Abstract
S-Adenosylmethionine (SAM, also known as AdoMet) radical enzymes use SAM and a [4Fe-4S] cluster to catalyze a diverse array of reactions. They adopt a partial triose-phosphate isomerase (TIM) barrel fold with N- and C-terminal extensions that tailor the structure of the enzyme to its specific function. One extension, termed a SPASM domain, binds two auxiliary [4Fe-4S] clusters and is present within peptide-modifying enzymes. The first structure of a SPASM-containing enzyme, anaerobic sulfatase-maturating enzyme (anSME), revealed unexpected similarities to two non-SPASM proteins, butirosin biosynthetic enzyme 2-deoxy-scyllo-inosamine dehydrogenase (BtrN) and molybdenum cofactor biosynthetic enzyme (MoaA). The latter two enzymes bind one auxiliary cluster and exhibit a partial SPASM motif, coined a Twitch domain. Here we review the structure and function of auxiliary cluster domains within the SAM radical enzyme superfamily.
Keywords: Iron-Sulfur Protein, Post-translational Modification (PTM), Radical, S-Adenosylmethionine (SAM), Structural Biology, AdoMet
Introduction
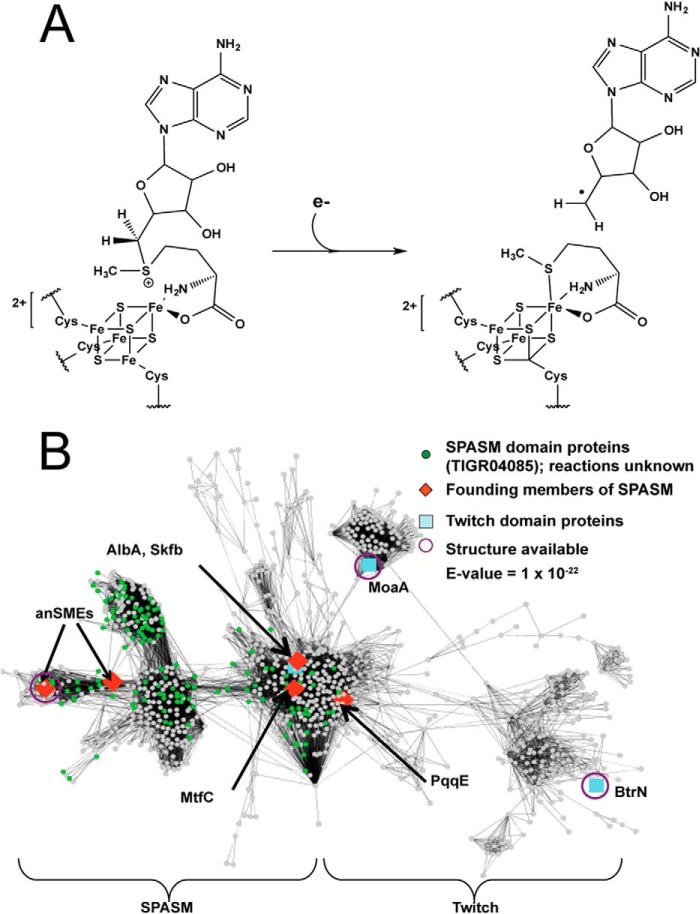
Members of the S-adenosylmethionine (SAM)2 radical superfamily catalyze a wide variety of radical-mediated reactions, including complex chemical transformations and rearrangements; modifications of peptides, DNA, and RNA; dehydrogenations; and sulfur insertions (1). Despite this diversity, there are unifying structural and mechanistic themes. For instance, SAM radical enzymes typically bind a [4Fe-4S] cluster using a conserved CX3CXφC motif (where φ is an aromatic residue). This motif provides three cysteine ligands to the iron atoms of the cluster, with the fourth ligand coming from the bidentate coordination of SAM to the unique iron (2, 3). Direct ligation of SAM to the cluster facilitates reductive cleavage of the C–S bond through an inner sphere electron transfer event, forming methionine and a 5′-deoxyadenosyl radical (5′-dAdo•) (Fig. 1A) (4). The abstraction of a hydrogen atom from the substrate by 5′-dAdo•, producing a substrate radical, ends the mechanistic similarity between enzymes of this superfamily; each enzyme utilizes a different mechanism to generate product. Structures of the first seven members of the SAM radical superfamily were used to define a core fold for binding SAM and for the generation of 5′-dAdo• species. This core consists of a partial (β/α)6 triose-phosphate isomerase (TIM) barrel (5). Outside of the core fold, the structure can vary greatly, with N- and C-terminal extensions that are functionalized for binding other cofactors or substrates. The SPASM subfamily is an example of a functionalized C-terminal extension for the binding of two auxiliary clusters.
FIGURE 1.
The SAM radical SPASM/Twitch subfamily similarity network. A, general mechanism for reductive cleavage of SAM by inner sphere electron transfer from the [4Fe-4S] to the C–S bond, forming a 5′-deoxyadenosyl radical and methionine. B, Protein Sequence Similarity Network (60) of the SPASM/Twitch domain containing proteins of the SAM radical superfamily. The nodes represent protein sequences with 40% sequence identity at an E-value (Expectation or Expect value) of 1 × 10−22. Proteins with SPASM domains map to 153 nodes (green dots). This figure was generated using Cytoscape (61).
Haft and Basu (6, 7) recognized that enzymes with this C-terminal extension appear to be involved in the modification of ribosomally translated peptides. This subclass is referred to as SPASM after the biochemically characterized members, AlbA, PqqE, anSMEs, and MftC, which are involved in subtilosin A, pyrroloquinoline quinone, anaerobic sulfatase, and mycofactocin maturation, respectively. The SPASM subfamily, accession TIGR04085, is composed of 281 sequences. However, recent similarity network analysis by Babbitt and co-workers (8) using the Structure Function Linkage Database identified additionally related sequences, expanding the number to 1,380 (Fig. 1B).
