the nucleolus for cancer therapy. However, IFI16, a multifunctional protein, remains relatively poorly characterized in HCC. Therefore, further studies are needed to examine the location-function relationship for IFI16 in HCC chemotherapy.
Author contribution
Xin-li SHI, Ming-yuan LI, and Bao-ning WANG designed the research; Xin-li SHI performed the experiments; Xin-li SHI wrote the manuscript with contribution from other authors.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No 81071362) and the Youth Foundation of Hebei Educational Committee, China (No QN2014012). We thank Prof Cong LIU (West China Second University Hospital) for kindly supplying us with HCC cell lines.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA-Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–48. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Liao JC, Lam R, Brazda V, Duan S, Ravichandran M, Ma J, et al. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure. 2011;19:418–29. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–18. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes. Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Wei W, Greenway A, Trapani JA. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene. 2000;19:6033–42. doi: 10.1038/sj.onc.1204005. [DOI] [PubMed] [Google Scholar]
- Dawson MJ, Trapani JA. HIN-200: a novel family of IFN-inducible nuclear proteins expressed in leukocytes. J Leukocyte Biol. 1996;60:310–6. doi: 10.1002/jlb.60.3.310. [DOI] [PubMed] [Google Scholar]
- Fujiuchi N, Aglipay JA, Ohtsuka T, Maehara N, Sahin F, Su GH, et al. Requirement of IFI16 for the maximal activation of p53 induced by ionizing radiation. J Biol Chem. 2004;279:20339–44. doi: 10.1074/jbc.M400344200. [DOI] [PubMed] [Google Scholar]
- Mazibrada J, Andrea MD, Rittà M, Borgogna C, Pfeffer U, Chiusa L, et al. In vivo growth inhibition of head and neck squamous cell carcinoma by the Interferon-inducible gene IFI16. Cancer Lett. 2010;287:33–43. doi: 10.1016/j.canlet.2009.05.035. [DOI] [PubMed] [Google Scholar]
- Alimirah F, Chen J, Davis FJ, Choubey D. IFI16 in human prostate cancer. Mol Cancer Res. 2007;5:251–9. doi: 10.1158/1541-7786.MCR-06-0269. [DOI] [PubMed] [Google Scholar]
- Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Prod Natl Acad Sci U S A. 2004;101:6669–74. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012;8:e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Bowie AG. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:351–3. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglipay JA, Lee SW, Okada S, Fujiuchi N, Ohtsuka T, Kwak JC, et al. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8. doi: 10.1038/sj.onc.1207057. [DOI] [PubMed] [Google Scholar]
- Duan X, Ponomareva L, Veeranki S, Choubey D. IFI16 induction by glucose restriction in human fibroblasts contributes to autophagy through activation of the ATM/AMPK/p53 pathway. PLoS One. 2011;6:e19532. doi: 10.1371/journal.pone.0019532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Pereira-Smith OM, Choubey D. Role of IFI 16 in cellular senescence of human fibroblasts. Oncogene. 2004;23:6209–17. doi: 10.1038/sj.onc.1207836. [DOI] [PubMed] [Google Scholar]
- Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–40. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- Costa S, Borgogna C, Mondini M, De Andrea M, Meroni P, Berti E, et al. Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. Brit J Dermatol. 2011;164:282–90. doi: 10.1111/j.1365-2133.2010.10097.x. [DOI] [PubMed] [Google Scholar]
- Khalid SS, Hamid S, Siddiqui AA, Qureshi A, Qureshi N. Gene profiling of early and advanced liver disease in chronic hepatitis C patients. Hepa Inter. 2011;5:782–8. doi: 10.1007/s12072-011-9252-4. [DOI] [PubMed] [Google Scholar]
- Shi X, Liu J, Liu Q, Li M. IFI16 mis-localization can be a contributing factor to hepatocellular carcinoma progression. Med Hypotheses. 2014;82:398–400. doi: 10.1016/j.mehy.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Wang J, Zheng T, Chen X, Song X, Meng X, Bhatta N, et al. MDM2 antagonist can inhibit tumor growth in hepatocellular carcinoma with different types of p53 in vitro. J Gastroen Hepatol. 2011;26:371–7. doi: 10.1111/j.1440-1746.2010.06440.x. [DOI] [PubMed] [Google Scholar]
- Shi X, Liu J, Ren L, Mao N, Tan F, Ding N, et al. Nutlin-3 downregulates p53 phosphorylation on serine392 and induces apoptosis in hepatocellular carcinoma cells. BMB Rep. 2014;46:221–6. doi: 10.5483/BMBRep.2014.47.4.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu Z, Yin J, Chou W, Kwok C, Qin Z, et al. Ursolic acid induces human hepatoma cell line SMMC-7721 apoptosis via p53-dependent pathway. Chin Med J. 2010;123:1915–23. [PubMed] [Google Scholar]
