Abstract
In this minireview, we describe the radical S-adenosylmethionine enzymes involved in the biosynthesis of thiamin, menaquinone, molybdopterin, coenzyme F420, and heme. Our focus is on the remarkably complex organic rearrangements involved, many of which have no precedent in organic or biological chemistry.
Keywords: Biosynthesis, Enzyme Mechanism, Heme, radical, S-Adenosylmethionine (SAM), Thiamin, Deazaflavin, Menaquinone, Molybdopterin, Rearrangement
Introduction
Radical S-adenosylmethionine (SAM)2 enzymes are among the most intriguing enzymes discovered over the past 15 years. In these enzymes, reduction of SAM by a [4Fe-4S]1+ cluster generates the adenosyl radical, which then abstracts a hydrogen atom from the substrate. The resulting substrate radical usually undergoes a rearrangement or fragmentation reaction to give the product radical (1). Sequence analysis suggests that a large number of enzymes use the radical SAM catalytic motif (2). We are still at an early stage of defining the catalytic mechanisms and the structural enzymology of this fascinating enzyme family.
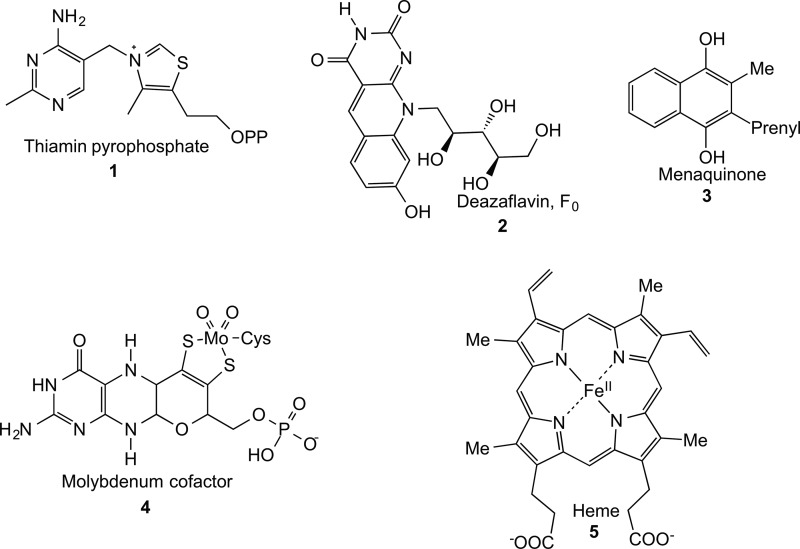
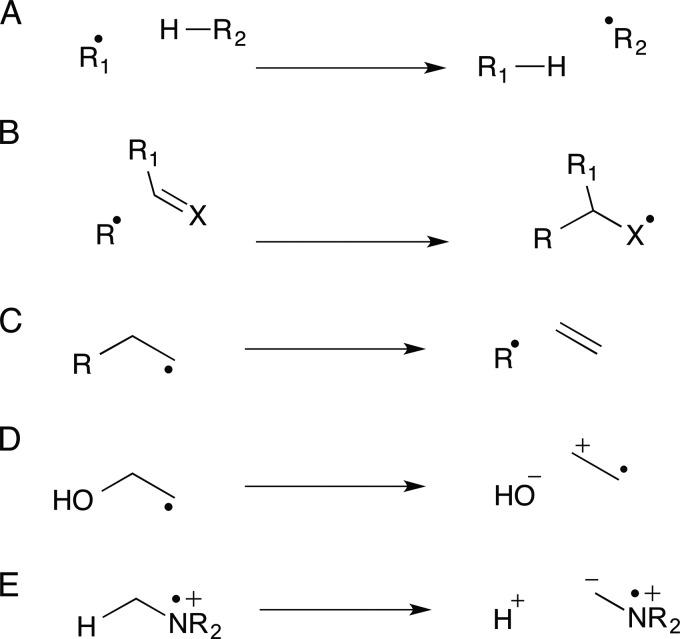
This minireview focuses on the organic chemistry of the radical SAM enzymes involved in cofactor biosynthesis: thiamin (1), F0 (2), menaquinone (3), molybdopterin (4), and heme (5) (Fig. 1). (Biotin and lipoic biosynthesis also uses radical SAM enzymology and is covered separately in another minireview in this series (42).) In contrast to iron(IV)-oxo-derived radicals, where the dominant chemistry involves radical recombination with the iron-bound oxygen (P450 rebound rate of >1010 s−1) (3), radicals formed by hydrogen atom transfer to the 5′-deoxyadenosyl (5′-dA) radical are more persistent because the reverse reaction is relatively slow. This allows time for complex rearrangements to occur. Radical SAM enzymes catalyze a remarkable range of reactions due to the high intrinsic reactivity of organic radicals, which can undergo rapid hydrogen atom abstraction, double-bond addition, and fragmentation reactions as shown in Fig. 2.