Enzymes in this subfamily were expected to use a seven-cysteine (9) motif to bind two auxiliary [4Fe-4S] clusters, leaving a unique iron for substrate binding (7, 10). However, the first, and thus far only, structure of a SPASM protein, the anaerobic sulfatase-maturating enzyme (anSME) from Clostridium perfringens, showed full cysteine ligation to both [4Fe-4S] clusters (11). anSME was also the first structure of a SAM radical enzyme with dehydrogenase activity, and it, along with the structures of butirosin biosynthetic enzyme BtrN from Bacillus circulans, and molybdenum cofactor biosynthetic enzyme MoaA (12) (Fig. 2), revealed unexpected structural homology between SPASM and non-SPASM enzymes (13). In particular, the comparison of anSME, BtrN, and MoaA led to the identification of a truncated SPASM domain used for binding a single auxiliary [4Fe-4S] cluster, which has been termed a Twitch domain (Fig. 1B) (11, 13). This review discusses the function and architecture of the newly defined SPASM/Twitch subclass and explores the roles of auxiliary clusters in radical SAM chemistry with particular focus on the auxiliary clusters of this subclass. We suggest that SPASM and Twitch domains will constitute a larger subfamily than anticipated.
FIGURE 2.
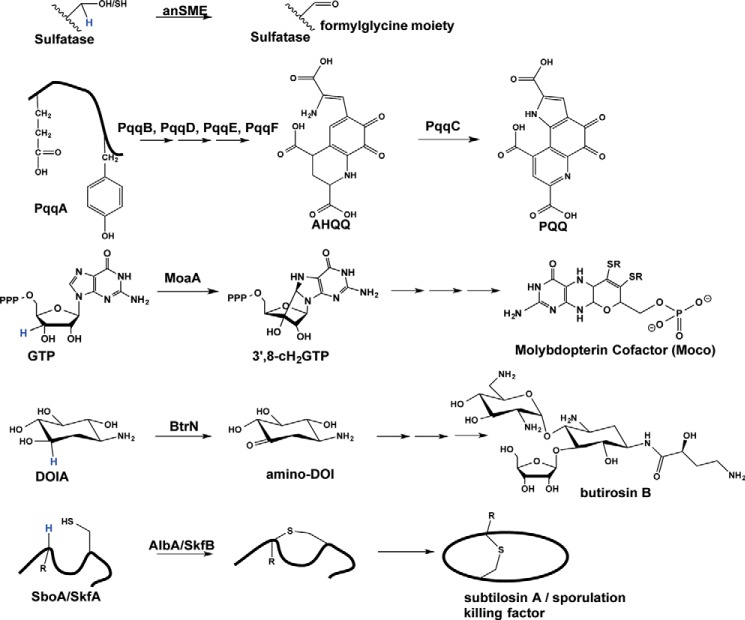
SPASM/Twitch subfamily reactions. The involvement of SPASM/Twitch domain containing enzymes, anSME, MoaA, BtrN, and AlbA/SkfB in the biosynthesis of their respective products is shown. The specific reaction catalyzed by SPASM enzyme PqqE is not indicated as its substrate is not known. For AlbA, only one of the three thioether bond formations is shown. Hydrogen atoms known or proposed to be abstracted by the 5′-deoxyadenosyl radical are shown in blue. DOIA, 2-deoxy-scyllo-inosamine; amino-DOI, 3-amino-2,3-dideoxy-scyllo-inosose.
Function of SPASM/Twitch Subfamily
A chief function of this subfamily appears to be in the post-translational modification of peptides. Post-translational modifications expand the chemical repertoire of enzymes by generating modified amino acids that are well suited to perform specific reactions, priming peptides for cofactor biosynthesis, or rigidifying proteins or peptides (Fig. 2).
anSME catalyzes the co- or post-translational modification of an arylsulfatase active site cysteine or serine residue to a catalytically essential formylglycine (FGly) moiety in anaerobic organisms (10, 14–17). The active site FGly allows sulfatases to perform their hydrolysis function, removing sulfate groups from a wide array of substrates (e.g. sulfated polysaccharides, sulfolipids, and steroid sulfates) (18–20). Sulfatase activity is important in humans and bacteria, with a lack of activity leading to disease in humans (21) and leading to an inability to colonize the mucosal layer of the host's gut upon inhibition in bacteria (22). anSMEs are known to use SAM radical chemistry, in addition to two auxiliary [4Fe-4S] clusters, to perform the dehydrogenation of a serine or cysteine residue to the FGly moiety (Fig. 2). Until recently, very little was known about the structure and mechanism of anSME in comparison with the related formylglycine-generating enzymes found in eukaryotes and aerobically living prokaryotes. With the structural and mechanistic information on anSME from C. perfringens (11, 17), our understanding of the anaerobic sulfatase-maturating enzyme family is on the rise.
AlbA is involved in the post-translational modification of a linear peptide into the cyclic peptide natural product subtilosin A. In particular, AlbA is responsible for forming three thioether bonds between the sulfur atoms of three cysteine residues and the α-carbons of two phenylalanines and one threonine, yielding a rigidified peptide (Fig. 2) (23). Mutation of the corresponding gene, albA, indicates that AlbA is essential for subtilosin A biosynthesis and shows that thioether bond formation is a critical aspect of subtilosin A maturation (24). Peptide-derived natural products that contain thioether bonds, such as subtilosin A, are collectively known as sactipeptides. In terms of the medical relevance of this class of compounds, subtilosin A, which is produced by the soil bacterium Bacillus subtilis, shows antimicrobial activity against both Gram-negative and Gram-positive bacteria, as well as some human pathogens (25). Another sactipeptide produced by B. subtilis, sporulation killing factor, is excreted under nutrient limitation to lyse neighboring cells in what can only be described as cannibalistic behavior (26–28). A different SAM radical enzyme, SkfB, is responsible for the formation of the thioether bond of this sactipeptide (29).
