Background: Arg/Abl2 has two actin-binding domains, but how they influence actin filaments was unknown.
Results: Arg and cortactin cooperatively stabilize actin filaments; Arg enhances Arp2/3 complex activation and stimulates severing by cofilin.
Conclusion: Arg directly regulates actin filament stability, branching, and severing, which are modulated by cortactin.
Significance: These activities may underlie the control of actin-based cellular structures by Arg.
Keywords: Actin, Arp2/3 Complex, Cofilin, Cortactin, Tyrosine-protein Kinase (Tyrosine Kinase)
Abstract
Both Arp2/3 complex and the Abl2/Arg nonreceptor tyrosine kinase are essential to form and maintain diverse actin-based structures in cells, including cell edge protrusions in fibroblasts and cancer cells and dendritic spines in neurons. The ability of Arg to promote cell edge protrusions in fibroblasts does not absolutely require kinase activity, raising the question of how Arg might modulate actin assembly and turnover in the absence of kinase function. Arg has two distinct actin-binding domains and interacts physically and functionally with cortactin, an activator of the Arp2/3 complex. However, it was not known whether and how Arg influences actin filament stability, actin branch formation, or cofilin-mediated actin severing or how cortactin influences these reactions of Arg with actin. Arg or cortactin bound to actin filaments stabilizes them from depolymerization. Low concentrations of Arg and cortactin cooperate to stabilize filaments by slowing depolymerization. Arg stimulates formation of actin filament branches by Arp2/3 complex and cortactin. An Arg mutant lacking the C-terminal calponin homology actin-binding domain stimulates actin branch formation by the Arp2/3 complex, indicative of autoinhibition. ArgΔCH can stimulate the Arp2/3 complex even in the absence of cortactin. Arg greatly potentiates cofilin severing of actin filaments, and cortactin attenuates this enhanced severing. The ability of Arg to stabilize filaments, promote branching, and increase severing requires the internal (I/L)WEQ actin-binding domain. These activities likely underlie important roles that Arg plays in the formation, dynamics, and stability of actin-based cellular structures.
Introduction
The precise regulation of actin polymerization and depolymerization is critical for the formation, maintenance, and dynamic behavior of actin-based subcellular structures. Cells use formins, Arp2/36 complex, and other proteins to nucleate new filaments, which grow by elongation of their barbed ends (1, 2). ADF/cofilin proteins sever actin filaments, producing a new barbed end at each severing site (3). However, extensive cofilin-mediated actin severing can also lead to loss of actin filaments and disruption of branched actin networks (4). Cross-linking proteins can organize actin filaments into bundles and other structures in cellular protrusions such as microvilli, filopodia, and neuronal dendritic spines (5).
The Abl family kinases, including vertebrate Abl1/Abl and Abl2/Arg, are important regulators of cellular morphogenesis and migration in developing metazoans. Abl2/Arg in particular is essential for the proper formation and stability of actin-based structures, including fibroblast cell edge protrusions, cancer cell invadopodia, and dendritic spines (6–8). In response to growth factor or adhesion receptor stimulation, Arg functions in part through phosphorylation of a number of substrates, including key cytoskeletal regulators (6). One important Arg substrate is cortactin, an actin-binding protein that stimulates the Arp2/3 complex to nucleate actin filament branches (9–11). Cortactin also stabilizes actin filament branches (9) and inhibits actin filament severing by cofilin (12, 13).
Abl family kinases also have kinase-independent functions. This was first noted by Hoffmann and co-workers (14), who found that many of the defects in cell morphogenesis in abl mutant Drosophila embryos could be rescued by a kinase-inactive Abl transgene. Similarly, a kinase-inactive Arg point mutant can largely rescue the deficiencies in cell edge protrusion of Arg knock-out fibroblasts (15). These data suggest that Abl family kinases also regulate actin-dependent processes in a kinase-independent manner, but the mechanisms by which they do so are unclear.
Abl family kinases are unique in that they contain C-terminal extensions with protein-protein interaction motifs and cytoskeletal binding domains. The Arg C-terminal extension uniquely contains multiple cytoskeletal binding domains, including an internal actin-binding (I/L)WEQ domain, a microtubule-binding domain, and a C-terminal actin-binding calponin homology (CH) domain (Fig. 1A) (16, 17). These domains allow Arg to bundle actin filaments and cross-link actin filaments and microtubules in vitro (16, 17). Arg binds cooperatively to actin filaments at a saturation binding density of one Arg to two actin subunits and induces a 30° twist in the actin filament (18, 19). Cortactin also binds actin filaments directly through its “cortactin repeat” region (Fig. 1A) (10, 20). Remarkably, cortactin binds Arg-coated actin filaments at twice the density (one cortactin to two actins) of naked actin filaments (one cortactin to four actins) (19). The capacity of Arg to recruit cortactin to polymerizing actin may be an important regulatory mechanism within cells. Consistent with this model, knocking down Arg in cultured neurons significantly reduces cortactin in dendritic spines, resulting in their destabilization (7).