FIGURE 1.
Cofactor biosynthetic pathways make extensive use of radical SAM enzymology. The structures of the five cofactors described in this minireview are shown.
FIGURE 2.
Examples of the major radical reactions found in organic chemistry. A, hydrogen atom abstraction, e.g. 6 to 10 and 14 to 15. B, addition to double bonds, e.g. 15 to 16. C–E, β-bond scission reactions, e.g. 10 to 11, 16 to 17, 17 to 18+19, 19 to 21, 22 to 23, 24 to 25, and 25 to 26. All enzymatic examples are taken from Fig. 3.
Prokaryotic Thiamin Pyrimidine Synthase (ThiC)
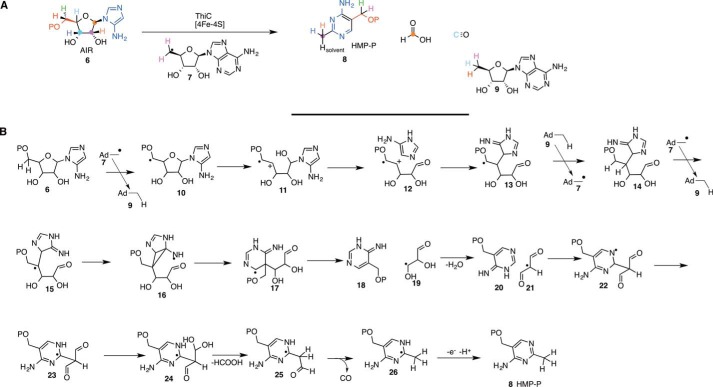
Thiamin pyrophosphate (1) is an important cofactor in carbohydrate metabolism and branched chain amino acid biosynthesis, where it plays a key role in stabilization of the acyl carbanion biosynthon. The thiamin pyrimidine synthase (ThiC) catalyzes the conversion of aminoimidazole ribotide (AIR; 6) to hydroxymethylpyrimidine phosphate (HMP-P; 8) (Fig. 3A) (4, 5). The origin of all of the atoms of the product and the fate of all of the atoms of the substrate have been determined using isotope labeling studies (6–8). This rearrangement, as far as we can tell, is the most complex unsolved rearrangement in primary metabolism.
FIGURE 3.
Radical SAM enzymology in thiamin pyrimidine biosynthesis. A, results of a comprehensive labeling study to determine the fate of all of the atoms of AIR (6) during the formation of the HMP-P (8). B, mechanistic proposal for the formation of HMP-P (8).
A mechanistic proposal for this reaction is shown in Fig. 3B (5). In this proposal, radical 7 abstracts a hydrogen atom from 6 to form 10. A β-scission followed by N-glycosyl bond cleavage gives 12. Electrophilic addition to the aminoimidazole followed by hydrogen atom transfer gives 14. The regenerated 5′-dA radical (7) abstracts a hydrogen atom from 14 to form 15. Radical addition to the imine followed by β-scission gives 17. A second β-scission followed by a diol dehydratase-like rearrangement gives 20 and 21. Radical addition to the pyrimidine followed by tautomerization gives 23. Loss of formate and CO followed by electron transfer back to the [4Fe-4S]2+ cluster completes the formation of HMP-P (8). This mechanism is supported by labeling studies (5–8), by the identification of CO and formate as reaction products (4, 5), and by a structure of the enzyme with desamino-AIR bound at the active site (4).
