Background: Cyclin E-CDK2 is a key protein kinase in the cell cycle.
Results: Phosphorylation of PHF8 by cyclin E-CDK2 enhances its demethylase activity toward H3K9me2 and promotes rDNA transcription as well as S phase progression.
Conclusion: Cyclin E-CDK2-dependent phosphorylation of PHF8 affects its function.
Significance: We unveiled the mechanism of how cyclin E-CDK2 regulates the function of PHF8 in cell cycle regulation.
Keywords: Cell Cycle; Cell Signaling; Cyclin; Cyclin-dependent Kinase (CDK); Phosphorylation; Cyclin E-CDK2, PHF8, Phosphorylation, rRNA Transcription
Abstract
Cyclin E-CDK2 is a key regulator in G1/S transition. Previously, we identified a number of CDK2-interacting proteins, including PHF8 (plant homeodomain finger protein 8). In this report, we confirmed that PHF8 is a novel cyclin E-CDK2 substrate. By taking the approach of mass spectrometry, we identified that PHF8 Ser-844 is phosphorylated by cyclin E-CDK2. Immunoblotting analysis indicated that WT PHF8 demethylates histone H3K9me2 more efficiently than the cyclin E-CDK2 phosphorylation-deficient PHF8-S844A mutant. Furthermore, flow cytometry analysis showed that WT PHF8 promotes S phase progression more robustly than PHF8-S844A. Real-time PCR results demonstrated that PHF8 increases transcription of cyclin E, E2F3, and E2F7 to significantly higher levels compared with PHF8-S844A. Further analysis by ChIP assay indicated that PHF8 binds to the cyclin E promoter stronger than PHF8-S844A and reduces the H3K9me2 level at the cyclin E promoter more efficiently than PHF8-S844A. In addition, we found that cyclin E-CDK2-mediated phosphorylation of PHF8 Ser-844 promotes PHF8-dependent rRNA transcription in luciferase reporter assays and real-time PCR. Taken together, these results indicate that cyclin E-CDK2 phosphorylates PHF8 to stimulate its demethylase activity to promote rRNA transcription and cell cycle progression.
Introduction
Cyclins and cyclin-dependent kinases play important roles during cell cycle progression (1). Different cyclin-CDK complexes exert their function at different cell cycle phases. Among them, cyclin E-CDK2 plays a critical role at the G1/S transition (2, 3) via phosphorylating a number of downstream signaling proteins (4–7). Multiple cyclin E-CDK2 substrates are cell division regulators. Cyclin E-CDK2 assists cyclin D-CDK4 in phosphorylating retinoblastoma protein, which leads to the release of E2F transcriptional factors from retinoblastoma protein-dependent inhibition to promote the expression of genes (such as cyclin E, myc, and DNA polymerase) that drive cells to enter S phase (8, 9). Cyclin E-CDK2 also phosphorylates the CDK inhibitor p27 to promote its degradation (10). Smad3 is a transcriptional factor that regulates the TGF-β signaling pathway. The phosphorylation of Smad3 by cyclin E-CDK2 inhibits its transcriptional activity (11). Cyclin E-CDK2 phosphorylates CREB-binding protein-p3002 to stimulate its histone acetyltransferase activity during cell cycle progression (12). NPAT is phosphorylated by the cyclin E-CDK2 complex to increase histone transcription required for S phase entry and progression (13). In addition to its function in promoting G1/S transition, cyclin E-CDK2 is also involved in DNA damage checkpoint control by phosphorylating the DNA helicase complex subunits MCM3 and MCM7 (14, 15).
As cyclins-CDKs are activated orderly during cell cycle progression, their phosphorylation substrates orchestrate the cellular changes that ultimately lead to cell division. However, many of these substrates remain unknown. To understand the molecular mechanisms of cyclin E-CDK2 and its associated proteins in cell cycle regulation, we employed the tandem affinity purification technique to identify 14 novel proteins interacting with CDK2, including the histone demethylase PHF8 (16). PHF8 belongs to a class of plant homeodomain-containing zinc finger proteins that play an important role in histone demethylation, gene transcription, and chromatin remodeling. PHF8 can demethylate H3K9me2/1, H3K27me2, or H4K20me1, relieve the transcriptional repression, and promote the transcription of related genes (17–20). It has been found that PHF8 can also bind to the promoter region of rDNA and regulate rDNA transcription (21, 22). As a histone demethylase, PHF8 regulates the retinoic acid response in acute promyelocytic leukemia (23). The germ line F279S mutation in PHF8 that disrupts the demethylase activity causes hereditary X-linked mental retardation (24, 25).
PHF8 is involved in the regulation of the cell cycle. PHF8 can interact with E2F1, HCF-1, and SET1A and promote cell cycle G1/S transition (26), whereas loss of PHF8 leads to prolonged G2 phase and defective mitosis (27). However, the mechanism of PHF8 regulation is not clear, and it remains to be elucidated whether phosphorylation influences PHF8 function. In this report, we demonstrate that cyclin E-CDK2 phosphorylates Ser-844 of the PHF8 demethylase and regulates its cellular distribution. We found that mutation of the cyclin E-CDK target site (Ser-844) negatively affects the ability of PHF8 to promote the S phase progression. Further analysis revealed that overexpression of PHF8 up-regulates cyclin E by affecting PHF8 and H3K9me2 levels at the cyclin E promoter. Additionally, we found that WT PHF8 promotes rDNA transcription more effectively than the PHF8-S844A mutant. We conclude that phosphorylation of PHF8 by cyclin E-CDK2 plays an important role in regulating gene transcription and cell cycle progression.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
pCMV-SPORT6-PHF8 was purchased from Open Biosystems. PHF8 and its mutant PHF8-S844A were cloned into pCMV FLAG. The truncated PHF8 (PHF8 (1–316), PHF8 (317–680), PHF8 (681–1024), and PHF8 mutants PHF8 (317–680) 381AGA, PHF8 (317–680) 641AKA, and PHF8 (681–1024) S844A and full-length CDK2 were cloned into pET41b. The mammalian expression vectors for cyclin E and CDK2 were generated as described previously (16).
The following primary antibodies were used. FLAG antibody (catalog no. M2) was purchased from Sigma. PHF8 (catalog no. P-15), myc (catalog no. 9E10), His (catalog no. H-3), CDK2 (catalog no. D-12), cyclin E (catalog no. E-4), β-actin catalog no. 1-19), and lamin B (catalog no. M-20) antibodies were purchased from Santa Cruz Biotechnology. Ser(P) antibody (catalog no. 2324S) was purchased from Cell Signaling Technology. Histone H3 (catalog no. ab1791) and H3K9me2 (catalog no. ab1220) antibodies were purchased from Abcam.
Coimmunoprecipitation and GST Pulldown
For immunoprecipitation, 293T cells were transfected with the indicated plasmids. The cell lysates were incubated with the indicated antibodies at 4 °C for 2 h, followed by addition of protein A-agarose beads. After 4 h of incubation at 4 °C, the beads were washed with lysis buffer. The immune complexes were separated by SDS-PAGE, followed by immunoblot analyses.
For the GST pulldown assay, 1 μg of GST-PHF8, GST-PHF8 (1–316), GST-PHF8 (317–680), GST-PHF8 (681–1024), or GST as a negative control was incubated with cell lysates from 293T cells overexpressing myc-tagged cyclin E and CDK2 or with His-CDK2 purified from BL21. The glutathione beads were then added and incubated for 2 h. The bound proteins were eluted with sample loading buffer and analyzed by immunoblotting with anti-myc or CDK2 antibodies.