PqqE is one of several enzymes that participate in the maturation of the bacterial redox cofactor pyrroloquinoline quinone (PQQ) from a peptide precursor (Fig. 2). In particular, PQQ is formed post-translationally from a skeleton peptide, PqqA, through complex rearrangements initiated by the cross-linking of glutamyl and tyrosyl side chains (30, 31). PQQ is then excised from PqqA and attached to its target enzyme. PqqE is known to contain two [4Fe-4S] clusters and to reductively cleave SAM (32), and although it is believed to be involved in an early step in PQQ biogenesis, the exact reaction catalyzed has not been identified (33).
BtrN, a 2-deoxy-scyllo-inosamine dehydrogenase, is a member of the Twitch structural subclass of SAM radical enzymes and is involved in the biosynthesis of the aminoglycoside antibiotic butirosin B. BtrN is also a member of a recently described functional subclass of the SAM radical enzymes, SAM radical dehydrogenases, with the sulfatase maturases anSME and AtsB being the only other biochemically characterized members (10, 17, 34). BtrN contains a Twitch domain (13), which houses one auxiliary [4Fe-4S] cluster. Using SAM radical chemistry, BtrN catalyzes the oxidation of the C3 hydroxyl group of 2-deoxy-scyllo-inosamine by a hydrogen atom abstraction, deprotonation, and one-electron oxidation to produce the ketone group in 3-amino-2,3-dideoxy-scyllo-inosose (amino-DOI) (Fig. 2) (35, 36).
The Twitch subclass enzyme MoaA is involved in the biosynthesis of molybdopterin (Moco). MoaA catalyzes the first step in Moco biosynthesis, the complex rearrangement of guanosine triphosphate to 3′-8-cH2GTP, which is then converted to Moco in subsequent steps (Fig. 2) (37, 38). In humans, defects in molybdenum cofactor biogenesis lead to death shortly after birth, and these patients show neurological abnormalities including untreatable seizures and attenuated brain growth (39).
The SAM Radical Core Fold
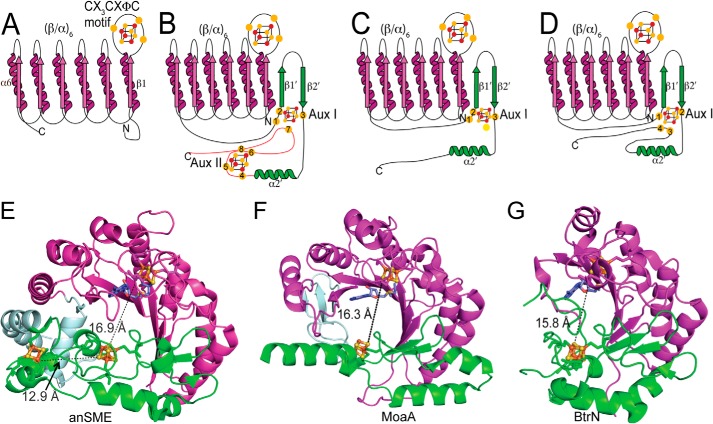
SAM radical enzymes adopt a core fold, with exceptions described in a recent review (40) and below. This core is responsible for SAM binding and radical generation and has been defined as a partial TIM barrel (β/α)6 with six α-helices making up the outside of the partial barrel and the six parallel β-sheets forming the inner face of the barrel (Fig. 3A) (5). The active site is located within the lateral opening of the partial TIM barrel and includes a [4Fe-4S] cluster, SAM, and substrate-binding sites. Although some SAM radical enzymes are composed of a complete TIM barrel (41, 42), most have the partial barrel architecture. Recently, the structural diversity of this enzyme superfamily was expanded when a significant deviation was reported for the queuosine biosynthetic enzyme 7-carboxy-7-deazaguanine synthase (QueE). QueE was found to have a hypermodified barrel fold in which three of the six α-helices are replaced by two loops and one 310 helix (43).
FIGURE 3.
Folds of SPASM/Twitch subfamily members. A, topology of SAM (β/α)6 core fold (magenta). Cluster cysteine ligands are indicated as yellow spheres. B, general topology of a SPASM domain-containing SAM radical enzyme. The SPASM domain (green), at the C terminus of the SAM radical core, binds two [4Fe-4S] clusters. C, topology of a Twitch domain-containing SAM radical enzyme with an open iron coordination site for substrate (yellow circle). D, topology of a Twitch domain-containing SAM radical enzyme with a fully ligated auxiliary cluster. E–G, ribbon representations of the crystal structures of anSME, MoaA, and BtrN, respectively. The SAM core is indicated in magenta followed by the SPASM or Twitch domains in green. The SAM to Aux I cluster distances for these enzymes, as well as the Aux I to Aux II distance for anSME, are shown as black dashed lines.
The [4Fe-4S] cluster motif, more popularly known as the CX3CXφC motif, binds the [4Fe-4S] cluster responsible for initiating radical chemistry. The three cysteines of this motif reside on the loop linking β1 to α1, termed the cluster-binding loop (Fig. 3A). This motif, which is largely but not absolutely conserved (42, 43), allows for binding of SAM to the cluster through the open coordination site on the unique iron atom of the cluster, as well as through hydrophobic interactions from the aromatic residue (φ).
With respect to cluster binding, the largest deviations observed through crystallographic studies thus far are found in the SAM radical enzymes QueE, mentioned above, and ThiC, which is involved in the biosynthesis of the thiamin pyrimidine moiety. In the former case, the cysteine motif has an 11-amino acid insertion between the first two cysteines (CX14CXφC) (43), and in the latter, the cluster-binding region is not part of the SAM radical core, but is instead found in a separate domain (42). For QueE, the 11-residue insert does not alter the way in which SAM binds and is instead believed to be important for binding of the physiological reductase (43). For ThiC, no structure is available of the fully reconstituted protein, and thus the exact mode of cluster and SAM binding remains to be determined.