Prokaryotic Thiamin Thiazole Biosynthesis (ThiH)
Tyrosine lyase (ThiH) catalyzes a tyrosine cleavage reaction to form dehydroglycine (28) and p-cresol (29) (9, 10). Dehydroglycine (28) is a substrate for the bacterial thiazole synthase involved in the formation of 32 (11). A mechanistic proposal for the lyase reaction is shown in Fig. 4 (9). Abstraction of the amine hydrogen atom by the 5′-dA radical gives 30. A β-scission gives dehydroglycine (28) and radical 31, which is quenched by protonation and reduction. This mechanism is supported by substrate analog studies and product characterization (9, 10). Radical SAM amino acid lyases are a growing family of enzymes and, in addition to ThiH, now include NosL (involved in nosiheptide biosynthesis) (12), the maturase HydG (involved in the biosynthesis of the cyanide and carbon monoxide ligands of the [FeFe]-hydrogenase cluster) (13, 14), and CofH (involved in deazaflavin biosynthesis; see below). The recent structural studies on tryptophan lyase (NosL) provides clear support for amine hydrogen atom abstraction and suggest that radical SAM amino acid lyases are likely to proceed by amine rather than phenol hydrogen atom abstraction (15).
FIGURE 4.
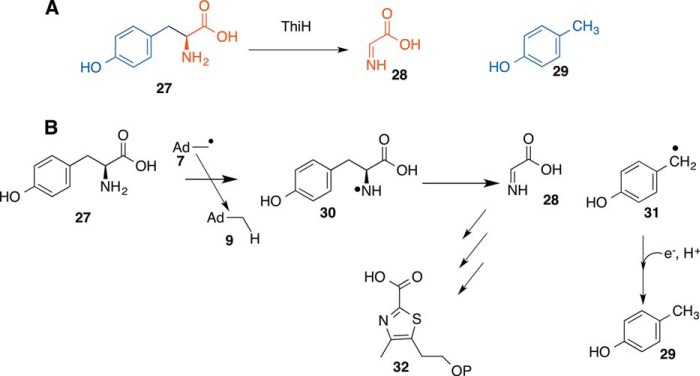
Radical SAM enzymology in thiamin thiazole biosynthesis. A, tyrosine lyase (ThiH)-catalyzed reaction. B, mechanistic proposal for this reaction.
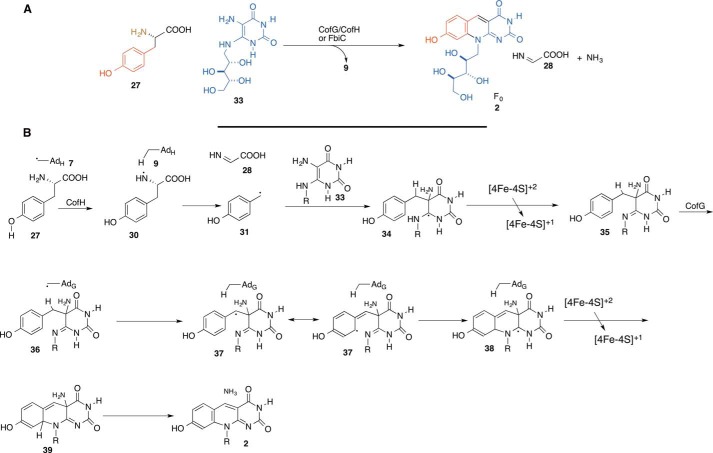
Coenzyme F420 Biosynthesis (CofG/CofH or FbiC)
F420 is a hydride transfer cofactor found in methanogens and in some actinomyces and cyanobacteria. Its precursor, F0 (2), is formed from tyrosine (27) and ribityldiaminouracil (33) (Fig. 5A). The F0-synthase-catalyzed reaction has been recently reconstituted using the CofG/CofH and FbiC systems (16). The formation of F0 is mediated by two separate radical SAM active sites, one in CofG and the other in CofH or both in separate domains of FbiC. F0-synthase is therefore an unusual radical SAM enzyme in that it uses two separate adenosyl radicals to catalyze F0 formation. The key mechanistic question posed by F0-synthase is how two adenosyl radicals cooperate in the assembly of the deazaflavin cofactor. The current mechanistic proposal for this enzyme is shown in Fig. 5B. In this mechanism, abstraction of the tyrosine amine hydrogen atom by the CofH adenosyl radical gives 30, which then undergoes fragmentation, leading to the formation of 28 and 31. Addition of 31 to diaminouracil (33) gives 34, which is oxidized to 35. A second hydrogen atom abstraction by the CofG adenosyl radical gives 37. Cyclization to 38, oxidation, and elimination of ammonia complete the formation of the deazaflavin (2). This mechanism is supported by the sequence similarity of CofH and ThiH (Fig. 4). The observation of 33 bound to CofH suggests that CofH catalyzes the first step of F0 formation.