Cell Culture and Synchronization
293T and HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum (PAA) and 100 μg/ml streptomycin and 100 units/ml penicillin at 37 °C with 5% CO2. HeLa cells were synchronized at early S phase by treatment with double thymidine in the same way as described for U2OS cells in a previous report (16). Briefly, cells were synchronized at early S transition by treatment with 2 mm thymidine for 16 h in complete medium, released in fresh medium for 8 h, and then incubated with 2 mm thymidine for another 16 h. Cells were synchronized to early S phase at this point.
In Vitro Kinase Assay
1 μg of GST-PHF8, GST-PHF8 (1–316), GST-PHF8 (317–680), and GST-PHF8 (681–1024) or GST-PHF8 mutants was incubated with GST-cyclin E and GST-CDK2 in kinase buffer (50 mm Tris-Cl (pH 7.5), 10 mm MgCl2, and 1 mm DTT) with 1 mm cold ATP and 0.5 μCi of [γ-32P]ATP at 30 °C for 30 min. The reaction was terminated with SDS-PAGE loading buffer. The samples were subjected to 8% SDS-PAGE gel and analyzed by autoradiography.
Mass Spectrometry
GST-PHF8 (681–1024) was incubated with GST-cyclin E and GST-CDK2 for the in vitro kinase assay as described above in the presence of cold ATP. Subsequently, GST-PHF8 (681–1024) was isolated using SDS-PAGE and then trypsinized. The tryptic peptides were analyzed by HPLC-ESI/MS/MS with a Thermo Finnigan LTQ adapted for nanospray ionization. All MS/MS spectra were processed using the SEQUEST (BioworksBrowser 3.3.1 SP1).
RNA Interference
RNA interference was carried out using double-stranded RNAs. The synthetic siRNA duplexes corresponding to the PHF8 and CDK2 mRNA sequences and the control siRNA sequence were obtained from Ribobio. 293T cells or HeLa cells were plated in a 6-well plate and transfected with 100 nm siRNA using Lipofectamine 2000 transfection reagent (Invitrogen).
Transient Transfection and Flow Cytometry Analysis
Cells were plated in 60-mm dishes and transfected with the indicated siRNAs or plasmids and pCMV GFP-H2B plasmid for the selection of transfected cells. Cells were harvested at the indicated time, fixed in ethanol, stained with propidium iodide, and then subjected to flow cytometry analysis.
Dual-Luciferase Reporter Activity Assay
293T cells in a 24-well plate were transfected with plasmids for expressing PHF8, PHF8-S844A, or siRNA targeting PHF8 or CDK2 and the rDNA luciferase reporter pHrD-IRES-Luc (28) and pRL-TK for 48 h. The cell lysates were harvested for the Dual-Luciferase assay according to the instructions of the manufacturer. Three independent experiments were performed.
RNA Extraction and Real-time PCR
Total RNA was extracted from cells by using TRIzol reagent (Invitrogen) according to the instructions of the manufacturer. Real-time PCR for cyclin E, E2F3, and E2F7 was performed using SYBR Green premix reagent (Toyobo, Japan) with β-actin as the internal control. Real-time PCR analysis of rRNA was also performed using SYBR Green premix reagent with GAPDH as the internal control. The relative amount of mRNA or rRNA was quantified using the comparative threshold cycle (CT) method. Primers used were as follows: cyclin E, 5′-CTCCAGGAAGAGGAAGGCAA-3′ (forward) and 5′-TCGATTTTGGCCATTTCTTCA-3′ (reverse); E2F3, 5′-AAGAAGAAGTCTAAAAACAACGTCCAA-3′ (forward) and 5′-CTTGACACTGGGCCAGCAT-3′ (reverse); E2F7, 5′-GGAAAGGCAACAGCAAACTCT-3′ (forward) and 5′-GGGAGAGCACCAAGAGTAGAAGA-3′ (reverse); β-actin, 5′-GTGAAGGTGACAGCAGTCGGTT-3′ (forward) and 5′-GAAGTGGGGTGGCTTTTAGGA-3′ (reverse); GAPDH, 5′-CGACCACTTTGTCAAGCTCA-3′ (forward) and 5′-AGGGGAGATTCAGTGTGGTG-3′ (reverse); and 47 S rRNA precursor, 5′-TGTCAGGCGTTCTCGTCTC-3′ (forward) and 5′-GAGAGCACGACGTCACCAC-3′ (reverse).
Chromatin Immunoprecipitation
HeLa Cells were treated with 1% formaldehyde for 10 min at room temperature. The cross-linking was stopped by the addition of 125 mm glycine. The cells were lysed, and the lysates were sonicated at 30 watt for 5 s and paused for 10 s up to 8 cycles to shear DNA to an average fragment size of 200–1000 bp. Immunoprecipitation was carried out with the indicated antibody. Normal mouse IgG was used as the negative control. After immunoprecipitation, the eluates were incubated at 65 °C for 4 h to reverse the cross-linking, followed by treatment with proteinase K (0.2 mg/ml) at 45 °C for 2 h. Then the DNA were precipitated and subjected to PCR with the indicated primers. The tested promoters were quantified by SYBR Green-based real-time quantitative PCR. All samples were subjected to PCR amplification with oligonucleotide primers specific for the indicated promoter DNA. Primers were as follows: cyclin E promoter, 5′-CCCCGTCCCTGCGCCTCGCTG-3′ (forward) and 5′-CGGCGGCGGCGACGGCAGTGG-3′ (reverse); rDNA promoter, 5′-AGAGGGGCTGCGTTTTCGGCC-3′ (forward) and 5′-CGAGACAGATCCGGCTGGCAG-3′ (reverse).
RESULTS
PHF8 Interacts with Cyclin E-CDK2 in an RXL-dependent Manner
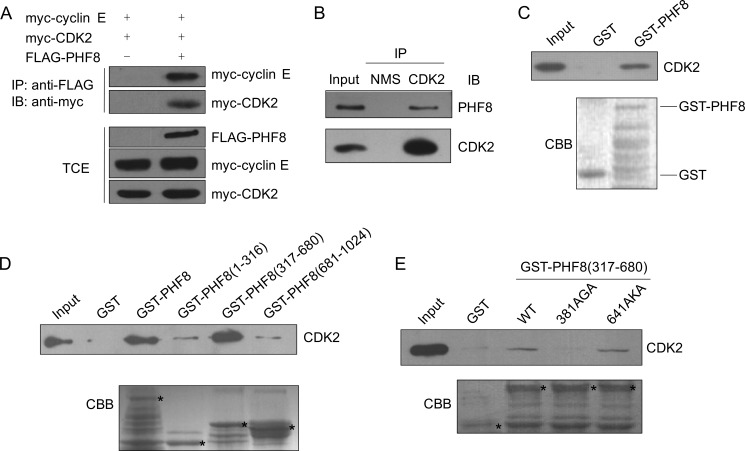
We previously identified a number of novel CDK2-associated proteins, including PHF8 (16). PHF8 is known as an H3K9me2 demethylase that consists of a plant homeodomain and a JmjC (Jumonji C) domain. To confirm whether PHF8 is a bona fide CDK2 interaction partner, we analyzed the association between PHF8 and CDK2 by coimmunoprecipitation and GST pulldown. pCMV FLAG-PHF8 and pCMV myc-cyclin E or CDK2 were cotransfected into 293T cells. The cell lysates were subjected to immunoprecipitation with FLAG antibody and immunoblotted with myc antibody. As shown in Fig. 1A, myc-cyclin E or myc-CDK2 coimmunoprecipitated with FLAG-PHF8. Next we examined whether endogenous PHF8 and CDK2 are associated with each other. The cell lysates from 293T cells were immunoprecipitated with CDK2 antibody and then immunoblotted with PHF8 antibody. As shown in Fig. 1B, PHF8 interacts with CDK2. These results confirm that PHF8 can interact with CDK2 in vivo. To examine whether there is a direct interaction between PHF8 and CDK2, GST-PHF8 and His-CDK2 were purified for the GST pulldown assay. As shown in Fig. 1C, PHF8 directly binds CDK2.
