Background: Polycomb repressive complex PRC1 is an epigenetic regulator of cellular differentiation. However, its function during adipogenesis is unknown.
Results: FBXL10/KDM2B recruited noncanonical PRC1 complex in F-box and LRR motifs dependent on cell cycle-related genes and Pparg genes and repressed 3T3-L1 adipogenesis.
Conclusion: Noncanonical PRC1 complex containing FBXL10/KDM2B regulates adipogenesis.
Significance: Our findings revealed a novel epigenetic mechanism of adipogenesis.
Keywords: Adipocyte, Adipogenesis, Cell Differentiation, Peroxisome Proliferator-activated Receptor (PPAR), Polycomb, 3T3-L1 cells, FBXL10/KDM2B/JHDM1B, Mitotic Clonal Expansion (MCE), Polycomb Repressive Complex 1 (PRC1), RING1B
Abstract
Polycomb repressive complex 1 (PRC1) plays an essential role in the epigenetic repression of gene expression during development and cellular differentiation via multiple effector mechanisms, including ubiquitination of H2A and chromatin compaction. However, whether it regulates the stepwise progression of adipogenesis is unknown. Here, we show that FBXL10/KDM2B is an anti-adipogenic factor that is up-regulated during the early phase of 3T3-L1 preadipocyte differentiation and in adipose tissue in a diet-induced model of obesity. Interestingly, inhibition of adipogenesis does not require the JmjC demethylase domain of FBXL10, but it does require the F-box and leucine-rich repeat domains, which we show recruit a noncanonical polycomb repressive complex 1 (PRC1) containing RING1B, SKP1, PCGF1, and BCOR. Knockdown of either RING1B or SKP1 prevented FBXL10-mediated repression of 3T3-L1 preadipocyte differentiation indicating that PRC1 formation mediates the inhibitory effect of FBXL10 on adipogenesis. Using ChIP-seq, we show that FBXL10 recruits RING1B to key specific genomic loci surrounding the key cell cycle and the adipogenic genes Cdk1, Uhrf1, Pparg1, and Pparg2 to repress adipogenesis. These results suggest that FBXL10 represses adipogenesis by targeting a noncanonical PRC1 complex to repress key genes (e.g. Pparg) that control conversion of pluripotent cells into the adipogenic lineage.
Introduction
The mouse 3T3-L1 cell line has been a paradigm for investigating molecular events that contribute to adipocyte differentiation (1). Following the growth arrest by contact inhibition, treatment of 3T3-L1 preadipocytes with an induction mixture comprised of insulin, glucocorticoids, and the phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (MDI4 mixture) induces two cycles of mitotic clonal expansion (MCE) followed by terminal differentiation to mature adipocytes. This adipogenic process is orchestrated by an elaborate cascade of sequentially acting transcription factors and chromatin modifiers that shape differentiation downstream of the hormones and signaling pathways that are targeted by the MDI mixture. During the very early stage of adipogenesis, there is a rapid induction of Cebpb and Cebpd within 24 h after induction that is required to complete the first round of MCE that is completed 24–36 h after the initiation of induction. This is followed by the second round of MCE that is completed 48–60 h post-induction and the activation of two critical pro-adipogenic transcription factors, Cebpa and Pparg expression (2, 3). These two factors then initiate positive feedback to induce their own expression and also coordinately activate transcription of many genes that define the terminally differentiated adipocyte phenotype (2, 4). A number of transcription factors and signaling pathways have been identified as regulators of adipogenesis (5, 6), and recent studies have also uncovered novel epigenetic processes that contribute to adipogenesis in 3T3-L1 cells (7–9).
Epigenetic regulation, including DNA methylation and post-translational covalent modifications of histones, modifies chromatin structure and is pivotal in cell lineage decisions by selectively activating or repressing transcription of subsets of tissue-specific genes. The broad range modification of histones in specific chromosomal regions has a dominant role in determining whether large areas of chromatin are “open” and actively transcribed or “closed” and transcriptionally repressed. Polycomb group (PcG) proteins are conserved chromatin proteins that contribute to gene silencing throughout higher eukaryotes (10, 11). PcGs are assembled into large multiprotein complexes that selectively occupy chromatin sites and modify chromatin activity. The two best characterized PcG complexes are polycomb repressive complex 1 (PRC1) and PRC2, and they are known to affect lysine 119 ubiquitination of histone H2A (H2AK119ub) and H3K27 tri-methylation (H3K27me3), respectively (12–14).
PRC1 complexes contain a central core RING1 protein that is the catalytically active ubiquitinating enzyme (15). Although it is clear that H2A ubiquitination plays an important role in PRC1-mediated silencing, exactly how H2A ubiquitination influences transcriptional repression is not fully understood. Other mechanisms, including chromatin compaction and interaction with the general transcription machinery, are considered to contribute to full PRC1 repressive capacity (16, 17). Studies on polynucleosome compaction suggest that chromatin dynamics, including nucleosome positioning, organization, and packing, all contribute to transcriptional activity (18).