FIGURE 5.
Radical SAM enzymology in deazaflavin biosynthesis. A, F0-synthase-catalyzed reaction. B, mechanistic proposal for F0-synthase.
Futalosine-dependent Menaquinone Biosynthesis (MqnC and MqnE)
Menaquinone (3) is a membrane-bound electron transfer cofactor that plays a key role in bacterial respiration, oxidative phosphorylation, and photosynthesis. In humans, menaquinone (vitamin K) is used for the remarkable carboxylation of glutamic acid residues in proteins involved in blood clotting and bone morphogenesis.
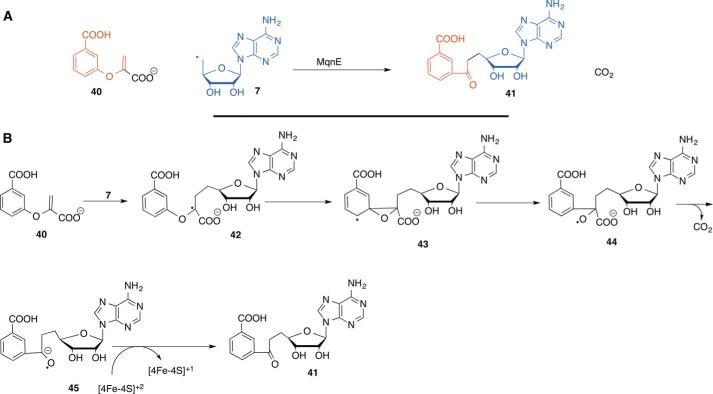
In the recently discovered futalosine-dependent pathway (17–20), MqnE catalyzes the conversion of 40 to aminofutalosine (41) (21). A mechanistic proposal is outlined in Fig. 6. In this proposal, the 5′-dA radical (7) adds to 40 to yield the captodative radical (42). Rearrangement of this radical via 43 gives 44. Such rearrangements have previously been proposed in nonenzymatic systems, but have not yet been studied in an enzyme-mediated reaction (22–24). Decarboxylation of 44, facilitated by the alkoxy radical, yields the ketyl radical (45). A final electron transfer, possibly to the [4Fe-4S]2+ cluster, completes the formation of aminofutalosine (41) (21). All previously characterized rearrangements mediated by radical SAM enzymes involve radical generation by hydrogen atom abstraction from the substrate (25–28). MqnE represents a new catalytic motif involving addition of the adenosyl radical to the substrate to generate the rearranging radical.
FIGURE 6.
Radical SAM enzymology in menaquinone biosynthesis. A, reaction catalyzed by aminofutalosine synthase (MqnE). B, mechanistic proposal for the MqnE-catalyzed reaction.
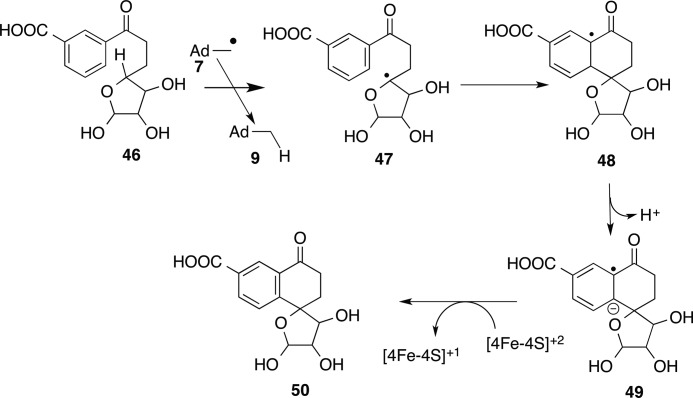
MqnC catalyzes the conversion of 46 to 50 (19, 29). A mechanistic proposal is outlined in Fig. 7. In this proposal, the 5′-dA radical abstracts a hydrogen atom from C4′ of 46 to give radical 47, which adds to the benzene ring to give radical 48. Deprotonation followed by oxidation completes the reaction. This mechanism is supported by the identification of the site of hydrogen atom abstraction and by the product structure (29).