FIGURE 1.
PHF8 interacts with cyclin E-CDK2. A, coimmunoprecipitation assay. 293T cells were transfected with myc-tagged cyclin E and CDK2 and FLAG-tagged PHF8. Cell lysates were harvested for immunoprecipitation (IP) with FLAG antibody, followed by immunoblotting (IB) with myc antibody. TCE, total cell extract. B, 293T cell lysates were immunoprecipitated with CDK2 antibody or normal mouse IgG and then immunoblotted with PHF8 antibody. C–E, GST, GST-PHF8, or GST-PHF8 mutants, as indicated, immobilized on glutathione beads were incubated with His-CDK2 in lysis buffer for 2 h. The associated protein was eluted with SDS loading buffer and immunoblotted with CDK2 antibody. The asterisks represent GST or GST fusion proteins. CBB, Coomassie Brilliant Blue. NMS, normal mouse serum.
To determine which region of PHF8 is critical for its binding with cyclin E-CDK2, we generated a set of truncated PHF8 mutants for GST pulldown assays. Our data showed that PHF8 (317–680) binds with CDK2 much stronger than PHF8 (1–316) or PHF8 (681–1024) (Fig. 1D). The RXL motif has been shown previously to be the essential cyclin-binding motif in a wide range of cyclin-CDK-interacting proteins (29). Therefore, we asked whether the interaction between PHF8 and cyclin E-CDK2 is dependent on the RXL motif of PHF8. There are two RXL motifs in PHF8 (317–680) (amino acids 381–383 and 641–643, respectively). To find out which RXL motif on PHF8 is important for its binding with cyclin E-CDK2, we generated the PHF8 (317–680) AXA mutants, in which RXL motifs were converted into non-functional AXA motifs by site-directed mutagenesis. The data in Fig. 1E show that the PHF8 (316–680) 381AGA mutant, but not PHF8 (316–680) 641AKA, greatly weakened the interaction between PHF8 and CDK2, indicating that the PHF8 (381–383) RXL motif is critical for its interaction with CDK2.
PHF8 Is Phosphorylated by Cyclin E-CDK2
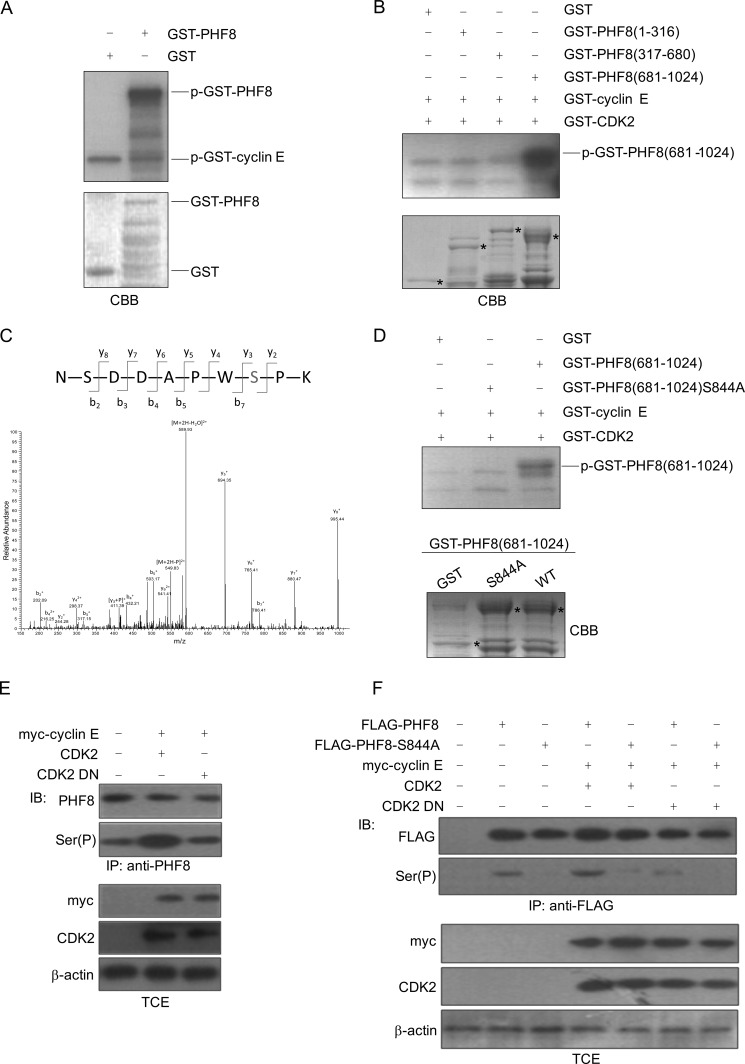
To examine whether PHF8 can be phosphorylated by cyclin E-CDK2, GST-PHF8 was used in in vitro kinase assays. As shown in Fig. 2A, PHF8 is phosphorylated by cyclin E-CDK2. To identify target cyclin E-CDK2 phosphorylation region(s) within PHF8, we performed in vitro kinase assays with truncated GST-PHF8 proteins. As shown in Fig. 2B, GST-PHF8 (681–1024) is phosphorylated much stronger than GST-PHF8 (1–316) or GST-PHF8 (317–680). To determine the cyclin E-CDK2 phosphorylation site(s) of PHF8, we employed mass spectrometry analysis of purified GST-PHF8 (681–1024) phosphorylated in vitro by the cyclin E-CDK2 complex. The data in Fig. 2C indicate that PHF8 Ser-844 is phosphorylated by cyclin E-CDK2. To further verify the phosphorylation site, we generated GST-PHF8 (681–1024) S844A and performed an in vitro kinase assay with cyclin E-CDK2. As shown in Fig. 2D, cyclin E-CDK2-dependent phosphorylation of the PHF8 (681–1024) S844A mutant is reduced significantly compared with wild-type PHF8 (681–1024). Then we transfected 293T cells with cyclin E-CDK2 or cyclin E-CDK2-DN (CDK2 dominant negative mutant) and analyzed the phosphorylation of endogenous PHF8 with Ser(P) antibody. As shown in Fig. 2E, phosphorylation of PHF8 at Ser was much stronger in CDK2-overexpressed cells than that in control or CDK2 DN-overexpressed cells, which indicates that endogenous PHF8 can be phosphorylated by cyclin E-CDK2. Furthermore, we transfected 293T cells with FLAG-PHF8 and FLAG-PHF8-S844A expression plasmids with cyclin E-CDK2 or cyclin E-CDK2 DN expression plasmids into 293T cells. Then the cell lysates were immunoprecipitated with FLAG antibody by immunoblotting with Ser(P) antibody. The data indicate that phosphorylation on serine of FLAG-PHF8, but not FLAG PHF8-S844A, can be enhanced by cyclin E-CDK2, whereas cyclin E-CDK2 DN did not affect the phosphorylation of FLAG-PHF8 (Fig. 2F), suggesting that cyclin E-CDK2 can phosphorylate PHF8-Ser-844 in cells. Taken together, these data suggest that PHF8 Ser-844 is the specific site phosphorylated by cyclin E-CDK2.