SAM binding is facilitated by four different motifs, some of which are more highly conserved than others. The “GGE motif,” named after residues in the pyruvate formate lyase-activating enzyme and MoaA, resides at the C-terminal end of β2 and interacts with the amino group of SAM, helping to orient the methionyl moiety of SAM molecule. Interactions between residues from β4, called the “ribose motif,” and the hydroxyls of the ribose moiety appear to play an important role in positioning of 5′-dAdo• with respect to substrate for hydrogen atom abstraction (44). The “GXIXGXXE motif,” which is only partially conserved in terms of sequence, is located on β5 and provides hydrophobic interactions with the adenine ring of SAM. Finally, the “β6 motif” is responsible for hydrogen bonding via backbone atoms to the adenine ring. Interestingly, the structure of BtrN shows that residues at the end of β5 can replace the function of the β6 motif. BtrN thus contains both the GXIXGXXE motif and the β6 motif on β5 (13). We suggest that the β6 motif be renamed the “adenine-binding motif.” Taken together, all of these motifs, which span across the SAM radical core, work together to ensure that SAM is positioned in the correct orientation for hydrogen atom abstraction from the substrate after homolytic cleavage.
SPASM/Twitch Domain Fold
In addition to the core fold, SAM radical enzymes contain N- or C-terminal extensions. The SPASM domain at the C-terminal end of the core fold binds two auxiliary [4Fe-4S] clusters using a conserved seven-cysteine motif, CX9–15GX4CXnCX2CX5CX3CXnC (9). Based on the reactions performed by the biochemically characterized SPASM members and the presence of only seven cysteine residues in the conserved cysteine motif, an open coordination site on one auxiliary [4Fe-4S] cluster has been proposed to ligate the peptide substrate (7, 10). Through the work of Goldman et al. (11), we now have a snapshot of one of these enzymes, anSME, both with and without substrate bound. This structure shows that both clusters of anSME are fully ligated by cysteine residues, with auxiliary cluster I (Aux I) 16.9 Å from the SAM radical cluster and 12.9 Å from the second auxiliary cluster (Aux II) (Fig. 3, B and E). Aux I is ligated by four cysteine residues: one before the predicted SPASM domain seven-cysteine motif, two within the motif, and one downstream cysteine. A conserved β-hairpin motif lies between the second and third coordination sites of Aux I, with an α-helix following (α2′). This helix leads to the second auxiliary cluster (Aux II) (Fig. 3B). Aux II is coordinated by four cysteine residues of the seven-cysteine motif, but not in the same order as the primary sequence. Unexpectedly, the protein chain travels back and forth between the two auxiliary clusters such that cysteines 4, 5, 6, and 8 coordinate Aux II, whereas cysteine 7 is the downstream cysteine ligand to Aux I (Fig. 3B). Based on sequence homology, it is predicted that this cluster-binding architecture will be a common feature of SPASM domain-containing enzymes (7, 11, 13), with the caveat that some SPASM proteins may not display full cysteine ligation.
The protein topology around Aux I of anSME shows structural similarity to the auxiliary cluster domain of the non-SPASM SAM radical enzyme MoaA. MoaA also displays a ∼16 Å distance between its SAM radical cluster and its auxiliary cluster and adopts an abridged SPASM domain architecture (Fig. 3, C and F). This abridged SPASM fold provides three cysteine ligands to bind one [4Fe-4S] cluster and contains the β-hairpin motif and α2′, all of which are found in the first half of the SPASM domain. Based on the similarity between anSME and MoaA, this abridged SPASM domain was subsequently coined the Twitch domain (11, 13). MoaA contains no cysteine ligands following α2′, and the resulting open coordination site on the [4Fe-4S] cluster is used for substrate binding (45), as was originally proposed for the SPASM enzymes.
BtrN also contains a Twitch domain that binds one [4Fe-4S] cluster similar to the auxiliary cluster-binding domain of MoaA. However, the auxiliary cluster in BtrN is fully ligated (13), showing the versatility of the Twitch domain. Like anSME and MoaA, the auxiliary cluster of BtrN is ∼16 Å from the SAM radical cluster (Fig. 3, D and G) and has a domain architecture made up of a β-hairpin followed by α2′, with cysteine ligands flanking both ends of the β-hairpin. Similar to anSME, cysteine residues following α2′ ligate an auxiliary cluster. Unlike anSME and MoaA, BtrN has only one cysteine ligand before the β-hairpin (Fig. 3D). Thus, both the Aux I of anSME and the Aux cluster of BtrN have full ligation by cysteine residues, although the cysteine positions in the primary sequence are different.
Overall, these structural snapshots show two different coordination modes of [4Fe-4S] clusters in Twitch domains, one in which the [4Fe-4S] cluster can bind substrate directly to an open coordination site (MoaA) and one in which it cannot (BtrN). They also reveal two different varieties of SAM radical dehydrogenases, one with one auxiliary [4Fe-4S] cluster (BtrN) and one with two auxiliary clusters (anSME).
Given the structural homology between anSME, BtrN, and MoaA, we propose that the entire area of sequence space shown in Fig. 1B be considered its own SAM radical enzyme subclass, encompassing ∼15% of the enzymes currently identified as belonging to the SAM radical superfamily in the Structure Function Linkage Database (8). The three available structures of SPASM/Twitch proteins represent three edges of sequence space and thus exemplify the structural diversity within this subclass.
Roles of Auxiliary Clusters in the SPASM/Twitch Subclass
Recent structural data have served to clarify the function(s) of auxiliary clusters in SPASM/Twitch enzymes (11, 13). For anSME and BtrN, crystal structures show that auxiliary clusters are fully ligated both in the presence and in the absence of substrates and that substrates are bound through protein-mediated hydrogen-bonding interactions at distances of 9–10 Å away from their auxiliary clusters (Fig. 4, A and B). These long distances also suggest that auxiliary clusters do not function in substrate deprotonation. For anSME, both crystallographic and mutagenesis data support Asp-277 as being responsible for the deprotonation step (11). In particular, Asp-277 is adjacent to the substrate, and its mutation to Asn results in a protein with only 0.8% of wild-type activity and increased uncoupling of SAM cleavage from product formation (11). For BtrN, Arg-152 has been proposed to play a role in substrate deprotonation based solely on its location in the active site (13). Instead of binding or deprotonating substrate, the observed 9–10 Å distances between cluster and substrate are consistent with a role for the auxiliary clusters as electron acceptors during substrate oxidation. There is diversity in the number of auxiliary clusters needed for substrate oxidation. anSME has two auxiliary clusters (12.9 Å apart) to provide a route for the electron from the buried active site to the protein surface, whereas BtrN only needs one auxiliary cluster for the electron to reach the protein surface, where it would be accessible to an external electron acceptor.