FIGURE 7.
Radical SAM enzymology in menaquinone biosynthesis. Shown is the mechanistic proposal for the MqnC-catalyzed conversion of 46 to 50.
MqnE and MqnC both catalyze radical additions to benzene rings. Such reactions are still poorly understood biotransformations.
Molybdopterin Biosynthesis (MoaA)
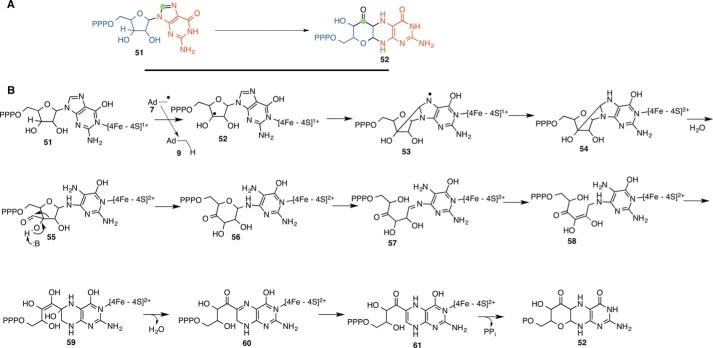
Molybdopterin (4) is a redox cofactor used by enzymes such as xanthine oxidase, sulfite oxidase, nitrate reductase, carbon monoxide dehydrogenase, and formate dehydrogenase. The first step in the biosynthesis of this cofactor involves the MoaA-catalyzed conversion of GTP (51) to the pterin (52) (Fig. 8A) (30, 31). This enzyme has two [4Fe-4S] clusters at the active site. The N-terminal cluster is involved in the reductive cleavage of SAM, and the C-terminal cluster is involved in substrate binding (32, 33). A mechanistic proposal is shown in Fig. 8B (34, 35). In this proposal, hydrogen atom abstraction occurs from C3′ of GTP (51), and the resulting radical adds to C8 of the purine to form 53. Reduction by the purine-liganded cluster gives 54, and aminal hydrolysis yields 55, followed by an α-ketol rearrangement to 56. Ring opening followed by tautomerization gives 58, which is converted to 52 by conjugate addition, water elimination, tautomerization, and ring closure sequence. As far as we can determine, analogous carbon insertion reactions have not been previously reported in carbohydrate chemistry.
FIGURE 8.
Radical SAM enzymology in molybdopterin biosynthesis. A, MoaA-catalyzed reaction. B, mechanistic proposal for the conversion of GTP (51) to the pterin (52).
This mechanism is supported by labeling studies demonstrating that C8 of GTP is inserted between C2′ and C3′ of the GTP ribose (36) and that the 5′-dA radical abstracts a hydrogen atom from C3′ of GTP (35). In addition, intermediate 54 has been trapped using native substrate 51 and the substrate analog 2′,3′-dideoxy-GTP (34, 37).
Oxygen-independent Coproporphyrinogen III Oxidase (HemN)
Heme (5) functions as an oxygen carrier, a redox cofactor, and a source of the iron-oxo intermediate in biochemical hydroxylation, epoxidation, and dehydrogenation reactions. HemN catalyzes the oxidative decarboxylation of coproporphyrinogen III (62) to form protoporphyrinogen IX (63) in heme biosynthesis. A mechanistic proposal for this reaction is shown in Fig. 9 (38). In this proposal, the 5′-dA radical abstracts a hydrogen atom from 62, generating 64. This then undergoes decarboxylation and oxidation to form 63. Repetition of this chemistry generates the B-ring vinyl group. This mechanism is supported by a structure of HemN (39), by the identification of the hydrogen atoms abstracted from coproporphyrinogen III (40), and by the detection of the substrate radical by EPR (41). This decarboxylation (as well as the conversion of 44 to 45 for MqnE) nicely demonstrates how an impossible decarboxylation in a closed shell species becomes possible through radical chemistry.