FIGURE 2.
Cyclin E-CDK2 phosphorylates PHF8 at Ser-844. A, GST or GST-PHF8 were incubated with GST-cyclin E and GST-CDK2 for the in vitro kinase assay. CBB, Coomassie Brilliant Blue. B, GST-PHF8 (1–316), GST-PHF8 (317–680), and GST-PHF8 (681–1024) were incubated with GST-CDK2 and GST-cyclin E for the in vitro kinase assay. The asterisks represent GST or GST fusion proteins. C, identification of phosphorylation site(s) of PHF8 (681–1024) by HPLC-ESI/MS/MS spectrometry. Purified GST-PHF8 (681–1024) was incubated with GST-CDK2 for the in vitro kinase assay in the presence of cold ATP and then subjected to mass spectrometry analysis. [y3+P] indicates phosphorylation with an increase of 80 mass unit. D, purified GST-PHF8 (681–1024) and GST-PHF8 (681–1024) S844A mutant were incubated with GST-CDK2 in the presence of [γ-32p]ATP for the in vitro kinase assay. E and F, 293T cells were transfected with the indicated plasmids. The cell lysates were harvested for immunoprecipitation (IP) with PHF8 antibody (E) and FLAG antibody (F), followed by immunoblotting (IB) with Ser(P) antibody. TCE, total cell extract.
Effects of PHF8 Knockdown on H3K9me2 Demethylation and Cell Cycle Progression
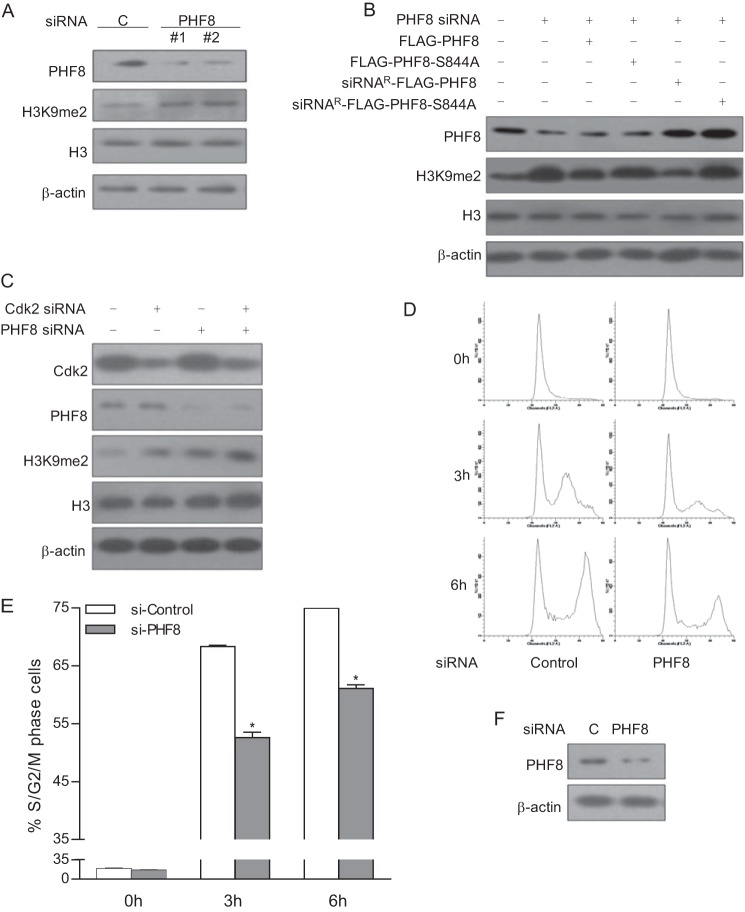
PHF8 has been reported to function as an H3K9me2 demethylase and to affect cell cycle progression (21, 26). Consistent with previous reports, we found that the level of H3K9me2 was elevated in 293T cells treated with two sets of PHF8 siRNAs (Fig. 3A). Also, we generated siRNA-resistant FLAG-PHF8 and FLAG-PHF8-S844A expression plasmids and transfected them with PHF8 siRNA into 293T cells. Then we analyzed the level of H3K9me2 in transfected cells by immunoblotting. As shown in Fig. 3B, only siRNA-resistant PHF8, but not the PHF8 S844A mutant, can reduce the level of H3K9me2. We also transfected HeLa cells with CDK2 siRNA with or without PHF8 siRNA and examined the level of PHF8 and H3K9me2 by immunoblotting. We found that knockdown of CDK2 can elevate the level of H3K9me2 and that further knockdown of PHF8 increased H3K9me2 to certain levels (Fig. 3C), which suggests that CDK2 may affect PHF8 demethylase activity. To examine the effect of PHF8 on cell cycle progression, we treated HeLa cells with PHF8 siRNA. The immunoblot data showed that PHF8 siRNA efficiently knocks down PHF8 expression in HeLa cells (Fig. 3D). The siRNA-transfected HeLa cells were synchronized at early S phase by double thymidine block, released for 3 and 6 h respectively, and subjected to flow cytometry analysis. The data indicated that knockdown of PHF8 significantly inhibited S phase progression (Fig. 3, E and F). These results suggest that knockdown of PHF8 affects H3K9me2 demethylation and cell cycle progression.
FIGURE 3.
Knockdown of PHF8 effects H3K9me2 demethylation and cell cycle progression. A, 293T cells were transfected with a control (C), PHF8 siRNA #1, or PHF8 siRNA #2. The cell lysates were harvested for immunoblotting with the indicated antibodies. B, 293T cells were transfected with FLAG-PHF8 and FLAG-PHF8 S844A or siRNA resistance PHF8 and PHF8 S844A together with PHF8 siRNA. The cell lysates were harvested for immunoblotting with the indicated antibodies. C, HeLa cells were transfected with a control, CDK2 siRNA, or PHF8 siRNA. The cell lysates were subjected to immunoblot analysis with the indicated antibodies. D–F, HeLa cells were transfected with control or PHF8 siRNA. Then the cells were synchronized to early S phase, released for 3 and 6 h, and subjected to flow cytometry analysis (D). The percentages of S/G2/M phase cells in different groups were calculated (E). The cell lysates were subjected to immunoblot analysis with the indicated antibodies (F). *, p < 0.01.