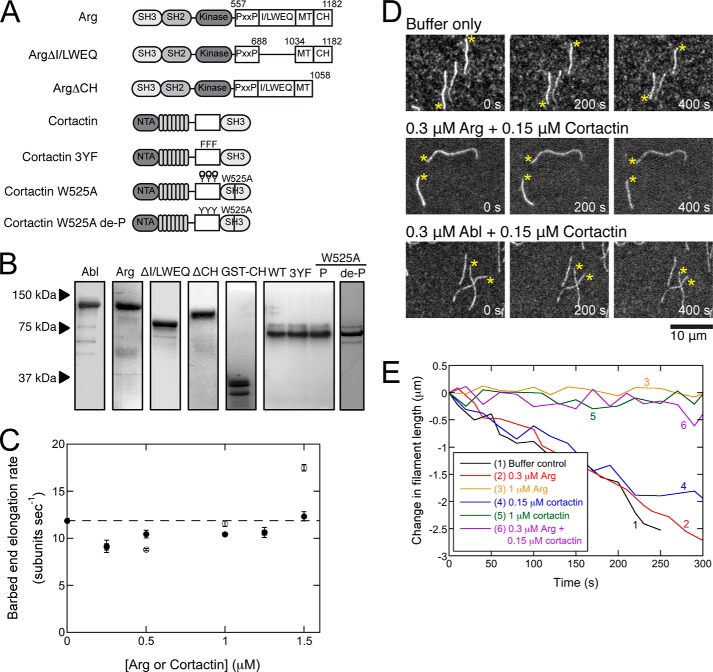
FIGURE 1.
Arg and cortactin cooperatively stabilize actin filaments. A, domain architecture of wild type and variant Arg and cortactin. B, Coomassie-stained SDS-polyacrylamide gel of 10 μg of purified wild type and variant Arg and cortactin constructs. C, dependence of barbed end elongation rates on the concentration of Arg or cortactin. Conditions are as follows: 1.5 μm Mg-ATP-actin monomers (20% labeled with Alexa 488), 35 mm KCl, 0.7 mm MgCl2, 0.7 mm EGTA, 7 mm imidazole (pH 7.0), 0.5% (4000 cP) methyl cellulose, 15 mm glucose, 200 μm ATP, 50 mm DTT, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 25 mm Tris-HCl (pH 8.0), 250 mm NaCl, 2.5% glycerol, 0.005% Triton X-100, 0.5 mm EDTA. Error bars are S.E. from the analysis of 10–20 filaments. D and E, actin depolymerization observed by TIRF microscopy. Actin was polymerized from 1.5 μm Mg-ATP-actin (20% labeled with Alexa 488) as in C, and then actin was washed out with the same buffer without actin but with other proteins. D, time series of fluorescence micrographs of pre-assembled actin filaments (20% Alexa 488-labeled) depolymerizing in the absence or presence of cortactin and Arg or Abl. The concentration of each protein is indicated. Asterisks denote barbed ends. E, time courses of the depolymerization of representative actin filaments in the absence (black) or presence of 0.3 μm Arg (red), 1 μm Arg (orange), 0.15 μm cortactin (blue), 1 μm cortactin (green), or both 0.3 μm Arg and 0.15 μm cortactin (purple).
Previous work demonstrated that Arg is a key regulator of subcellular structures composed of actin filaments, but it was not known whether Arg alone or in combination with cortactin controls actin filament polymerization, depolymerization, branching by the Arp2/3 complex, or severing by cofilin. We used total internal reflection fluorescence (TIRF) microscopy to investigate each of these reactions. We found that neither Arg nor cortactin alters actin elongation, but both slow dissociation of subunits from the barbed end of the filament. Low concentrations of Arg and cortactin cooperate to stabilize actin filaments. Arg potentiates actin filament branching by the Arp2/3 complex with cortactin and the verprolin-connecting acidic domain (VCA) of N-WASp. Arg also stimulates filament severing by cofilin in a concentration-dependent manner, but cortactin attenuates this stimulation. Together, our results illustrate that Arg directly regulates actin filament dynamics in several ways and that cortactin modulates these activities.
EXPERIMENTAL PROCEDURES
Molecular Cloning, Expression, and Purification of Recombinant Proteins
Murine Arg and cortactin cDNAs were cloned with an engineered His6 tag into the pFastbac1 vector (Invitrogen). Arg deletion constructs and fragments as well as cortactin 3YF and W525A mutants were generated using PCR-based mutagenesis (15, 17). Recombinant baculoviruses expressing these proteins were generated using the Bac-to-Bac baculovirus expression system (21). Cells were lysed in lysis buffer (50 mm Tris (pH 7.5), 500 mm NaCl, 0.01% Triton X-100, 5% glycerol, 20 mm imidazole, 1 mm DTT, 1 mm EDTA, and protease inhibitors (benzamidine, aprotinin, leupeptin, and chymostatin)). His6-tagged proteins were purified on a Ni2+ affinity column and eluted with 250 mm imidazole, followed by an SP-Sepharose ion exchange column with a 100 mm NaCl wash. Final elution buffer was 50 mm Tris (pH 8.0), 500 mm NaCl, 5% glycerol, 0.01% Triton X-100, 1 mm DTT, 1 mm EDTA. De-P W525A was dephosphorylated as described previously (22) by incubating with 1.0 unit of calf intestinal phosphatase (New England Biolabs, Inc.) for each microgram of protein for 4 h at room temperature and was re-purified as described above (19).
Actin was purified from an acetone powder of frozen chicken breast muscle (Trader Joe's) by one cycle of polymerization and depolymerization (23), followed by gel filtration in Buffer G (2 mm Tris-HCl (pH 8.0), 0.5 mm ATP, 0.5 mm DTT, 0.1 mm CaCl2, and 1 mm NaN3) and storage in Buffer G at 4 °C.
Actin monomers were polymerized by dialyzing in 50 mm PIPES/Tris (pH 6.8), 50 mm KCl, 0.2 mm ATP, 0.2 mm CaCl2 and labeled on lysines by incubating overnight at 4 °C with a 1:13 molar ratio of actin to Alexa Fluor 488 succinimidyl ester (A-20000, Invitrogen). Labeled actin was then pelleted by ultracentrifugation at 120,000 × g, depolymerized, clarified, and gel-filtered on Sephacryl S-300 resin in Buffer G. Purified Alexa 488 monomers were typically labeled 30–50%. Prior to each experiment, Ca2+-ATP actin monomers (20% Alexa 488-labeled) were converted to Mg2+-ATP actin by addition of 0.05 mm MgCl2 and 0.2 mm EGTA followed by a 5-min incubation. Bovine Arp2/3 complex and GST-VCA were purified from thymus as described previously by Higgs et al. (24).