- Miao R, Han Y, An L, Yang J, Wang Q. Seleno-podophyllotoxin derivatives induce hepatoma SMMC-7721 cell apoptosis through Bax pathway. Cell Biol Int. 2008;32:217–23. doi: 10.1016/j.cellbi.2007.08.034. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Gene Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, Karagiannis TC. γH2AX as a molecular marker of aging and disease. Epigenetics. 2010;5:129–36. doi: 10.4161/epi.5.2.11080. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Suzuki K, Kodama S, Watanabe M. Phosphorylated histone H2AX foci persist on rejoined mitotic chromosomes in normal human diploid cells exposed to ionizing radiation. Radiat Res. 2006;165:269–76. doi: 10.1667/rr3508.1. [DOI] [PubMed] [Google Scholar]
- Clarke C, Apostolidis V, Hii L, Gough D, Trapani J, Johnstone R. Critical role of the transcription factor AP-1 for the constitutive and interferon-induced expression of IFI 16. J Cell Biochem. 2003;89:80–93. doi: 10.1002/jcb.10475. [DOI] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–7. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Yu F, Hao X, Zhao H, Ge C, Yao M, Yang S, et al. Delta-like 1 contributes to cell growth by increasing the interferon-inducible protein 16 expression in hepatocellular carcinoma. Liver Int. 2010;30:703–14. doi: 10.1111/j.1478-3231.2010.02214.x. [DOI] [PubMed] [Google Scholar]
- Gugliesi F, Bawadekar M, De Andrea M, Dell'Oste V, Caneparo V, Tincani A, et al. Nuclear DNA sensor IFI16 as circulating protein in autoimmune diseases is a signal of damage that impairs endothelial cells through high-affinity membrane binding. PLoS One. 2013;8:e 63045. doi: 10.1371/journal.pone.0063045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87:5005–18. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. P Natl Acad Sci U S A. 2012;109:10558–63. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, et al. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem. 1999;274:1883–6. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- Verma R, Rigatti MJ, Belinsky GS, Godman CA, Giardina C. DNA damage response to the Mdm2 inhibitor Nutlin-3. Biochem Pharmacol. 2010;79:565–74. doi: 10.1016/j.bcp.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, et al. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123–33. doi: 10.1053/j.gastro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- Borgogna C, Toniutto P, Smirne C, Azzimonti B, Rittà M, Avellini C, et al. Expression of the interferon-inducible proteins MxA and IFI16 in liver allografts. Histopathology. 2009;54:837–46. doi: 10.1111/j.1365-2559.2009.03311.x. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–94. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, et al. Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–71. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brázda V, Coufal J, Liao JC, Arrowsmith CH. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem Biophys Res Commun. 2012;422:716–20. doi: 10.1016/j.bbrc.2012.05.065. [DOI] [PubMed] [Google Scholar]
- O'Neill LA. Sensing the dark side of DNA. Science. 2013;339:763–4. doi: 10.1126/science.1234724. [DOI] [PubMed] [Google Scholar]
- Fontana MF, Vance RE. Two signal models in innate immunity. Immunol Rev. 2011;243:26–39. doi: 10.1111/j.1600-065X.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- Ansari MA, Singh VV, Dutta S, Veettil MV, Dutta D, Chikoti L, et al. Constitutive interferon-inducible protein 16-inflammasome activation during epstein-barr virus latency I, II and III in B and epithelial cells. J Virol. 2013;87:8606–23. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land WG. Transfusion-related acute lung injury: The Work of DAMPs. Transfus Med Hemother. 2013;40:3–13. doi: 10.1159/000345688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Pulskens WP, Sadler JJ, Butter LM, Teske GJ, Ulland TK, et al. Necrotic cells trigger a sterile inflammatory response through the Nlrp3 inflammasome. Proc Natl Acad Sci U S A. 2009;106:20388–93. doi: 10.1073/pnas.0908698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–7. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Nagai K, Nakahata S, Saito Y, Ichikawa T, Suekane A, et al. Overexpression of the DNA sensor proteins, absent in melanoma 2 and interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation. Cancer Sci. 2012;103:782–90. doi: 10.1111/j.1349-7006.2012.02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]