FBXL10 is a paralog of the first identified JmjC domain containing histone demethylase FBXL11. FBXL10 contains several functional domains, including the catalytic JmjC, a DNA-binding CXXC zinc finger, an F-box domain, a PHD finger, and eight leucine-rich repeats (LRRs). The JmjC domain is required for demethylation of FBXL10 targets, including H3K36 (19, 20) and H3K4me3 (21, 22), which contribute to cellular senescence (20), apoptosis (23), repression of ribosomal RNA expression (22), hematopoietic stem cell self-renewal (24), pluripotent stem cell generation (iPS) (19), and the proper generation of the neural tube in vivo (25).
Besides regulation of histone demethylation, FBXL10 silences gene expression by interacting with a noncanonical PRC1 complex (26–28) that contains RING1B E3 ubiquitin ligase and additional proteins such as SKP1 and PCGF1/NSPC1 (a paralog of BMI1) (26–29). In this complex, FBXL10 influences gene repression by modulating H2AK119 ubiquitination. The mechanism includes FBXL10 recruiting the PRC1 complex to nonmethylated CpG islands through its CXXC DNA binding domain (26, 29, 30), which is important in embryonic stem (ES) cells. This mechanism has been proposed to account for ∼43% of RING1B/PRC1 repression (30). It was recently reported that FBXL10 maintains embryonic stem cell status, importantly, not through normal demethylation activity but through noncanonical PRC1 recruiting (31). Thus, the noncanonical PRC1 complex containing FBXL10 plays a major role in regulating cell differentiation status, but its role in adipogenesis has not been investigated. In this study, we show that FBXL10 regulates 3T3-L1 preadipocyte differentiation independent of its demethylase activity but through noncanonical PRC1 complex formation.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit polyclonal anti-SKP1 (catalog no. 2156), rabbit monoclonal anti-RING1B (catalog no. 5694, clone D22F2, used for immunoblot), and anti-H2Aub (catalog no. 8240, Clone D27C4, used for immunoblot) were from Cell Signaling Technology (Danvers, MA); mouse monoclonal anti-RING1B (catalog no. 39663, used for ChIP) was from Active Motif (Carlsbad, CA); mouse monoclonal anti-TATA-binding protein (1TBP18) (NB500-700) was from Novus Biologicals (Littleton, CO); rabbit polyclonal anti-BCOR (SAB4502272), mouse monoclonal anti-β-actin (clone AC74, A5316), mouse monoclonal anti-FLAG M2-peroxidase (HRP) antibody (A8592), horseradish peroxidase conjugated affinity-purified goat anti-mouse IgG (A4416), and anti-rabbit IgG (A0545) were from Sigma; mouse monoclonal anti-V5 tag (R960-25) was from Invitrogen; control mouse IgG (0107-01) was from Southern Biotechnology (Birmingham, AL); rabbit polyclonal anti-histone H3K4me3 (catalog no. 07-473), rabbit polyclonal anti-Histone H3K27me3 (catalog no. 07449), and mouse polyclonal anti-histone H2Aub (catalog no. 05-678, clone E6C5, used for ChIP) were from Millipore (Billerica, MA); rabbit polyclonal anti-mouse IgM (catalog no. 31196, used for ChIP) was from Thermo Fisher Scientific (Waltham, MA); mouse monoclonal anti-PPARγ (IgG-A3409) were raised in our laboratory as described previously (9).
Cell Culture and Staining
3T3-L1 mouse preadipocytes (CL-173, American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. For adipocyte differentiation, 2 days after reaching confluence (day 0), 3T3-L1 preadipocytes were treated with differentiation medium containing insulin (1 μg/ml), 0.25 μm dexamethasone, and 0.5 mm isobutylmethylxanthine (MDI mixture) for 48 h followed by treatment with insulin (1 μg/ml) alone with medium replacement every 2 days as described previously (8, 9). To induce differentiation by PPARγ ligand, troglitazone (1 μm) (Cayman, Ann Arbor, MI) was added to the medium on the day of induction (day 0) in addition to the MDI mixture for 48 h followed by treatment with insulin (1 μg/ml) and troglitazone (1 μm) with medium replacement every 2 days. To inhibit adipogenesis, 3T3-L1 preadipocytes were exposed to 20 ng/ml Wnt3a (R&D Systems, Minneapolis, MN) in addition to MDI mixture for 48 h from the 1st day of induction. On day 8 of differentiation, the cells were stained with Oil Red O (ORO) as described previously (8, 9). TT2 embryonic stem (ES) cells were cultured as described (32).