Human cofilin 1 was expressed in pLysS cells with 1 mm isopropyl β-d-1-thioglactopyranoside over a 4-h incubation period at 37 °C. Cell pellets were resuspended in 10 mm Tris-HCl (pH 7.5), 1 mm EGTA, and 2 mm DTT and lysed by sonication. Cell lysates were clarified by centrifugation at 21,200 × g for 45 min. The lysate supernatant was applied to a DEAE-Sepharose column, and the flow-through was dialyzed in 150 mm NaCl, 10 mm Tris-HCl (pH 7.5), 2 mm DTT. The protein was then concentrated to 25 ml using a centrifuge filter device (Corning Glass, 10-kDa molecular mass cutoff), gel-filtered on a 400-ml column of Sephacryl S-200 resin, and dialyzed in KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole (pH 7.0)), flash-frozen, and stored at −80 °C.
Total Internal Reflection Fluorescence Microscopy Experiments
Open-ended glass flow chambers were prepared (25–27). Prior to each reaction, the chamber was incubated for 1 min with two 8-μl washes of each of the following solutions: 1) 0.5% Tween 80 in high salt TBS (HS-TBS: 50 mm Tris-HCl (pH 7.5), 600 mm KCl); 2) 250 nm N-ethylmaleimide-inactivated chicken skeletal muscle myosin in HS-TBS; and 3) 10% BSA in HS-TBS. The chamber was washed twice with HS-TBS between each incubation step and with KMEI buffer immediately before introducing actin.
Equal parts of 2× microscopy buffer (70 mm KCl, 1.4 mm MgCl2, 1.4 mm EGTA, 14 mm imidazole (pH 7.0), 1% (4000 cP) methyl cellulose, 30 mm glucose, 400 μm ATP, 100 mm DTT, 40 μg/ml catalase, 200 μg/ml glucose oxidase) and elution buffer (see above) were mixed and added to the actin monomers in Buffer G to induce polymerization. The resulting buffer contained 250 mm NaCl and 35 mm KCl. To compensate for the effect of salt on the affinity of cofilin for actin filaments (28), we used 30 μm cofilin in the severing experiments.
For depolymerization, branching, and severing experiments, actin was polymerized in the chamber until filaments were visible. Then the actin monomer solution was replaced with a fresh sample of proteins (i.e. Arg and/or cortactin, plus 10 nm bovine Arp2/3 complex, 100 nm GST-VCA, and 1 μm ATP-actin monomers for branching experiments, or 30 μm human cofilin 1 for severing experiments) in microscopy buffer, and imaging was continued.
Fluorescence micrographs were collected every 5–10 s using prism-style total internal reflection microscopy on an Olympus IX-70 inverted microscope and a Hamamatsu C4747-95 CCD (Orca-ER) camera controlled by MetaMorph software (Molecular Devices, Union City, CA). Changes in filament length were measured using ImageJ software (National Institutes of Health) for at least 10 filaments in each condition over 10–20 frames, which covered a span of at least 200 s. We assumed 370 actin subunits per μm of filament (29).
Kinase Assays
Kinase reactions were performed using 50 nm Arg and 1 μm substrate (cofilin and GST-CrkII as a control) in 25 mm Hepes (pH 7.25), 100 mm NaCl, 1 mm DTT, 1 mm sodium orthovanadate, 5% glycerol, 5 mm MgCl2, 5 mm MnCl2, 5 μm ATP, 0.75 μCi of [γ-32P]ATP. Reactions were terminated at 5, 30, and 60 min, separated on SDS-polyacrylamide gels, and exposed to phosphorimaging screens.
Co-sedimentation Assays
Two micromolar actin filaments were incubated with either 50 μm cofilin or 1 μm cortactin or both and centrifuged at 100,000 × g for 1 h. Pellet samples were analyzed by SDS-PAGE and stained with Coomassie Blue. The intensities of the cofilin and actin bands were quantified with ImageJ, and the relative filament saturation by cofilin was obtained by dividing the intensity of the cofilin band by the intensity of the actin band in each lane.
RESULTS
Arg and Cortactin Inhibit Actin Depolymerization
We used total internal reflection fluorescence microscopy to extend our studies of the effects of Arg and cortactin on actin polymerization (19) by measuring rates of barbed end elongation and shortening. Arg binds actin filaments cooperatively with a KD, (app) ≈0.3 μm (17), whereas cortactin binds with a KD = 0.4 μm (31). Neither protein significantly affected elongation of Mg-ATP-actin (20% labeled with Alexa 488) at concentrations up to 1.5 μm (Fig. 1C).
We confirmed our previous observation that 1 μm Arg slows depolymerization of actin filaments ∼6-fold (Fig. 1E). At 1 μm, Arg saturates ∼91% of the binding sites on the actin filaments. Experiments with deletion mutants (Fig. 1, A and B) showed that inhibition of depolymerization by 1 μm Arg depends much more on the internal Arg (I/L)WEQ actin-binding site than the C-terminal CH domain (Table 1). Thus the ability of the CH domain to change the twist in the actin filament (18) is not required to slow depolymerization. The Arg paralog Abl and cortactin also slowed depolymerization (Table 1).
TABLE 1.