Phosphorylation of PHF8 Promotes H3K9me2 Demethylation and S Phase Progression
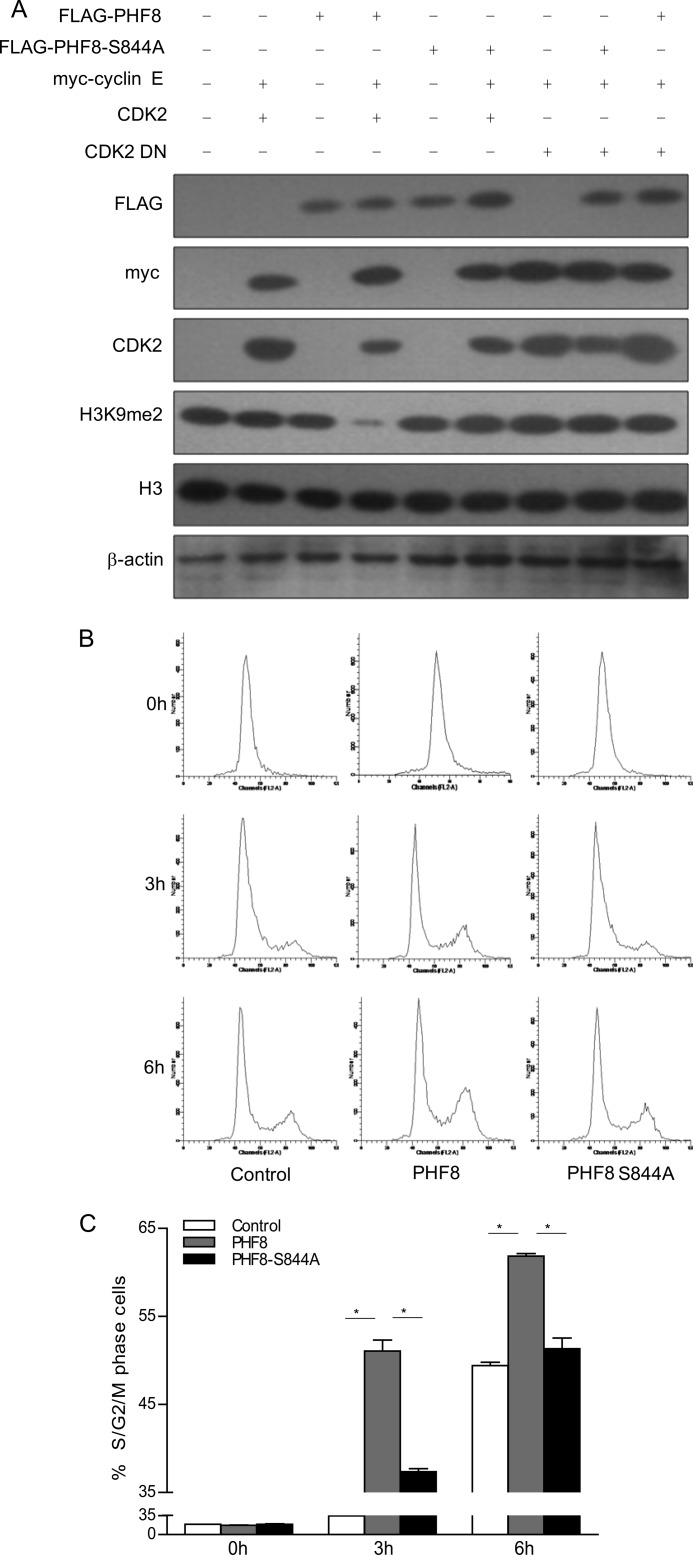
Because we found that PHF8 Ser-844 is phosphorylated by cyclin E-CDK2, we wondered whether phosphorylation of PHF8 by cyclin E-CDK2 would affect its function. To determine whether the demethylation of H3K9me2 is influenced by CDK2, we transfected 293T cells with FLAG-PHF8 and FLAG-PHF8-S844A expression plasmids with CDK2 or CDK2 DN expression plasmids. Then we analyzed the level of H3K9me2 in transfected cells. The data showed that overexpression of PHF8, but not the PHF8-S844A mutant, led to the reduction of H3K9me2. More importantly, overexpression of FLAG-PHF8 with cyclin E and CDK2, but not with CDK2 DN, demethylates H3K9me2 much more efficiently than overexpression of FLAG-PHF8 alone (Fig. 4A). These data suggest that phosphorylation of PHF8 at Ser-844 by cyclin E-CDK2 enhances PHF8 demethylase activity and promotes H3K9me2 demethylation.
FIGURE 4.
Phosphorylation of PHF8 Ser-844 promotes H3K9me2 demethylation and S phase progression. A, 293T cells were transfected with the indicated plasmids. The cell lysates were harvested and subjected to immunoblotting analysis. B and C, HeLa cells were transfected with control, pCMV FLAG-PHF8, or pCMV FLAG-PHF8-S844A and pCMV GFP-H2B as a sorting marker. The cells were synchronized at early S phase by double thymidine treatment, released for 3 and 6 h, and subjected to flow cytometry analysis (B). The percentages of S/G2/M phase cells in different groups were calculated (C). *, p < 0.05.
To determine the effect of PHF8 phosphorylation on the cell cycle, we transfected pCMV FLAG-PHF8 or pCMV FLAG-PHF8-S844A into HeLa cells. The cells were synchronized at early S phase by double thymidine treatment, released for 3 and 6 h, and subjected to flow cytometry analysis. As shown in Fig. 4, B and C, 6 h after release, 61% of PHF8-overexpressing cells entered S phase, (10% more compared with PHF8-S844A-overexpressed cells). These results indicate that phosphorylation of PHF8 at Ser-844 positively regulates its ability to promote S phase entry.
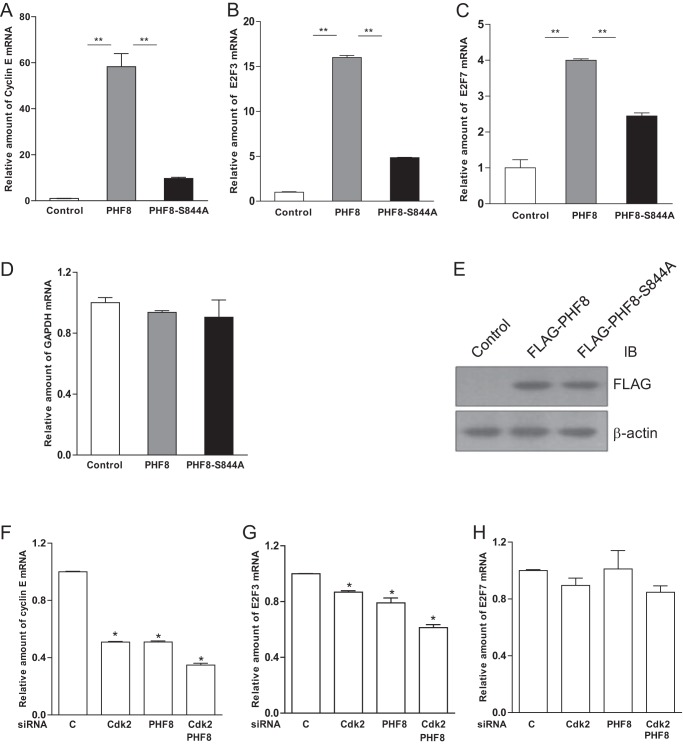
PHF8 Regulates Transcription of Cyclin E, E2F3, and E2F7
To explore how phosphorylation of PHF8 by cyclin E-CDK2 mechanistically regulates cell cycle progression, we transfected HeLa cells with FLAG-PHF8- or FLAG-PHF8-S844A-expressing plasmids and examined the mRNA levels of cyclin E, E2F3, E2F7, and GAPDH by real-time PCR. As shown in Fig. 5A–D, the amount of mRNAs (cyclin E, E2F3, and E2F7) in PHF8-overexpressed cells is significantly higher compared with PHF8-S844A-overexpressed cells, although PHF8-S844A also modestly up-regulated cyclin E, E2F3, and E2F7 mRNAs. However, there is no change in the level of GAPDH. The levels of PHF8 and PHF8-S844A are comparable (Fig. 5E). In addition, we performed similar experiments with knockdown of CDK2 and PHF8. We found that knockdown of CDK2 and PHF8 can reduce the expression levels of cyclin E and E2F3, respectively (Fig. 5, F and G), but it did not affect the level of E2F7 (Fig. 5H). Moreover, we noticed that knockdown of CDK2 and PHF8 together made the trend of cyclin E and E2F3 reduction more apparent. These results indicate that phosphorylation of PHF8 by cyclin E-CDK2 enhances its ability to promote transcription of cyclin E and E2F3, which may contribute to the ability of PHF8 to promote cell cycle progression. It is worth pointing out that overexpression of PHF8 causes the increase of E2F7 expression. It is known that induction of E2F7 during G0-G1/S phase causes cell cycle arrest, whereas expression of E2F7 during G2/M phase failed to disturb cell cycle progression. In addition, it has been reported that E2F7 is highly expressed during mid- to late S phase and represses transcription of G1/S-regulated genes such as E2F1 (30). Together with our data, we postulate that E2F7 may play a positive role in promoting S phase progression, whereas PHF8 could promote its function.