Retroviral Transduction and Cell Proliferation
To construct retroviral expression vectors for FBXL10-1 and -2 containing the V5 epitope tag at the COOH terminus, we amplified coding sequences of mouse FBXL10-1 (IMAGE clone ID 6851421/AV133-d1) and mouse FBXL10-2 (cDNA prepared from 3T3-L1 preadipocytes) and cloned them into the EcoRI/NotI site of pMXs-puro vector driven by weak LTR promoter (a kind gift from Dr. Toshio Kitamura, University of Tokyo) (33). We also constructed their internal deletion mutants (ΔJmjC, ΔCXXC, ΔPHD, ΔF-box, and ΔLRR) by using KOD-Plus mutagenesis kit (TOYOBO, Osaka, Japan). To construct retroviral expression vector for FBXL11 containing the FLAG epitope tag at the amino terminus, we amplified coding sequences of mouse Fbxl11 by PCR using the cDNA prepared from white adipose tissue of C57BL/6J mice and ligated into the NotI site of pMXs-IRES-puro vector LTR promoter. All PCR-generated constructs were verified by sequencing. Using these vectors, retroviral infection to 3T3-L1 preadipocytes was performed as described (8, 9). For growth curve analysis, retrovirally transduced 3T3-L1 preadipocytes on the indicated time points were counted on the indicated days.
Cell Cycle Analysis Using Fluorescence-activated Cell Sorting (FACS)
The cell cycle analysis was performed using APC BrdU flow kit (BD Biosciences) as described elsewhere (34). Briefly, cultured cells were labeled with 10 μm bromodeoxyuridine (BrdU) for 1 h, washed, collected, fixed, and permeabilized with Cytofix/Cytoperm buffer (BD Biosciences) containing 3–5% formaldehyde. Then the cells were treated with DNase (300 μg/ml) to expose BrdU epitope for 1 h at 37 °C, washed, and then stained with FITC-conjugated anti-BrdU antibody for 20 min at room temperature, washed again, and centrifuged. Staining buffer containing 7-aminoactinomycin D (7-AAD) was added to each tube to resuspend the cells, and the stained cells were analyzed by flow cytometry using FACScan II (BD Biosciences). Acquired multiparameter data were analyzed using CellQuest software (BD Biosciences) with the combination of BrdU and 7-AAD; two color flow cytometric analysis permits the enumeration and characterization of cells that are actively synthesizing DNA (BrdU incorporation) in terms of their cell cycle position. As shown by the region gates applied to the 7-AAD versus BrdU dot plot, flow cytometric analysis of cells stained with the reagents allowed the discrimination of cell subsets that were in the S phase (green), G0/G1 phase (purple), and G2+M phase (blue).
Immunoprecipitation and Immunoblot Analysis
The methods for cell fractionation and immunoprecipitation were described previously (35–38). Immunoblots were visualized by chemiluminescence using Super Signal West Dura Extended Duration Substrate (Thermo Fisher Scientific), and luminescent images were analyzed by ImageQuant LAS 4000mini (GE Healthcare).
Identification of FBXL10-interacting Proteins
Retrovirally transduced 3T3-L1 preadipocytes were grown in a 15-cm dish, harvested, and centrifuged at 1,000 × g for 5 min. Then the pellet of cells was washed once with PBS and frozen at −80 °C until use. All subsequent operations were carried out on ice or at 4 °C. Each cell pellet was thawed out and allowed to swell in hypotonic buffer B (10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, and a mixture of protease inhibitors (Complete, Mini, EDTA-free; Roche Applied Science)) for 30 min, passed through a 25-gauge needle five times, and centrifuged at 20,000 × g for 1 min. The pellet was resuspended in 0.2 ml of buffer C (10 mm HEPES (pH 7.9), 10 mm KCl, 1 mm MgCl2, 0.5% Nonidet P-40, and a mixture of protease inhibitors) and sonicated on ice 10 times using 10-s pulses using a Sonifier cell disruptor model 250 (Branson Ultrasonics), and then the debris was removed by centrifugation. Supernatants were collected, and buffer was exchanged to 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 0.1 mm EDTA, 5% glycerol, 0.1% Nonidet P-40 by Econo-Pac 10DG (Bio-Rad), ultrafiltrated by Amicon Ultra-4 MWCO 30K (Millipore), and used for immunoprecipitation. The samples were incubated with control IgG or anti-V5 epitope antibody cross-linked with Dynabeads® protein G (Invitrogen) and rotated for 18 h at 4 °C. The beads were washed three times with buffer containing 20 mm HEPES (pH 7.9), 200 mm KCl, 2 mm EDTA, 0.1% Nonidet P-40, after which the protein complexes were eluted with buffer containing 62.5 mm Tris-HCl (pH 6.8), 2% SDS, and 25% glycerol for 5 min at 95 °C. Each eluate was precipitated with methanol and chloroform, washed with ice-cold acetone, and centrifuged at 2,000 × g for 10 min as described (39). Each pellet was air-dried and resuspended in 25 mm NH4HCO3 buffer containing 25% (v/v) CH3CN at room temperature. The samples were reduced in 1.2 mm tris(2-carboxyethyl)phosphine for 15 min at 50 °C and alkylated in 3 mm iodoacetamide for 30 min at room temperature, respectively. The samples were digested overnight with 100 ng of trypsin (Promega) at 37 °C as described (40). Aliquots of trypsinized samples were analyzed by liquid chromatography-tandem mass spectrometry (LC/MS/MS) as we described previously (40). Protein quantification was done by processing the acquired LC-MS data with the software Progenesis LC-MS (version 2.6; Nonlinear Dynamics, Newcastle, UK). MS/MS spectra from features with charge +2, +3, and +4 were exported and searched against the Swiss-Prot database using Mascot (version 2.3; Matrix Science).