Barbed end depolymerization rates mediated by Arg and Abl
| Arg concentrations | Arg | ArgΔ(I/L)WEQ | ArgΔCH | Abl |
|---|---|---|---|---|
| μm | ||||
| 0 (control) | −3.3 ± 0.16a | |||
| 0.3 | −3.2 ± 0.17 | −2.7 ± 0.11 | −3.8 ± 0.26 | −2.7 ± 0.17 |
| 1 | −0.45 ± 0.09 | −2.3 ± 0.13 | −0.37 ± 0.08 | −0.43 ± 0.07 |
| 0.3 + 0.15 μm cortactin | −0.6 ± 0.13 | −0.84 ± 0.13 | −0.62 ± 0.13 | −2.6 ± 0.19 |
a Polymerization rates are in subunits/s. Errors are standard error from the analysis of 10–20 filaments.
Neither Arg nor cortactin inhibited depolymerization effectively at concentrations below their KD values, so high densities of either protein on filaments are required to slow depolymerization (Fig. 1E and Tables 1 and 2). The binding densities are only about 50% saturation with 0.3 μm Arg and 27% saturation with 0.15 μm cortactin, where neither alone inhibited depolymerization (Fig. 1E and Table 1).
TABLE 2.
Barbed end depolymerization rates mediated by cortactin
| Cortactin concentrations | Cortactin | Cortactin 3YF | Cortactin W525A | Cortactin de-P W525A |
|---|---|---|---|---|
| μm | ||||
| 0 (control) | −3.3 ± 0.16a | |||
| 0.15 | −2.6 ± 0.15 | −2.9 ± 0.20 | −3.0 ± 0.23 | −1.94 ± 0.16 |
| 1 | −0.41 ± 0.07 | −0.67 ± 0.04 | −0.47 ± 0.09 | −0.31 ± 0.08 |
| 0.15 + 0.3 μm Arg | −0.6 ± 0.13 | −0.6 ± 0.11 | −0.6 ± 0.30 | −0.49 ± 0.08 |
a Polymerization rates are in subunits/s. Errors are standard error from the analysis of 10–20 filaments.
Although neither Arg nor cortactin inhibited depolymerization at low concentrations, a combination of 0.3 μm Arg plus 0.15 μm cortactin slowed depolymerization to the same extent as 1 μm of either protein (Fig. 1, D and E, and Table 1). However, combining 0.15 μm cortactin with 0.3 μm Abl did not slow depolymerization (Fig. 1D and Table 1).
Experiments with Arg deletion mutants demonstrated that recruitment of cortactin by the Arg CH domain is not necessary, but it can be sufficient for synergism between Arg and cortactin under some conditions. First, even though ArgΔCH does not recruit cortactin to actin filaments (19), 0.3 μm ArgΔCH allowed 0.15 μm cortactin to slow barbed end depolymerization (Table 1). Second, 1 μm ArgΔ(I/L)WEQ promotes cortactin binding to filaments (19), but it did not slow polymerization by itself. Yet 0.3 μm ArgΔ(I/L)WEQ synergized with 0.15 μm cortactin to slow depolymerization (Table 1). These observations suggest that loading filaments with cortactin by the Arg CH domain can compensate for the loss of the (I/L)WEQ domain in protecting filaments from depolymerization.
We used several mutants of cortactin (Fig. 1A) to test whether direct interaction with Arg is required to stabilize actin filaments. First, a point mutation in the cortactin Src homology 3 domain (W525A) disrupted binding to the Arg PYLPRLP motif, which is required for Arg to phosphorylate several tyrosines in cortactin (15, 19). Second, replacement of the phosphorylatable tyrosine residues Tyr-421, Tyr-466, and Tyr-482 with phenylalanine in the 3YF mutant prevented phosphorylation and subsequent Arg Src homology 2 domain binding (33). Third, we treated W525A with calf intestinal phosphatase, which we have previously shown to remove all tyrosine phosphorylation, to produce dephosphorylated (de-P)-W525A (19). All three cortactin variants have wild type affinity for actin filaments (33). Published experiments with these mutated proteins showed that these reciprocal interactions between Arg and cortactin contribute to cell edge protrusion in motile cells and stabilize the dendritic spines of neurons (7, 15, 34).
Like wild type cortactin, 1 μm cortactin mutants slowed depolymerization of actin filaments, and 0.15 μm of the variants in combination with 0.3 μm Arg also slowed depolymerization (Table 2). Thus, neither of the defined Arg-cortactin binding interfaces is required for cooperative filament stabilization.
Effects of Arg on Branch Formation by Arp2/3 Complex
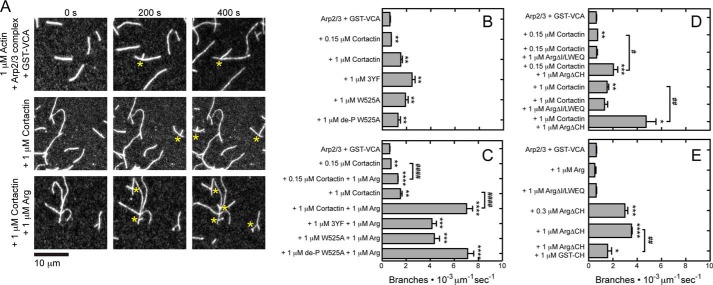
Cortactin weakly activates branch formation by the Arp2/3 complex on its own, but it robustly stimulates branch formation when combined with the nucleation-promoting factor N-WASp (9, 35–37). We confirmed that 1 μm cortactin increased branching by Arp2/3 complex and N-WASp GST-VCA ∼3-fold (Fig. 2, A and B) (38). Arg alone did not affect the rate of branch formation at any concentration tested, but combining 1 μm Arg with 0.15 or 1 μm cortactin increased the rate of branch formation above that observed for either concentration of cortactin alone (Fig. 2, A, C, and E). This stimulation depended on the presence of the internal (I/L)WEQ actin-binding domain of Arg (Fig. 2D). Cortactin mutants bearing disruptions of either or both interactions with Arg showed synergistic activation of branching with 1 μm Arg. The magnitude of this effect was reduced for the 3YF and W525A mutants but not for de-P-W525A (Fig. 2, B and C).