FIGURE 9.
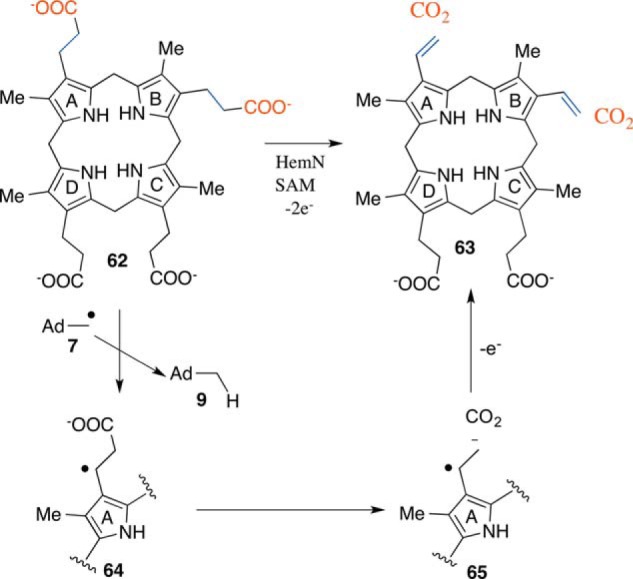
Radical SAM enzymology in heme biosynthesis. Shown is the mechanistic proposal for the coproporphyrinogen III oxidase (HemN)-catalyzed formation of protoporphyrinogen IX (63).
Conclusions
Radical SAM enzymes are surprisingly prevalent in cofactor biosynthesis and play key roles in the biosynthesis of thiamin, menaquinone, molybdopterin, F420, heme, biotin, and lipoic acid. The novelty of the chemistry demonstrates that, in the protected and catalytic environment of the enzyme active site, organic radicals can undergo complex rearrangements never before found in organic chemistry. As most of these enzymes have only recently become available, mechanistic analysis is still in its infancy, and much remains to be discovered in this new and exciting area of cofactor biosynthesis.
This research was supported by Robert A. Welch Foundation Grant A-0034 and National Institutes of Health Grant DK44083 (to T. P. B.). This is the third article in the Thematic Minireview Series on Radical SAM Enzymes.
- SAM
- S-adenosylmethionine
- 5′-dA
- 5′-deoxyadenosyl
- AIR
- aminoimidazole ribotide
- HMP-P
- hydroxymethylpyrimidine phosphate.
REFERENCES
- 1. Broderick J. B., Duffus B. R., Duschene K. S., Shepard E. M. (2014) Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atkinson J. K., Ingold K. U. (1993) Cytochrome P450 hydroxylation of hydrocarbons: variation in the rate of oxygen rebound using cyclopropyl radical clocks including two new ultrafast probes. Biochemistry 32, 9209–9214 [DOI] [PubMed] [Google Scholar]
- 4. Chatterjee A., Li Y., Zhang Y., Grove T. L., Lee M., Krebs C., Booker S. J., Begley T. P., Ealick S. E. (2008) Reconstitution of ThiC in thiamin pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chatterjee A., Hazra A. B., Abdelwahed S., Hilmey D. G., Begley T. P. (2010) A radical dance in thiamin biosynthesis: mechanistic analysis of the bacterial hydroxymethylpyrimidine phosphate synthase. Angew. Chem. Int. Ed. Engl. 49, 8653–8656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tazuya K., Tanaka K., Yamada K., Kumaoka H. (1999) The origin of the nitrogen atoms of the pyrimidine moiety of thiamin under anaerobic conditions in Escherichia coli. Bitamin 73, 373–378 [Google Scholar]
- 7. Yamada K., Kumaoka H. (1982) Biosynthesis of thiamin. Incorporation of a two-carbon fragment derived from ribose of 5-aminoimidazole ribotide into the pyrimidine moiety of thiamin. Biochem. Int. 5, 771–776 [Google Scholar]
- 8. Lawhorn B. G., Mehl R. A., Begley T. P. (2004) Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Org. Biomol. Chem. 2, 2538–2546 [DOI] [PubMed] [Google Scholar]
- 9. Challand M. R., Martins F. T., Roach P. L. (2010) Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli. J. Biol. Chem. 285, 5240–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kriek M., Martins F., Challand M. R., Croft A., Roach P. L. (2007) Thiamine biosynthesis in Escherichia coli: identification of the intermediate and by-product derived from tyrosine. Angew. Chem. Int. Ed. Engl. 46, 9223–9226 [DOI] [PubMed] [Google Scholar]