FIGURE 5.
PHF8 regulates the transcription of cyclin E, E2F3, and E2F7. A–E, HeLa cells were transfected with pCMV FLAG-PHF8, pCMV FLAG-PHF8-S844A, or empty vector as a control. Total RNAs from transfected cells were extracted and subjected to real-time PCR for cyclin E (A), E2F3 (B), E2F7 (C), and GAPDH (D) with β-actin as an internal control. The cell lysates were subjected to immunoblot (IB) analysis with the indicated antibodies (E). F–H, HeLa cells were knocked down with siRNA control, CDK2, PHF8, and CDK2 + PHF8. Total RNAs from transfected cells were extracted and subjected to real-time PCR for cyclin E (F), E2F3 (G), and E2F7 (H). *, p < 0.05; **, p < 0.01.
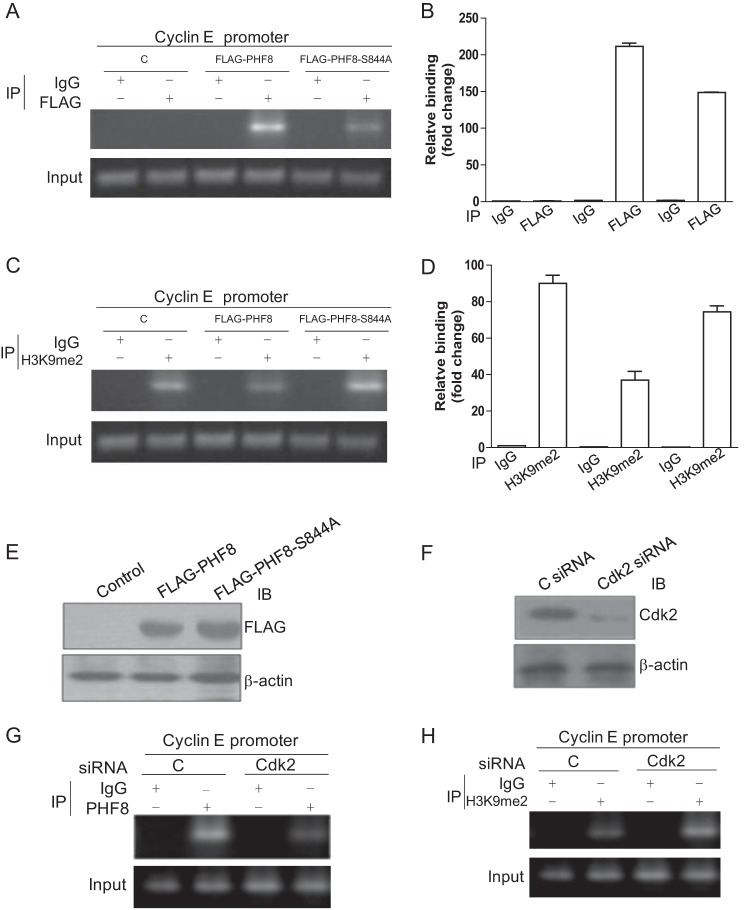
To understand how phosphorylation of PHF8 up-regulates transcription of cyclin E, we transfected HeLa cells with FLAG-PHF8- or FLAG-PHF8-S844A-expressing plasmids and examined the relative amount of PHF8 and H3K9me2 on cyclin E promoters by ChIP assays. The ChIP assays demonstrated that the amount of WT PHF8 associated with cyclin E promoters is significantly higher compared with PHF8-S844A (Fig. 6, A and B), whereas the amount of H3K9me2 on cyclin E promoters was reduced in PHF8-overexpressing cells compared with controls and PHF8-S844A-overexpressing cells (Fig. 6, C and D). The immunoblot data indicate that expression levels of PHF8 and PHF8-S844A are similar (Fig. 6E). In addition, we performed experiments with knockdown of CDK2 by ChIP assay. The data indicated that knockdown of CDK2 reduced the binding of PHF8 with the cyclin E promoter and, correspondingly, increased the level of H3K9me2 on the cyclin E promoter (Fig. 6, F and G). The efficiency of knockdown of CDK2 is shown in Fig. 6H. These results demonstrate that phosphorylation of PHF8 by cyclin E-CDK2 influences the levels of PHF8 and H3K9me2 at the promoter regions of cyclin E, which, in turn, promotes cyclin E transcription.
FIGURE 6.
Phosphorylation of PHF8 influences its binding to the cyclin E promoter. A–E, HeLa cells were transfected with pCMV FLAG-PHF8, pCMV FLAG-PHF8-S844A, or empty vector as a control (C). The cells were harvested and subjected to ChIP assays. Briefly, the cells were treated with formaldehyde and sonicated. The sheared DNAs were immunoprecipitated (IP) with FLAG or H3K9me2 antibodies and then subjected to PCR with primers for the cyclin E promoter (A and C) and a quantitative PCR assay on the cyclin E promoter (B and D). E, the cell lysates were subjected to immunoblot (IB) analysis. F–H, HeLa cells were transfected with CDK2 siRNA. CDK2 knockdown efficiency was confirmed by immunoblot analysis (F). The cells were harvested and subjected to ChIP assays with primers for the cyclin E promoter as above (G and H).