FIGURE 2.
Arg (I/L)WEQ motif stimulates actin filament branching by the Arp2/3 complex and cortactin. Conditions are as follows: 35 mm KCl, 0.7 mm MgCl2, 0.7 mm EGTA, 7 mm imidazole (pH 7.0), 0.5% (4000 cP) methyl cellulose, 15 mm glucose, 200 μm ATP, 50 mm DTT, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 25 mm Tris-HCl (pH 8.0), 250 mm NaCl, 2.5% glycerol, 0.005% Triton X-100, 0.5 mm EDTA. Actin filaments (20% Alexa 488-labeled) were pre-assembled in the flow chamber before introducing 0.5 μm actin monomers, 10 nm Arp2/3 complex, 100 nm GST-VCA, and wild type or variant Arg and/or cortactin. A, time series of fluorescence micrographs of actin filament branching mediated by the Arp2/3 complex in the presence of GST-VCA and Arg deletion variants. Asterisks denote branches. B–E, rate of actin filament branch formation mediated by the Arp2/3 complex in the presence of GST-VCA and wild type or variant Arg in the absence or presence of cortactin or cortactin mutants. The units of branch formation are branches per micrometer of actin filament/s. Error bars are the S.E. for a minimum of three regions of each acquired time period. Asterisks represent a statistically significant difference in comparison with the control of 0.5 μm actin monomers, 10 nm Arp2/3, 100 nm GST-VCA. * or #, p < 0.05; ** or ##, p < 0.01; ***, p < 0.001; **** or ####, p < 0.0001.
Surprisingly, Arg lacking the CH domain promoted branching by the Arp2/3 complex even in the absence of cortactin, provided that GST-VCA was present. Thus, the CH domain seems to autoinhibit the ability of full-length Arg to enhance the nucleation promoting activity of GST-VCA. A sub-saturating concentration of ArgΔCH also strongly stimulated branching, indicating that this mechanism does not require saturated actin binding by Arg (Fig. 2E). The presence of 1 μm GST-tagged Arg CH domain partially inhibits the ability of ArgΔCH to stimulate actin branch formation (Fig. 2E).
Arg Promotes Severing by Cofilin, but Cortactin Inhibits This Activity
We used TIRF microscopy to measure filament severing by cofilin (Fig. 3) (39). For these assays, we had to balance the insolubility of Arg at low salt concentrations (21) with the salt sensitivity of cofilin binding to actin filaments (40). As a compromise, we used a high cofilin concentration, 30 μm, to obtain binding and severing in the higher salt buffer.
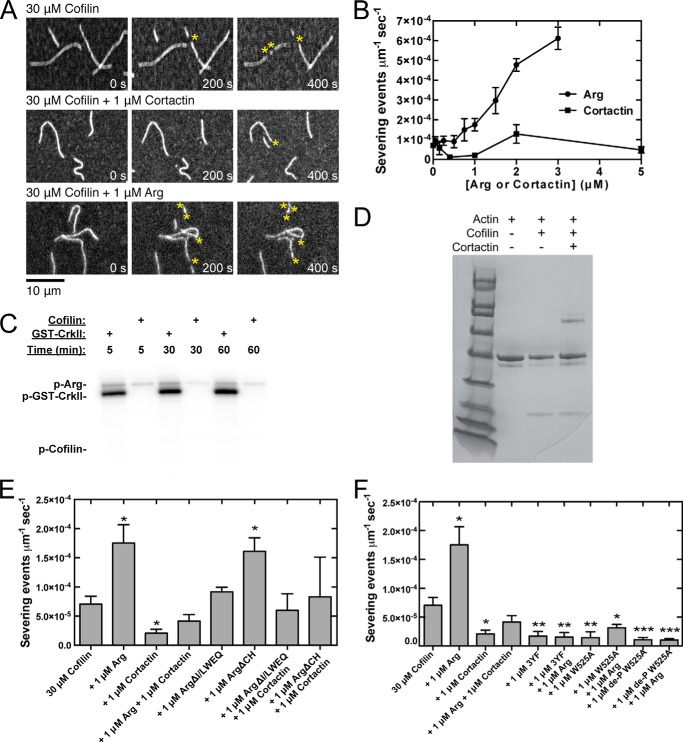
FIGURE 3.