- 11. Kriek M., Martins F., Leonardi R., Fairhurst S. A., Lowe D. J., Roach P. L. (2007) Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro. J. Biol. Chem. 282, 17413–17423 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Q., Li Y.-X., Chen D.-D., Yu Y., Duan L., Shen B., Liu W. (2011) Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat. Chem. Biol. 7, 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stich T. A., Myers W. K., Britt R. D. (2014) Paramagnetic intermediates generated by radical S-adenosylmethionine (SAM) enzymes. Acc. Chem. Res. 47, 2235–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuchenreuther J. M., Myers W. K., Suess D. L., Stich T. A., Pelmenschikov V., Shiigi S. A., Cramer S. P., Swartz J. R., Britt R. D., George S. J. (2014) The HydG enzyme generates an Fe(CO)2(CN) synthon in assembly of the FeFe hydrogenase H-cluster. Science 343, 424–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nicolet Y., Zeppieri L., Amara P., Fontecilla-Camps J. C. (2014) Crystal structure of tryptophan lyase (NosL): evidence for radical formation at the amino group of tryptophan. Angew. Chem. Int. Ed. Engl. 53, 11840–11844 [DOI] [PubMed] [Google Scholar]
- 16. Decamps L., Philmus B., Benjdia A., White R., Begley T. P., Berteau O. (2012) Biosynthesis of F0, precursor of the F420 cofactor, requires a unique two radical-SAM domain enzyme and tyrosine as substrate. J. Am. Chem. Soc. 134, 18173–18176 [DOI] [PubMed] [Google Scholar]
- 17. Dairi T. (2012) Menaquinone biosyntheses in microorganisms. Methods Enzymol. 515, 107–122 [DOI] [PubMed] [Google Scholar]
- 18. Dairi T., Kuzuyama T., Nishiyama M., Fujii I. (2011) Convergent strategies in biosynthesis. Nat. Prod. Rep. 28, 1054–1086 [DOI] [PubMed] [Google Scholar]
- 19. Hiratsuka T., Furihata K., Ishikawa J., Yamashita H., Itoh N., Seto H., Dairi T. (2008) An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321, 1670–1673 [DOI] [PubMed] [Google Scholar]
- 20. Seto H., Jinnai Y., Hiratsuka T., Fukawa M., Furihata K., Itoh N., Dairi T. (2008) Studies on a new biosynthetic pathway for menaquinone. J. Am. Chem. Soc. 130, 5614–5615 [DOI] [PubMed] [Google Scholar]
- 21. Mahanta N., Fedoseyenko D., Dairi T., Begley T. P. (2013) Menaquinone biosynthesis: formation of aminofutalosine requires a unique radical SAM enzyme. J. Am. Chem. Soc. 135, 15318–15321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bietti M., Calcagni A., Cicero D. O., Martella R., Salamone M. (2010) The O-neophyl rearrangement of 1,1-diarylalkoxyl radicals. Experimental evidence for the formation of an intermediate 1-oxaspiro[2,5]octadienyl radical. Tetrahedron Lett. 51, 4129–4131 [Google Scholar]
- 23. Baroudi A., Alicea J., Flack P., Kirincich J., Alabugin I. V. (2011) Radical O→C transposition: a metal-free process for conversion of phenols into benzoates and benzamides. J. Org. Chem. 76, 1521–1537 [DOI] [PubMed] [Google Scholar]
- 24. Baroudi A., Flack P., Alabugin I. V. (2010) Metal-free transformation of phenols into substituted benzamides: a highly selective radical 1,2-O→C transposition in O-aryl-N-phenylthiocarbamates. Chem. Eur. J. 16, 12316–12320 [DOI] [PubMed] [Google Scholar]
- 25. Frey P. A., Hegeman A. D., Ruzicka F. J. (2008) The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 [DOI] [PubMed] [Google Scholar]
- 26. Roach P. L. (2011) Radicals from S-adenosylmethionine and their application to biosynthesis. Curr. Opin. Chem. Biol. 15, 267–275 [DOI] [PubMed] [Google Scholar]