Phosphorylation of PHF8 at Ser-844 Enhances Its Ability to Promote Transcription of rDNA
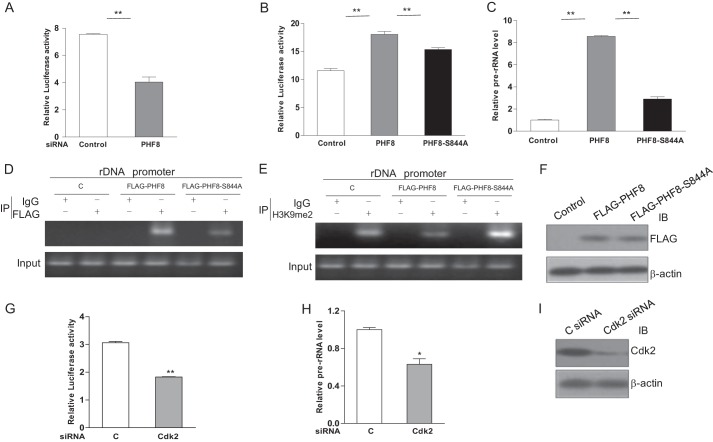
It has been reported that PHF8 promotes transcription of rDNA through binding with rDNA promoter (21). Consistently, we observed that knockdown of PHF8 causes a reduction of transcription of rDNA by rDNA luciferase reporter assay (Fig. 7A). To know whether phosphorylation of PHF8 regulates its ability to promote rDNA transcription, we transfected 293T cells with PHF8- or PHF8-S844A-expressing plasmids together with an rDNA luciferase reporter and then performed luciferase assays. Our data showed that overexpression of wild-type PHF8 activates rDNA promoters much more significantly compared with the phosphorylation-deficient PHF8-S844A mutant (Fig. 7B). Then we measured the levels of pre-rRNA by quantitative PCR. The result showed that the relative amount of pre-rRNA in PHF8-overexpressing cells is significantly higher compared with PHF8-S844A-overexpressing cells (Fig. 7C). Next we transfected HeLa cells with PHF8- or PHF8-S844A-expressing plasmids and examined the relative amount of PHF8 and H3K9me2 on rDNA promoters by ChIP assay. The data demonstrated that the amount of WT PHF8 associated with rDNA promoters is significantly higher compared with PHF8-S844A (Fig. 7D), whereas the amount of H3K9me2 on rDNA promoters was reduced in PHF8-overexpressing cells compared with controls and PHF8-S844A-overexpressing cells (Fig. 7E). The immunoblot data indicate that expression levels of PHF8 and PHF8-S844A are similar (Fig. 7F). In addition, we analyzed the rDNA promoter activity and pre-rRNA level in CDK2 knockdown 293T cells. The data show that knockdown of CDK2 causes a reduction of rDNA promoter activity and pre-rRNA transcription (Fig. 7, G–I). These results indicate that phosphorylation of PHF8 at Ser-844 by cyclin E-CDK2 increases its binding with the rDNA promoter and enhances its ability to promote rDNA transcription. Taken together, we propose that phosphorylation of PHF8 by cyclin E-CDK2 facilitates its loading onto chromatin and stimulates its demethylase activity, which enhances the expression of genes essential for the cell cycle progression, including cyclin E, E2Fs, and rDNAs.
FIGURE 7.
Phosphorylation of PHF8 promotes rRNA transcription. A, 293T cells were transfected with PHF8 siRNA or control siRNA and pHrD-IRES-Luc together with pRL-TK as an internal control. The cell lysates were harvested and subjected to Dual-Luciferase assay. B and C, 293T cells were transfected with pCMV FLAG-PHF8, pCMV FLAG-PHF8-S844A, or empty vector and pHrD-IRES-Luc together with pRL-TK as an internal control. The cell lysates were harvested and subjected to Dual-Luciferase assay (B). The pre-rRNA in transfected cells was quantified by RT-PCR, with GAPDH as an internal control (C). D–F, HeLa cells were transfected with pCMV FLAG-PHF8, pCMV FLAG-PHF8-S844A, or empty vector as a control. The cells were harvested and subjected to ChIP assays with primers for the rDNA promoter. D and E, immunoblot analysis was performed to check the expression of PHF8 and PHF8-S844A (F). G–I, 293T cells were transfected with CDK2 siRNA or control siRNA and rDNA luciferase reporter. The cell lysates were subjected to the luciferase assay (G). The total RNAs from transfected cells were extracted and used for real-time PCR for pre-rRNA (H). Immunoblot analysis was performed to check the expression of CDK2 (I). C, control. *, p < 0.05; **, p < 0.01.
DISCUSSION
The cyclin E-CDK2 serine/threonine kinase complex plays a critical role in coordinating both the onset of S phase and centrosome duplication during cell cycle progression. Cyclin E-CDK2 activates multiple signaling networks by phosphorylating multiple downstream signal transducers. For example, cyclin E-CDK2 phosphorylates NPAT to promote histone transcription (13, 31–33). CREB-binding protein-p300 is phosphorylated by cyclin E-CDK2 at G1/S transition to activate its histone acetyltransferase activity (34). We previously identified PHF8 as a novel CDK2-interacting protein using tandem affinity purification (16). PHF8 is a H3K9me2 demethylase and can regulate rDNA transcription and cell cycle progression. It has been reported that PHF8 is phosphorylated by CDK1 and regulates G2/M progression (26). However, the role of CDK2 in regulating the function of PHF8 is unclear. Here we demonstrated that PHF8 Ser-844 is phosphorylated by cyclin E-CDK2 and that phosphorylation of PHF8 affects its ability to demethylate H3K9me2 and regulate S Phase progression. These results suggest that the function of PHF8 in the cell cycle may be precisely controlled by cyclin-CDKs.
PHF8 is a H3K9me2 demethylase. Consistent with a previous report (21), we showed that the H3K9me2 level is reduced in PHF8-overexpressing cells. Interestingly, we found that cyclin E-CDK2 enhances the ability of PHF8 to promote H3K9me2 demethylation, which suggests that phosphorylation of PHF8 by cyclin E-CDK2 directs its demethylase activity toward H3K9me2. However, it remains to be determined whether phosphorylation of PHF8 influences the demethylation of other histones.
Multiple key regulators of G1/S transition include cyclin E and the E2F3 and E2F7 transcription factors. Their expression levels are precisely controlled during the cell cycle. Our results demonstrate that overexpression of PHF8 causes a significant increase of their mRNA levels compared with overexpression of the phosphorylation-deficient PHF8-S844A PHF8 mutant. Meanwhile, ChIP analysis showed that WT PHF8 is recruited to the cyclin E promoter more efficiently than the PHF8-S844A mutant. WT PHF8 reduces H3K9me2 levels at the cyclin E promoters more effectively than PHF8-S844A. These results indicate that phosphorylation of PHF8 by cyclin E-CDK2 enhances its binding to the cyclin E promoter and directs its demethylase activity toward H3K9me2, which, in turn, promotes the transcription of cyclin E to enhance the positive feedback loop promoting cell cycle progression. Therefore, it will be worth investigating whether PHF8 has similar effect on other genes involved in cell cycle regulation and whether phosphorylation of PHF8 also has a direct impact on transcription of these candidate genes. In addition, comparing the transcriptome of PHF8- and PHF8-S884A-overexpressing cells would generate valuable data to further our understanding of the signaling pathways regulated by cyclin E-CDK2.
It has been reported that PHF6, as a plant homeodomain family member and tumor suppressor, directly interacts with the upstream binding factor through its plant homeodomain and suppresses rRNA transcription (35). Our data show that overexpression of PHF8 promotes rRNA synthesis. These results indicate that PHF family members function as both positive and negative regulators of rRNA transcription to control the overall level of rRNA. However, it needs to be elucidated whether PHF8 binds the upstream binding factor to reverse the inhibitory effect of PHF6 on rRNA synthesis.