Arg promotes severing of actin filaments by cofilin, but this activity is inhibited by cortactin. Conditions are as follows: 35 mm KCl, 0.7 mm MgCl2, 0.7 mm EGTA, 7 mm imidazole (pH 7.0), 0.5% (4000 cP) methyl cellulose, 15 mm glucose, 200 μm ATP, 50 mm DTT, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 25 mm Tris-HCl (pH 8.0), 250 mm NaCl, 2.5% glycerol, 0.005% Triton X-100, 0.5 mm EDTA. Actin filaments (20% Alexa 488-labeled) were pre-assembled in the flow chamber before introducing 30 μm human cofilin and Arg or cortactin. A, time series of fluorescence micrographs of actin filament severing mediated by 30 μm cofilin, Arg, and cortactin. Concentrations of Arg and cortactin are indicated. B, dependence of cofilin severing activity on the concentration of Arg and cortactin. The units of severing activity are severing events per μm of actin filament per second. C, phosphorylation of 1 μm Crk-II and 1 μm cofilin by 50 nm Arg in 25 mm Hepes (pH 7.25), 100 mm NaCl, 1 mm DTT, 1 mm sodium orthovanadate, 5% glycerol, 5 mm MgCl2, 5 mm MnCl2, 5 μm ATP, 0.75 μCi of [γ-32P]ATP over 5, 30, and 60 min. D, pelleting assay. Actin filaments (2 μm) were incubated with either 50 μm cofilin or 1 μm cortactin or both and centrifuged at 100,000 × g for 1 h. Pellet samples were analyzed by SDS-PAGE and stained with Coomassie Blue. The intensities of the cofilin and actin bands were quantified with ImageJ. E and F, cofilin severing activity measured in the presence of wild type and mutant Arg constructs and wild type and mutant cortactin. Error bars are the standard error of the mean for a minimum of three regions of each acquired time period. Asterisks represent a statistically significant difference in comparison with the control of 30 μm human cofilin. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The rate of severing by cofilin increased with the concentration of Arg up to the highest concentration tested (Fig. 3, A and B). Arg does not phosphorylate cofilin (Fig. 3C), indicating that activation of severing does not involve phosphorylation. ArgΔ(I/L)WEQ did not stimulate severing, although ArgΔCH stimulated cofilin severing similar to wild type Arg (Fig. 3E). Thus, the previously observed ability of the Arg CH domain to alter actin filament twist (18) and recruit cortactin (19) is not part of the severing mechanism.
Conversely, cortactin had a biphasic effect on actin filament severing by cofilin: 1 μm cortactin inhibited severing (13), but higher concentrations had no effect (Fig. 3, A and B). Cofilin binds actin filaments in the presence of 1 μm cortactin (Fig. 3D), suggesting that cortactin inhibits severing by affecting the filament structure or flexibility. However, cortactin reduces the extent of filament saturation by cofilin by ∼20%, suggesting that higher concentrations of cortactin may compete with cofilin for binding and therefore promote severing. Arg, ArgΔCH, and ArgΔ(I/L)WEQ each overcame the inhibition of severing by 1 μm cortactin and restored the severing rate to that observed for cofilin alone (Fig. 3E). In contrast, mutants of cortactin disrupting one or both Arg interaction interfaces inhibited cofilin severing both in the presence and absence of Arg (Fig. 3F), suggesting that Arg-mediated phosphorylation, and possibly binding, of cortactin is required to overcome the inhibition of severing by cortactin.
DISCUSSION
The Abl2/Arg nonreceptor tyrosine kinase is an important regulator of actin-driven cellular processes in a variety of cellular contexts (15). Abl family kinases use both kinase-dependent and kinase-independent mechanisms to regulate the formation of actin-based cellular structures. Previous work described how Arg phosphorylates discrete cytoskeletal regulatory proteins to control changes in cell structure and function (8, 11, 15–17, 41–45).
Here, we report that Arg and its binding partner cortactin interact directly with actin filaments to affect their stability, branch formation, and severing. Arg alone has no effect on actin filament elongation but can stabilize actin filaments. Low concentrations of Arg can cooperate with cortactin to stabilize actin filaments, and this ability does not require the defined Arg-cortactin binding interfaces. Arg increases actin filament branching by cortactin and Arp2/3 complex. An Arg mutant lacking its CH domain potently stimulates branching by Arp2/3 complex and GST-VCA. Arg and cortactin have opposite effects on actin filament severing by cofilin; Arg promotes and cortactin inhibits severing. Together, they can cancel each other's effects on severing by interacting directly. In the absence of Arg-cortactin interactions, the severing rate is similar to that observed in the presence of wild type cortactin alone. Arg does not phosphorylate actin, Arp2/3 complex (45), or cofilin (Fig. 3C). Together, these findings strongly suggest that the effects of Arg on actin filament stability and branching do not require Arg kinase activity.
The concentrations of Arg and cortactin in cells are high enough to make the reactions described here physiologically relevant. For example, Arg is present at 0.4–0.6 μm in dendritic spines, and cortactin concentrations may be as high as 4 μm within breast cancer cell invadopodia (12, 17, 41). Dendritic spines in Arg-deficient neurons exhibit a loss of cortactin, reduced actin content, morphological changes, and loss of stability both in culture and in vivo (7, 32). The cooperative binding of Arg and cortactin to actin filaments that we have described may help to concentrate Arg and cortactin binding to individual subsets of actin filaments (17–19) at levels sufficient to influence filament stability, branching, and severing in key subcellular compartments. Interestingly, the Arg C-terminal domain alone (residues 557–1182), which lacks the kinase domain, nearly completely rescues the defects in cell edge protrusion of Arg knock-out fibroblasts (16). Our results are consistent with this observation and support our model that Arg and cortactin can regulate actin by a mechanism independent of phosphorylation.
Individually, Arg or cortactin bound to the filament slows the dissociation of actin subunits from barbed ends. This is likely to be a direct effect on the dissociating subunit rather than a change in filament conformation induced by the CH domain of Arg, because Arg without the CH domain has the same activity. Rather, the internal (I/L)WEQ domain mediates Arg's stabilization activity. Neither Arg nor cortactin influence association of actin monomers, another indication that they do not alter the conformation of the filament end. During polymerization, actin monomer binding is likely to stay ahead of the binding of Arg and cortactin near the barbed end, whereas direct interactions between Arg/cortactin bound near the barbed end are more likely during depolymerization. This mechanism would explain the selective effects of Arg and cortactin on actin filament depolymerization.