- 27. Vey J. L., Drennan C. L. (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Zhu X., Torelli A. T., Lee M., Dzikovski B., Koralewski R. M., Wang E., Freed J., Krebs C., Ealick S. E., Lin H. (2010) Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature 465, 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cooper L. E., Fedoseyenko D., Abdelwahed S. H., Kim S.-H., Dairi T., Begley T. P. (2013) In vitro reconstitution of the radical S-adenosylmethionine enzyme MqnC involved in the biosynthesis of futalosine-derived menaquinone. Biochemistry 52, 4592–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leimkühler S., Wuebbens M. M., Rajagopalan K. V. (2011) The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord. Chem. Rev. 255, 1129–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mehta A. P., Abdelwahed S. H., Xu H., Begley T. P. (2014) Molybdopterin biosynthesis: trapping of intermediates for the MoaA-catalyzed reaction using 2′-deoxyGTP and 2′-chloroGTP as substrate analogues. J. Am. Chem. Soc. 136, 10609–10614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hänzelmann P., Schindelin H. (2006) Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. U.S.A. 103, 6829–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lees N. S., Hänzelmann P., Hernandez H. L., Subramanian S., Schindelin H., Johnson M. K., Hoffman B. M. (2009) ENDOR spectroscopy shows that guanine N1 binds to [4Fe-4S] cluster II of the S-adenosylmethionine-dependent enzyme MoaA: mechanistic implications. J. Am. Chem. Soc. 131, 9184–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mehta A. P., Abdelwahed S. H., Begley T. P. (2013) Molybdopterin biosynthesis: trapping an unusual purine ribose adduct in the MoaA-catalyzed reaction. J Am Chem Soc 135, 10883–10885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mehta A. P., Hanes J. W., Abdelwahed S. H., Hilmey D. G., Hänzelmann P., Begley T. P. (2013) Catalysis of a new ribose carbon-insertion reaction by the molybdenum cofactor biosynthetic enzyme MoaA. Biochemistry 52, 1134–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rieder C., Eisenreich W., O'Brien J., Richter G., Götze E., Boyle P., Blanchard S., Bacher A., Simon H. (1998) Rearrangement reactions in the biosynthesis of molybdopterin-an NMR study with multiply 13C/15N labelled precursors. Eur. J. Biochem. 255, 24–36 [DOI] [PubMed] [Google Scholar]
- 37. Hover B. M., Loksztejn A., Ribeiro A. A., Yokoyama K. (2013) Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 135, 7019–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Layer G., Kervio E., Morlock G., Heinz D. W., Jahn D., Retey J., Schubert W. (2005) Structural and functional comparison of HemN to other radical SAM enzymes. Biol. Chem. 386, 971–980 [DOI] [PubMed] [Google Scholar]
- 39. Layer G., Moser J., Heinz D. W., Jahn D., Schubert W. (2003) Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 22, 6214–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seehra J. S., Jordan P. M., Akhtar M. (1983) Anaerobic and aerobic coproporphyrinogen III oxidases of Rhodopseudomonas spheroides. Mechanism and stereochemistry of vinyl group formation. Biochem. J. 209, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Layer G., Pierik A. J., Trost M., Rigby S. E., Leech H. K., Grage K., Breckau D., Astner I., Jänsch L., Heathcote P., Warren M. J., Heinz D. W., Jahn D. (2006) The substrate radical of Escherichia coli oxygen-independent coproporphyrinogen III oxidase HemN. J. Biol. Chem. 281, 15727–15734 [DOI] [PubMed] [Google Scholar]
- 42. Jarrett J. T. (2015) The biosynthesis of thiol- and thioether-containing cofactors and secondary metabolites catalyzed by radical S-adenosylmethionine enzymes. J. Biol. Chem. 290, 3972–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]