FIGURE 4.
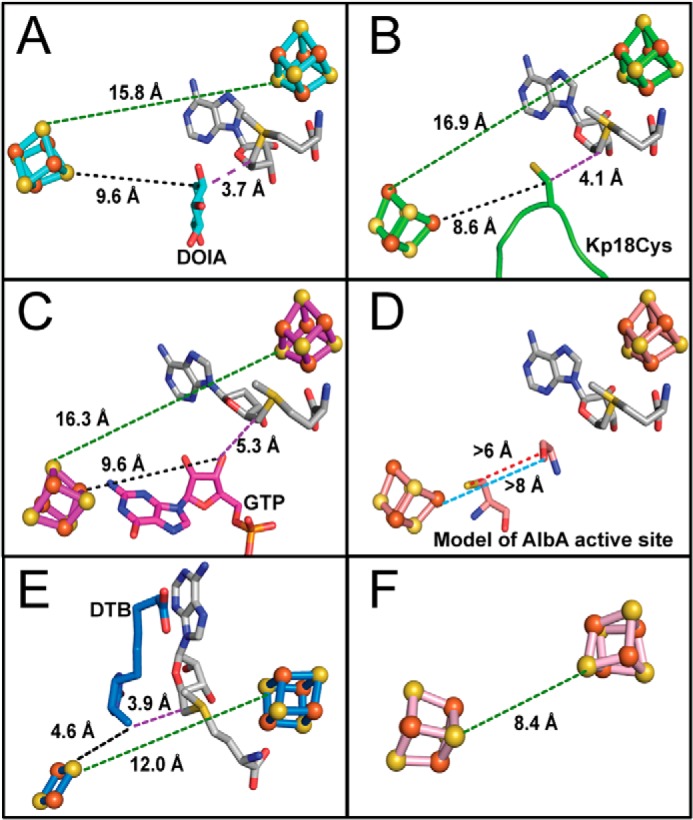
Auxiliary cluster positions in SPASM/Twitch enzymes. Distances are shown as follows: SAM cluster to Aux cluster (green dashed lines); hydrogen atom abstraction position on substrate to Aux cluster (black dashed lines); and hydrogen atom abstraction position to C5′ of SAM (purple dashed lines). A, BtrN. DOIA, 2-deoxy-scyllo-inosamine. B, anSME with peptide bound (green). C, MoaA. D, hypothetical AlbA model estimating distances between the site of hydrogen atom abstraction on a peptide glycine to the site of thioether bond formation on the sulfur of a peptide cysteine (red dashed line) and to the Aux cluster (blue dashed line). E, BioB with DTB bound. F, RimO.
The extent to which SPASM/Twitch enzymes will display full ligation of their auxiliary clusters is not known at this time. Current sequence data suggest that variations will occur. AlbA, for example, does not contain an eighth cysteine in its C-terminal auxiliary cluster domain. As more structures become available, it will be very interesting to discover how many of these enzymes, if any, (i) do indeed have an open iron coordination site for substrate ligation; (ii) use a non-cysteine residue for ligation of an auxiliary cluster, as in biotin synthase (BioB), which has an arginine ligand (41); and/or (iii) use a cysteine that is distal in primary sequence from its SPASM motif to provide the last auxiliary cluster ligand.
Roles of Auxiliary Clusters in Other SAM Radical Enzymes
A number of auxiliary cluster-containing non-SPASM/non-Twitch SAM radical enzymes have been identified, and the roles of their auxiliary clusters are under investigation. BioB, for example, has been proposed to use its auxiliary [2Fe-2S] in a sacrificial manner, donating one of its sulfur atoms to perform a sulfur insertion reaction during biotin formation (41, 46–48). Similarly, lipoyl synthase (LipA) contains an auxiliary [4Fe-4S] cluster that is proposed to be the source of the two sulfur atoms needed to form lipoic acid (49–51). Methylthioltransferases (MTTases), as their name suggests, transfer a methylthiol group to macromolecular substrates including the S12 ribosomal protein (catalyzed by RimO) and tRNA-A37 (catalyzed by MiaB). Methylthioltransferases also have auxiliary clusters (52, 53), which, unlike for BioB and lipoyl synthase, are not currently believed to serve as the source of sulfur for generation of the methylthiol group (54). Instead, a recent crystal structure of RimO has led to the proposal that its auxiliary cluster is responsible for binding a polysulfide moiety that serves as the sulfur source (55). Other auxiliary cluster-containing SAM radical enzymes have been reviewed recently (56) and include a subset of glycyl radical-activating enzymes and hydrogenase cofactor-maturating enzymes (HydE and HydG). Our understanding of this latter set of enzymes is still in its infancy, although hydrogenase cofactor maturation is an active area of research (57).
Predictions
The structures of the SPASM protein anSME and of the Twitch proteins BtrN and MoaA allow us to make predictions about other enzymes in the SPASM/Twitch subclass for which there are no structures. As mentioned above, AlbA and SkfB catalyze the internal peptide linkage of a cysteine side chain sulfur to a Cα position in the maturation of two natural products generated from ribosomally translated peptide scaffolds, subtilosin A and sporulation killing factor (Fig. 2). Following hydrogen atom abstraction from the Cα position, formation of a covalent bond with cysteine requires a 1-electron oxidation and deprotonation of its sulfur. The parallels in this system to the dehydrogenases (deprotonation and 1-electron oxidation) again made the direct ligation of substrate to an auxiliary cluster an attractive hypothesis (23, 29). AlbA contains seven cysteines in a SPASM motif, whereas SkfB contains five cysteine residues in a C-terminal domain. Using anSME as a model for SPASM proteins and BtrN for Twitch proteins, we predict that the structure of AlbA will parallel that of anSME, and SkfB will contain a Twitch domain similar to MoaA and BtrN. If these predictions are correct, then both SAM radical dehydrogenases and thioether bond-forming enzymes will use both the SPASM and Twitch enzyme architectures to catalyze analogous reactions.