Although Abl stabilizes actin filaments on its own, it does not cooperate with cortactin like Arg. Arg differs from Abl in having an internal (I/L)WEQ actin-binding domain, but this domain does not account for their differences. ArgΔ(I/L)WEQ, which lacks this domain, does not itself stabilize filaments, yet it cooperates with cortactin to stabilize actin filaments. Without this domain, Abl alone can stabilize actin filaments, even though it cannot cooperate with cortactin.
Cortactin stimulates GST-VCA-mediated activation of the Arp2/3 complex by binding branch junctions and facilitating GST-VCA release (38). We observed that Arg further increases Arp2/3 complex-mediated actin branching induced by cortactin and GST-VCA. Cortactin has a higher affinity for branch junctions than the sides of naked filaments (38). We propose that Arg enhances these interactions of cortactin with the Arp2/3 complex at branch junctions either directly or indirectly, explaining how Arg cooperates with cortactin at both low and high saturation densities of Arg.
Arg does not increase Arp2/3 complex-mediated actin branching by GST-VCA alone unless the CH domain is deleted. The CH domain is likely to autoinhibit the nucleation promoting activity of Arg, because the GST-CH domain inhibits actin filament branching by ArgΔCH. Because this activity depends on the (I/L)WEQ domain, the CH domain may mask the ability of the (I/L)WEQ domain to stimulate branching or interfere with Arp2/3 complex binding to actin filaments. Similar to cofilin, the CH domain induces a twist in the actin filament, which may alter or impair the binding of the (I/L)WEQ domain or Arp2/3 complex (18).
Cofilin severs actin filaments optimally when a low density of cofilin is bound along the filament, whereas filaments saturated with cofilin are stable (28). The (I/L)WEQ domain is required for Arg to promote severing by cofilin (Fig. 3E). The simplest explanation is that Arg competes with cofilin for binding filaments, keeping the density of bound cofilin low. However, the mechanism is likely to be more complicated, given that we did not observe lower rates of severing at Arg concentrations (2–3 μm) that saturate >97% of binding sites on the filaments. Low concentrations of cortactin inhibit actin filament severing by cofilin (Fig. 3B), likely by changing the filament conformation (30). Higher concentrations of cortactin have less effect on severing, perhaps by competing with cofilin for binding to the filament (Fig. 3D), so these competing effects balance out. Arg with or without the (I/L)WEQ and CH domains can overcome the inhibition of severing by cortactin (Fig. 3E). Cortactin binding to cofilin inhibits cofilin severing of actin filaments (12, 13). We observe a similar effect, but with significantly higher concentrations of cofilin than cortactin suggesting an alternative mechanism, possibly mediated through structural changes to the actin filament. Arg and cortactin do not compete for binding to filaments and Arg actually increases this interaction between cortactin and actin (19). It is possible that Arg-induced alterations in the mode of cortactin binding to filaments affect cortactin's ability to inhibit severing.
Arg and cortactin act together to mediate the formation, function, and stability of breast cancer cell invadopodia, fibroblast membrane ruffles, and cell edge protrusions and neuronal dendritic spines (8, 16, 32, 42), processes that depend on actin filament networks. We present multiple mechanisms by which Arg stabilizes filaments in a cooperative manner with its substrate, cortactin, and promotes actin nucleation through increased branching and severing. In combination, we expect these mechanisms to promote actin filament polymerization and stability. These activities may underlie the dependence of these diverse actin-based structures on Arg and cortactin.
Acknowledgment
We thank Christopher Jurgenson for providing purified bovine Arp2/3 complex and GST-VCA.
This work was supported, in whole or in part, by National Institutes of Health Grants CA-133346 and GM-100411 (to A. J. K.), GM-026338 (to T. D. P.), and NS-089439 (to A. J. K. and T. D. P.).
- Arp
- actin-related protein
- Abl
- Abelson
- Arg
- Abl-related gene
- CH
- calponin homology
- VCA
- verprolin-cofilin-acidic
- cP
- centipoise
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 2. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bamburg J. R., Harris H. E., Weeds A. G. (1980) Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 121, 178–182 [DOI] [PubMed] [Google Scholar]
- 4. Chan C., Beltzner C. C., Pollard T. D. (2009) Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Revenu C., Athman R., Robine S., Louvard D. (2004) The co-workers of actin filaments: from cell structures to signals. Nat. Rev. Mol. Cell Biol. 5, 635–646 [DOI] [PubMed] [Google Scholar]
- 6. Bradley W. D., Koleske A. J. (2009) Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J. Cell Sci. 122, 3441–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin Y. C., Yeckel M. F., Koleske A. J. (2013) Abl2/Arg controls dendritic spine and dendrite arbor stability via distinct cytoskeletal control pathways. J. Neurosci. 33, 1846–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mader C. C., Oser M., Magalhaes M. A., Bravo-Cordero J. J., Condeelis J., Koleske A. J., Gil-Henn H. (2011) An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71, 1730–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weaver A. M., Karginov A. V., Kinley A. W., Weed S. A., Li Y., Parsons J. T., Cooper J. A. (2001) Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374 [DOI] [PubMed] [Google Scholar]
- 10. Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000) Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyle S. N., Michaud G. A., Schweitzer B., Predki P. F., Koleske A. J. (2007) A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr. Biol. 17, 445–451 [DOI] [PubMed] [Google Scholar]
- 12. Magalhaes M. A., Larson D. R., Mader C. C., Bravo-Cordero J. J., Gil-Henn H., Oser M., Chen X., Koleske A. J., Condeelis J. (2011) Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195, 903–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oser M., Yamaguchi H., Mader C. C., Bravo-Cordero J. J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A. J., Condeelis J. (2009) Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186, 571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henkemeyer M., West S. R., Gertler F. B., Hoffmann F. M. (1990) A novel tyrosine kinase-independent function of Drosophila abl correlates with proper subcellular localization. Cell 63, 949–960 [DOI] [PubMed] [Google Scholar]