In summary, we identified PHF8 as a novel substrate of cyclin E-CDK2. We confirmed the essential role of cyclin E-CDK2-mediated Ser-844 phosphorylation in regulating PHF8 function. Our studies revealed that cyclin E-CDK2 regulates cell cycle progression through phosphorylation of PHF8, which consequently influences the H3K9me2 level on promoters of specific G1/S-regulating genes and rDNA transcription. These findings unveil new roles of cyclin E-CDK2 and PHF8 in cell cycle control.
Acknowledgments
We thank Dr. Y. Li (Institute of Psychology, Chinese Academy of Sciences) and Dr. Haiteng Deng (Tsinghua University) for help with flow cytometry and mass spectrometry, respectively.
This work was supported by National Natural Science Foundation of China Grant 81272272 and by Ministry of Science and Technology of China Grants 2013ZX10004611, 2012CB519003, and 2011CB504705.
- CREB
- cAMP response element-binding protein
- NPAT
- nuclear protein mapped to the AT locus
- ESI
- electrospray ionization
- DN
- dominant negative.
REFERENCES
- 1. Nigg E. A. (1995) Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays 17, 471–480 [DOI] [PubMed] [Google Scholar]
- 2. Hwang H. C., Clurman B. E. (2005) Cyclin E in normal and neoplastic cell cycles. Oncogene 24, 2776–2786 [DOI] [PubMed] [Google Scholar]
- 3. Sherr C. J. (1994) G1 phase progression: Cycling on cue. Cell 79, 551–555 [DOI] [PubMed] [Google Scholar]
- 4. Brown N. R., Noble M. E., Endicott J. A., Johnson L. N. (1999) The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1, 438–443 [DOI] [PubMed] [Google Scholar]
- 5. Schulman B. A., Lindstrom D. L., Harlow E. (1998) Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. U.S.A. 95, 10453–10458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hall C., Nelson D. M., Ye X., Baker K., DeCaprio J. A., Seeholzer S., Lipinski M., Adams P. D. (2001) HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-Cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21, 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adams P. D., Sellers W. R., Sharma S. K., Wu A. D., Nalin C. M., Kaelin W. G., Jr. (1996) Identification of a cyclin-Cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16, 6623–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dick F. A., Rubin S. M. (2013) Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 14, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bertoli C., Skotheim J. M., de Bruin R. A. (2013) Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14, 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hara T., Kamura T., Nakayama K., Oshikawa K., Hatakeyama S., Nakayama K. (2001) Degradation of p27 (Kip1) at the G0-G1 transition mediated by a Skp2-independent ubiquitination pathway. J. Biol. Chem. 276, 48937–48943 [DOI] [PubMed] [Google Scholar]
- 11. Cooley A., Zelivianski S., Jeruss J. S. (2010) Impact of cyclin E overexpression on Smad3 activity in breast cancer cell lines. Cell Cycle 9, 4900–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morris L., Allen K. E., La Thangue N. B. (2000) Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2, 232–239 [DOI] [PubMed] [Google Scholar]
- 13. Ma T., Van Tine B. A., Wei Y., Garrett M. D., Nelson D., Adams P. D., Wang J., Qin J., Chow L. T., Harper J. W. (2000) Cell cycle-regulated phosphorylation of p220 (NPAT) by cyclin E-Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14, 2298–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei Q., Li J., Liu T., Tong X., Ye X. (2013) Phosphorylation of minichromosome maintenance protein 7 (MCM7) by cyclin/cyclin-dependent kinase affects its function in cell cycle regulation. J. Biol. Chem. 288, 19715–19725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J., Deng M., Wei Q., Liu T., Tong X., Ye X. (2011) Phosphorylation of MCM3 protein by cyclin E/cyclin-dependent kinase 2 (CDK2) regulates its function in cell cycle. J. Biol. Chem. 286, 39776–39785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng M., Li F., Ballif B. A., Li S., Chen X., Guo L., Ye X. (2009) Identification and functional analysis of a novel cyclin E-Cdk2 substrate ankrd17. J. Biol. Chem. 284, 7875–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fortschegger K., Shiekhattar R. (2011) Plant homeodomain fingers form a helping hand for transcription. Epigenetics 6, 4–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fortschegger K., de Graaf P., Outchkourov N. S., van Schaik F. M., Timmers H. T., Shiekhattar R. (2010) PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell. Biol. 30, 3286–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu J., Shi G., Jia Y., Li J., Wu M., Li J., Dong S., Wong J. (2010) The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 20, 908–918 [DOI] [PubMed] [Google Scholar]
- 20. Krishnan S., Horowitz S., Trievel R. C. (2011) Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chem. Bio. Chem. 12, 254–263 [DOI] [PubMed] [Google Scholar]
- 21. Zhu Z., Wang Y., Li X., Wang Y., Xu L., Wang X., Sun T., Dong X., Chen L., Mao H., Yu Y., Li J., Li J., Chen P. A., Chen C. D. (2010) PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 20, 794–801 [DOI] [PubMed] [Google Scholar]
- 22. Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. (2010) PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 17, 445–450 [DOI] [PubMed] [Google Scholar]
- 23. Arteaga M. F., Mikesch J. H., Qiu J., Christensen J., Helin K., Kogan S. C., Dong S., So C. W. (2013) The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell 23, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., Tommerup N., Shi X., Gozani O., Rappsilber J., Salcini A. E., Helin K. (2010) A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laumonnier F., Holbert S., Ronce N., Faravelli F., Lenzner S., Schwartz C. E., Lespinasse J., Van Esch H., Lacombe D., Goizet C., Phan-Dinh Tuy F., van Bokhoven H., Fryns J. P., Chelly J., Ropers H. H., Moraine C., Hamel B. C., Briault S. (2005) Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42, 780–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu W., Tanasa B., Tyurina O. V., Zhou T. Y., Gassmann R., Liu W. T., Ohgi K. A., Benner C., Garcia-Bassets I., Aggarwal A. K., Desai A., Dorrestein P. C., Glass C. K., Rosenfeld M. G. (2010) PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lim H. J., Dimova N. V., Tan M. K., Sigoillot F. D., King R. W., Shi Y. (2013) The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20. Mol. Cell. Biol. 33, 4166–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghoshal K., Majumder S., Datta J., Motiwala T., Bai S., Sharma S. M., Frankel W., Jacob S. T. (2004) Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J. Biol. Chem. 279, 6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sørensen C. S., Lukas C., Kramer E. R., Peters J. M., Bartek J., Lukas J. (2001) A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol. Cell. Biol. 21, 3692–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Westendorp B., Mokry M., Groot Koerkamp M. J., Holstege F. C., Cuppen E., de Bruin A. (2012) E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 40, 3511–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao J., Kennedy B. K., Lawrence B. D., Barbie D. A., Matera A. G., Fletcher J. A., Harlow E. (2000) NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14, 2283–2297 [PMC free article] [PubMed] [Google Scholar]
- 32. Wei Y., Jin J., Harper J. W. (2003) The cyclin E-Cdk2 substrate and Cajal body component p220 (NPAT) activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 23, 3669–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeRan M., Pulvino M., Greene E., Su C., Zhao J. (2008) Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol. Cell. Biol. 28, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ait-Si-Ali S., Polesskaya A., Filleur S., Ferreira R., Duquet A., Robin P., Vervish A., Trouche D., Cabon F., Harel-Bellan A. (2000) CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene 19, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 35. Wang J., Leung J. W., Gong Z., Feng L., Shi X., Chen J. (2013) PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J. Biol. Chem. 288, 3174–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]