We can also use the recent structural data on this enzyme subclass to consider whether AlbA and SkfB will use an auxiliary cluster to bind their substrate. Because the distances between the SAM cluster and the Aux cluster (∼16 Å) as well as the SAM-binding motifs appear to be conserved in this subclass, we can predict for AlbA and SkfB that the distance between clusters will be ∼16 Å and that SAM will bind in a similar fashion as observed previously (5). The structures of anSME, BtrN, and MoaA also reveal similar distances (9.3 Å on average) between the hydrogen atom abstraction site on the substrate and the Aux cluster (9.6, 8.6, and 9.6 Å in BtrN, anSME, and MoaA, respectively) (Fig. 4, A–C). For BtrN and anSME, these long distances were used in part to make the argument that the Aux cluster was unlikely to be involved in the substrate deprotonation step that follows the hydrogen atom abstraction (11, 13). In MoaA, where a molecule of GTP is both the hydrogen atom abstraction site (3′-hydroxyl) (37) and the auxiliary cluster ligand (N1 of guanosine base) (58), this separation (7.4 Å) is both intramolecular and distal. Based on these numbers, if AlbA or SkfB peptidyl substrate binds such that one substrate residue (glycine in Fig. 4D) is close enough to SAM for hydrogen atom abstraction and a substrate cysteine is close enough to the auxiliary cluster for ligation, the distance between the glycine Cα and cysteine S would be at least 6 Å, which is too long for covalent bond formation. Although structures of AlbA and SkfB will be essential to resolve this question, our current structural data on anSME and BtrN predict that the auxiliary cluster of AlbA and SkfB will play a role in oxidation of substrate and not in its binding. Intriguingly, the peptidyl substrate of anSME binds in a tight turn (Fig. 4B), perhaps foreshadowing how AlbA and SkfB bind their peptidyl substrate to afford covalent linkage between two residues of the same peptide.
We can contrast these predictions with auxiliary cluster-containing SAM radical enzymes in different subfamilies where auxiliary clusters are believed to be involved in more than accepting an electron. For example, in the sulfur insertion enzyme BioB, the auxiliary [2Fe-2S] cluster is thought to play a direct role in the conversion of dethiobiotin (DTB) to biotin by donating a sulfur atom and accepting an electron from the resulting mercapto intermediate (59). This cluster is highly unusual in that it is positioned in the middle of a TIM barrel, ligated by three cysteines residing from β2, β3, and β5 strands of the SAM radical core fold, and an unprecedented arginine residue contributed by the C-terminal region (41). The distance between this Aux cluster and the SAM radical cluster in BioB is 12 Å, ∼4 Å closer than for BtrN, anSME, and MoaA. Also, the distance between the hydrogen atom abstraction site at the C9 position of DTB and the Aux cluster is 4.6 Å, ∼5 Å closer than for the SPASM/Twitch proteins (Fig. 4E). The structure of the methylthioltransferase RimO was also recently solved (55). As mentioned above, this structure has led to the hypothesis that the Aux cluster is not the direct sulfur source but may bind the sulfur source (55). Interestingly, the cluster-to-cluster distance in RimO is 8.4 Å, which is even closer than in BioB (12.0 Å), and twice as close as in the SPASM/Twitch architecture (Fig. 4F). Thus, the structures of the Aux cluster subset of SAM radical enzymes that are emerging show variation in cluster-to-cluster distance, in function (sulfur source (BioB), substrate binding (MoaA), and substrate oxidation (anSME, BtrN)), and in fold (both SPASM/Twitch and non-SPASM/Twitch). We have just scratched the surface of the structural exploration of this amazing superfamily, and more surprises likely await us.
Acknowledgments
We thank Dr. Jennifer Bridwell-Rabb, Dr. Dan Dowling, and Marco Jost for critical reading of the manuscript.
This work was supported by an MIT Office of the Dean of Graduate Education Fellowship (to T. A. J. G.) and National Science Foundation Grant MCB-0543833 (to C. L. D.). This is the first article in the Thematic Minireview series Radical SAM Enzymes.
- SAM
- S-adenosylmethionine
- 5′-dAdo•
- 5′-deoxyadenosyl radical
- TIM
- triose-phosphate isomerase
- anSME
- anaerobic sulfatase-maturating enzyme
- FGly
- formylglycine
- PQQ
- pyrroloquinoline quinone
- BtrN
- 2-deoxy-scyllo-inosamine dehydrogenase
- MoaA
- molybdenum cofactor biosynthetic enzyme
- 3′-8-cH2GTP
- (8S)-3′,8-cyclo-7,8-dihydroguanosine 5′-triphosphate
- QueE
- 7-carboxy-7-deazaguanine synthase
- DTB
- dethiobiotin
- BioB
- biotin synthase
- Moco
- molybdopterin
- Aux
- auxiliary cluster.