- 15. Lapetina S., Mader C. C., Machida K., Mayer B. J., Koleske A. J. (2009) Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J. Cell Biol. 185, 503–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller A. L., Wang Y., Mooseker M. S., Koleske A. J. (2004) The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 165, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Miller A. L., Mooseker M. S., Koleske A. J. (2001) The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc. Natl. Acad. Sci. U.S.A. 98, 14865–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galkin V. E., Orlova A., Koleske A. J., Egelman E. H. (2005) The Arg non-receptor tyrosine kinase modifies F-actin structure. J. Mol. Biol. 346, 565–575 [DOI] [PubMed] [Google Scholar]
- 19. MacGrath S. M., Koleske A. J. (2012) Arg/Abl2 modulates the affinity and stoichiometry of binding of cortactin to F-actin. Biochemistry 51, 6644–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu H., Parsons J. T. (1993) Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J. Cell Biol. 120, 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanis K. Q., Veach D., Duewel H. S., Bornmann W. G., Koleske A. J. (2003) Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol. Cell. Biol. 23, 3884–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu W., MacGrath S. M., Koleske A. J., Boggon T. J. (2012) Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. MacLean-Fletcher S., Pollard T. D. (1980) Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96, 18–27 [DOI] [PubMed] [Google Scholar]
- 24. Higgs H. N., Blanchoin L., Pollard T. D. (1999) Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38, 15212–15222 [DOI] [PubMed] [Google Scholar]
- 25. Kuhn J. R., Pollard T. D. (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Courtemanche N., Pollard T. D. (2012) Determinants of Formin homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem. 287, 7812–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paul A. S., Paul A., Pollard T. D., Pollard T. (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De La Cruz E. M. (2009) How cofilin severs an actin filament. Biophys. Rev. 1, 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huxley H. E., Brown W. (1967) Low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 30, 383–434 [DOI] [PubMed] [Google Scholar]
- 30. Pant K., Chereau D., Hatch V., Dominguez R., Lehman W. (2006) Cortactin binding to F-actin revealed by electron microscopy and 3D reconstruction. J. Mol. Biol. 359, 840–847 [DOI] [PubMed] [Google Scholar]
- 31. van Rossum A. G., de Graaf J. H., Schuuring-Scholtes E., Kluin P. M., Fan Y. X., Zhan X., Moolenaar W. H., Schuuring E. (2003) Alternative splicing of the actin binding domain of human cortactin affects cell migration. J. Biol. Chem. 278, 45672–45679 [DOI] [PubMed] [Google Scholar]
- 32. Sfakianos M. K., Eisman A., Gourley S. L., Bradley W. D., Scheetz A. J., Settleman J., Taylor J. R., Greer C. A., Williamson A., Koleske A. J. (2007) Inhibition of rho via arg and p190RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J. Neurosci. 27, 10982–10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gifford S. M., Liu W., Mader C. C., Halo T. L., Machida K., Boggon T. J., Koleske A. J. (2014) Two amino acid residues confer different binding affinities of Abelson family kinase Src homology 2 domains for phosphorylated cortactin. J. Biol. Chem. 289, 19704–19713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oser M., Mader C. C., Gil-Henn H., Magalhaes M., Bravo-Cordero J. J., Koleske A. J., Condeelis J. (2010) Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell Sci. 123, 3662–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weaver A. M., Heuser J. E., Karginov A. V., Lee W. L., Parsons J. T., Cooper J. A. (2002) Interaction of cortactin and N-WASp with Arp2/3 complex. Curr. Biol. 12, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 36. Kempiak S. J., Yamaguchi H., Sarmiento C., Sidani M., Ghosh M., Eddy R. J., Desmarais V., Way M., Condeelis J., Segall J. E. (2005) A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin. J. Biol. Chem. 280, 5836–5842 [DOI] [PubMed] [Google Scholar]
- 37. Kowalski J. R., Egile C., Gil S., Snapper S. B., Li R., Thomas S. M. (2005) Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 118, 79–87 [DOI] [PubMed] [Google Scholar]
- 38. Helgeson L. A., Nolen B. J. (2013) Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. Elife 2, e00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ichetovkin I., Grant W., Condeelis J. (2002) Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12, 79–84 [DOI] [PubMed] [Google Scholar]
- 40. Cao W., Goodarzi J. P., De La Cruz E. M. (2006) Energetics and kinetics of cooperative cofilin-actin filament interactions. J. Mol. Biol. 361, 257–267 [DOI] [PubMed] [Google Scholar]
- 41. Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. (1998) Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21, 1259–1272 [DOI] [PubMed] [Google Scholar]
- 42. Moresco E. M., Donaldson S., Williamson A., Koleske A. J. (2005) Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J. Neurosci. 25, 6105–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bradley W. D., Hernández S. E., Settleman J., Koleske A. J. (2006) Integrin signaling through arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol. Biol. Cell 17, 4827–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peacock J. G., Couch B. A., Koleske A. J. (2010) The Abl and Arg non-receptor tyrosine kinases regulate different zones of stress fiber, focal adhesion, and contractile network localization in spreading fibroblasts. Cytoskeleton 67, 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller M. M., Lapetina S., MacGrath S. M., Sfakianos M. K., Pollard T. D., Koleske A. J. (2010) Regulation of actin polymerization and adhesion-dependent cell edge protrusion by the Abl-related gene (Arg) tyrosine kinase and N-WASp. Biochemistry 49, 2227–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]