REFERENCES
- 1. Shisler K. A., Broderick J. B. (2012) Emerging themes in radical SAM chemistry. Curr. Opin. Struct. Biol. 22, 701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsby C. J., Hong W., Broderick W. E., Cheek J., Ortillo D., Broderick J. B., Hoffman B. M. (2002) Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 3143–3151 [DOI] [PubMed] [Google Scholar]
- 3. Walsby C. J., Ortillo D., Broderick W. E., Broderick J. B., Hoffman B. M. (2002) An anchoring role for FeS clusters: chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 11270–11271 [DOI] [PubMed] [Google Scholar]
- 4. Nicolet Y., Amara P., Mouesca J.-M., Fontecilla-Camps J. C. (2009) Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vey J. L., Drennan C. L. (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haft D. H. (2011) Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics 12, 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haft D. H., Basu M. K. (2011) Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 193, 2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akiva E., Brown S., Almonacid D. E., Barber A. E., 2nd, Custer A. F., Hicks M. A., Huang C. C., Lauck F., Mashiyama S. T., Meng E. C., Mischel D., Morris J. H., Ojha S., Schnoes A. M., Stryke D., Yunes J. M., Ferrin T. E., Holliday G. L., Babbitt P. C. (2014) The Structure-Function Linkage Database. Nucleic Acids Res. 42, D521–D530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benjdia A., Subramanian S., Leprince J., Vaudry H., Johnson M. K., Berteau O. (2010) Anaerobic sulfatase-maturating enzyme: A mechanistic link with glycyl radical-activating enzymes? FEBS J. 277, 1906–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grove T. L., Lee K.-H., St Clair J., Krebs C., Booker S. J. (2008) In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry 47, 7523–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldman P. J., Grove T. L., Sites L. A., McLaughlin M. I., Booker S. J., Drennan C. L. (2013) X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl. Acad. Sci. U.S.A. 110, 8519–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hänzelmann P., Schindelin H. (2004) Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 12870–12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goldman P. J., Grove T. L., Booker S. J., Drennan C. L. (2013) X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc. Natl. Acad. Sci. U.S.A. 110, 15949–15954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang Q., Peng J., Dierks T. (2004) Post-translational formylglycine modification of bacterial sulfatases by the radical S-adenosylmethionine protein AtsB. J. Biol. Chem. 279, 14570–14578 [DOI] [PubMed] [Google Scholar]
- 15. Benjdia A., Leprince J., Guillot A., Vaudry H., Rabot S., Berteau O. (2007) Anaerobic sulfatase-maturating enzymes: radical SAM enzymes able to catalyze in vitro sulfatase post-translational modification. J. Am. Chem. Soc. 129, 3462–3463 [DOI] [PubMed] [Google Scholar]
- 16. Berteau O., Guillot A., Benjdia A., Rabot S. (2006) A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes. J. Biol. Chem. 281, 22464–22470 [DOI] [PubMed] [Google Scholar]
- 17. Grove T. L., Ahlum J. H., Qin R. M., Lanz N. D., Radle M. I., Krebs C., Booker S. J. (2013) Further characterization of Cys-type and Ser-type anaerobic sulfatase maturating enzymes suggests a commonality in the mechanism of catalysis. Biochemistry 52, 2874–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt B., Selmer T., Ingendoh A., von Figura K. (1995) A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell 82, 271–278 [DOI] [PubMed] [Google Scholar]
- 19. Ghosh D. (2007) Human sulfatases: a structural perspective to catalysis. Cell. Mol. Life Sci. 64, 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bojarová P., Williams S. J. (2008) Sulfotransferases, sulfatases and formylglycine-generating enzymes: a sulfation fascination. Curr. Opin. Chem. Biol. 12, 573–581 [DOI] [PubMed] [Google Scholar]
- 21. Dierks T., Schmidt B., Borissenko L. V., Peng J., Preusser A., Mariappan M., von Figura K. (2003) Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα formylglycine generating enzyme. Cell 113, 435–444 [DOI] [PubMed] [Google Scholar]
- 22. Benjdia A., Martens E. C., Gordon J. I., Berteau O. (2011) Sulfatases and a radical S-adenosyl-l-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J. Biol. Chem. 286, 25973–25982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flühe L., Knappe T. A., Gattner M. J., Schäfer A., Burghaus O., Linne U., Marahiel M. A. (2012) The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 [DOI] [PubMed] [Google Scholar]
- 24. Zheng G., Hehn R., Zuber P. (2000) Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182, 3266–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shelburne C. E., An F. Y., Dholpe V., Ramamoorthy A., Lopatin D. E., Lantz M. S. (2007) The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J. Antimicrob. Chemother. 59, 297–300 [DOI] [PubMed] [Google Scholar]
- 26. Engelberg-Kulka H., Hazan R. (2003) Cannibals defy starvation and avoid sporulation. Science 301, 467–468 [DOI] [PubMed] [Google Scholar]
- 27. González-Pastor J. E., Hobbs E. C., Losick R. (2003) Cannibalism by sporulating bacteria. Science 301, 510–513 [DOI] [PubMed] [Google Scholar]
- 28. Liu W.-T., Yang Y.-L., Xu Y., Lamsa A., Haste N. M., Yang J. Y., Ng J., Gonzalez D., Ellermeier C. D., Straight P. D., Pevzner P. A., Pogliano J., Nizet V., Pogliano K., Dorrestein P. C. (2010) Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 107, 16286–16290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flühe L., Burghaus O., Wieckowski B. M., Giessen T. W., Linne U., Marahiel M. A. (2013) Two [4Fe-4S] clusters containing radical SAM enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor. J. Am. Chem. Soc. 135, 959–962 [DOI] [PubMed] [Google Scholar]
- 30. Velterop J. S., Sellink E., Meulenberg J. J. M., David S., Bulder I., Postma P. W. (1995) Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway. J. Bacteriol. 177, 5088–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Houck D. R., Hanners J. L., Unkefer C. J. (1991) Biosynthesis of pyrroloquinoline quinone. 2. Biosynthetic assembly from glutamate and tyrosine. J. Am. Chem. Soc. 113, 3162–3166 [Google Scholar]
- 32. Wecksler S. R., Stoll S., Tran H., Magnusson O. T., Wu S.-P., King D., Britt R. D., Klinman J. P. (2009) Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-l-methionine enzyme. Biochemistry 48, 10151–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wecksler S. R., Stoll S., Iavarone A. T., Imsand E. M., Tran H., Britt R. D., Klinman J. P. (2010) Interaction of PqqE and PqqD in the pyrroloquinoline quinone (PQQ) biosynthetic pathway links PqqD to the radical SAM superfamily. Chem. Commun. (Camb.) 46, 7031–7033 [DOI] [PubMed] [Google Scholar]
- 34. Benjdia A., Subramanian S., Leprince J., Vaudry H., Johnson M. K., Berteau O. (2008) Anaerobic sulfatase-maturating enzymes, first dual substrate radical S-adenosylmethionine enzymes. J. Biol. Chem. 283, 17815–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yokoyama K., Numakura M., Kudo F., Ohmori D., Eguchi T. (2007) Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J. Am. Chem. Soc. 129, 15147–15155 [DOI] [PubMed] [Google Scholar]
- 36. Yokoyama K., Ohmori D., Kudo F., Eguchi T. (2008) Mechanistic study on the reaction of a radical SAM dehydrogenase BtrN by electron paramagnetic resonance spectroscopy. Biochemistry 47, 8950–8960 [DOI] [PubMed] [Google Scholar]
- 37. Mehta A. P., Hanes J. W., Abdelwahed S. H., Hilmey D. G., Hänzelmann P., Begley T. P. (2013) Catalysis of a new ribose carbon-insertion reaction by the molybdenum cofactor biosynthetic enzyme MoaA. Biochemistry 52, 1134–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hover B. M., Loksztejn A., Ribeiro A. A., Yokoyama K. (2013) Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 135, 7019–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson J. L., Rajagopalan K. V., Wadman S. K. (1993) Human molybdenum cofactor deficiency. in Chemistry and Biology of Pteridines and Folates (Ayling J. E., Nair G., Baugh C. M., eds), pp. 373–378, Springer US New York Inc., New York: [DOI] [PubMed] [Google Scholar]
- 40. Dowling D. P., Vey J. L., Croft A. K., Drennan C. L. (2012) Structural diversity in the AdoMet radical enzyme superfamily. Biochim. Biophys. Acta 1824, 1178–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berkovitch F., Nicolet Y., Wan J. T., Jarrett J. T., Drennan C. L. (2004) Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chatterjee A., Li Y., Zhang Y., Grove T. L., Lee M., Krebs C., Booker S. J., Begley T. P., Ealick S. E. (2008) Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dowling D. P., Bruender N. A., Young A. P., McCarty R. M., Bandarian V., Drennan C. L. (2014) Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat. Chem. Biol. 10, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Farrar C. E., Jarrett J. T. (2009) Protein residues that control the reaction trajectory in S-adenosylmethionine radical enzymes: mutagenesis of asparagine 153 and aspartate 155 in Escherichia coli biotin synthase. Biochemistry 48, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hänzelmann P., Schindelin H. (2006) Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. U.S.A. 103, 6829–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ugulava N. B., Sacanell C. J., Jarrett J. T. (2001) Spectroscopic changes during a single turnover of biotin synthase: destruction of a [2Fe-2S] cluster accompanies sulfur insertion. Biochemistry 40, 8352–8358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jameson G. N. L., Cosper M. M., Hernández H. L., Johnson M. K., Huynh B. H. (2004) Role of the 2Fe-2S cluster in recombinant Escherichia coli biotin synthase. Biochemistry 43, 2022–2031 [DOI] [PubMed] [Google Scholar]
- 48. Tse Sum Bui B., Benda R., Schünemann V., Florentin D., Trautwein A. X., Marquet A. (2003) Fate of the (2Fe-2S)2+ cluster of Escherichia coli biotin synthase during reaction: a Mossbauer characterization. Biochemistry 42, 8791–8798 [DOI] [PubMed] [Google Scholar]
- 49. Cicchillo R. M., Lee K.-H., Baleanu-Gogonea C., Nesbitt N. M., Krebs C., Booker S. J. (2004) Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry 43, 11770–11781 [DOI] [PubMed] [Google Scholar]
- 50. Cicchillo R. M., Booker S. J. (2005) Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J. Am. Chem. Soc. 127, 2860–2861 [DOI] [PubMed] [Google Scholar]
- 51. Miller J. R., Busby R. W., Jordan S. W., Cheek J., Henshaw T. F., Ashley G. W., Broderick J. B., Cronan J. E., Jr., Marletta M. A. (2000) Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39, 15166–15178 [DOI] [PubMed] [Google Scholar]
- 52. Hernández H. L., Pierrel F., Elleingand E., García-Serres R., Huynh B. H., Johnson M. K., Fontecave M., Atta M. (2007) MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry 46, 5140–5147 [DOI] [PubMed] [Google Scholar]
- 53. Lee K.-H., Saleh L., Anton B. P., Madinger C. L., Benner J. S., Iwig D. F., Roberts R. J., Krebs C., Booker S. J. (2009) Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry 48, 10162–10174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Landgraf B. J., Arcinas A. J., Lee K. H., Booker S. J. (2013) Identification of an intermediate methyl carrier in the radical S-adenosylmethionine methylthiotransferases RimO and MiaB. J. Am. Chem. Soc. 135, 15404–15416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Forouhar F., Arragain S., Atta M., Gambarelli S., Mouesca J.-M., Hussain M., Xiao R., Kieffer-Jaquinod S., Seetharaman J., Acton T. B., Montelione G. T., Mulliez E., Hunt J. F., Fontecave M. (2013) Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat. Chem. Biol. 9, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lanz N. D., Booker S. J. (2012) Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes. Biochim. Biophys. Acta 1824, 1196–1212 [DOI] [PubMed] [Google Scholar]
- 57. Kuchenreuther J. M., Myers W. K., Suess D. L. M., Stich T. A., Pelmenschikov V., Shiigi S. A., Cramer S. P., Swartz J. R., Britt R. D., George S. J. (2014) The HydG enzyme generates an Fe(CO)2(CN) synthon in assembly of the FeFe hydrogenase H-cluster. Science 343, 424–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lees N. S., Hänzelmann P., Hernandez H. L., Subramanian S., Schindelin H., Johnson M. K., Hoffman B. M. (2009) ENDOR spectroscopy shows that guanine N1 binds to [4Fe-4S] cluster II of the S-adenosylmethionine-dependent enzyme MoaA: mechanistic implications. J. Am. Chem. Soc. 131, 9184–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fugate C. J., Stich T. A., Kim E. G., Myers W. K., Britt R. D., Jarrett J. T. (2012) 9-Mercaptodethiobiotin is generated as a ligand to the [2Fe-2S]+ cluster during the reaction catalyzed by biotin synthase from Escherichia coli. J. Am. Chem. Soc. 134, 9042–9045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Atkinson H. J., Morris J. H., Ferrin T. E., Babbitt P. C. (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PloS one 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]