Small Interfering RNA (siRNA)
The duplexes of each small interfering RNA (siRNA), targeting murine Fbxl10 mRNA (MSS219951 and MSS219952; Invitrogen), Skp1 (sc-36498A; Santa Cruz Biotechnology), Cul1 (MSS219122 and MSS219123; Invitrogen), or Ring1B (sc-62947A and sc-62947C; Santa Cruz Biotechnology) and control siRNA (Santa Cruz Biotechnology sc-44230 or Invitrogen Stealth RNAi, Negative Control Low GC Duplexes 12935-200) were transfected into cells using Lipofectamine RNAi MAX reagent (Invitrogen) as described (8, 9).
Isoform-specific Fbxl10 mRNA Quantitation
The expression levels of each isoform of Fbxl10 mRNAs were quantitatively analyzed by means of qPCR. As the standard template for Fbxl10-1 and -2, we generated the chimeric plasmid where the nucleotides from 1622 to 1651 of mouse Fbxl10-1 relative to the translation initiation site were replaced with the sequences from 1622 to 1651 of mouse Fbxl10-2 and used (see Fig. 1D). All reactions were done in triplicate.
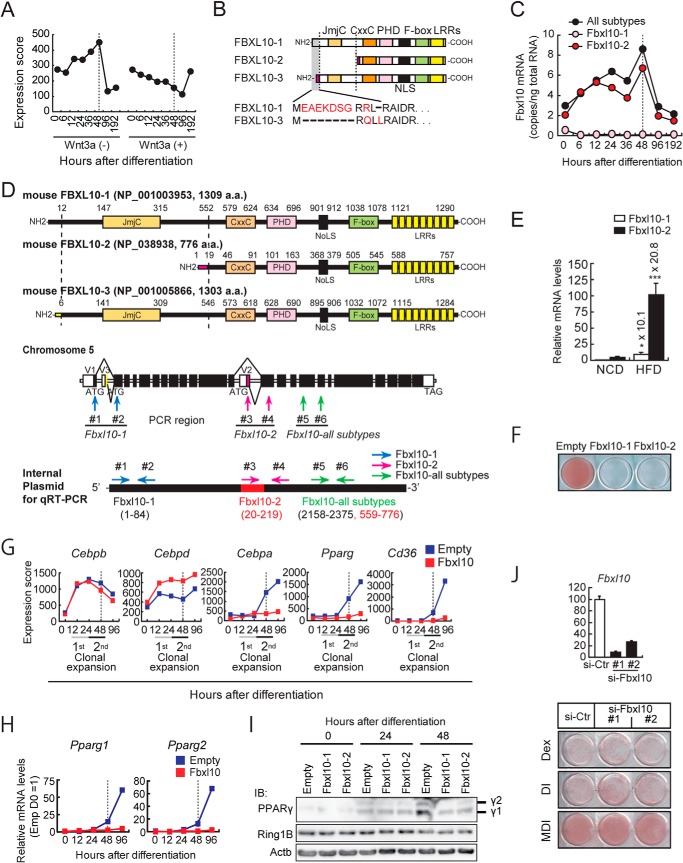
FIGURE 1.
FBXL10 is a novel anti-adipogenic factor. A, Fbxl10 gene expression during 3T3-L1 adipogenesis. Total RNA was extracted from 3T3-L1 cells at indicated time points of differentiation (0, 12, 24, 48 and 96 h after induction of differentiation), and transcriptional analyses were carried out using a microarray. B, schematic representations of three FBXL10 isoforms (FBXL10-1; NP_001003953, FBXL10-2; NP_038938, and FBXL10-3; NP_001005866). C and D, expression of Fbxl10 isoforms during 3T3-L1 preadipocytes differentiation (C). The PCR data were quantified based on standard curves generated by using serial dilutions of internal plasmid DNA containing both unique sequences of FBXL10-1 and FBXL10-2 and common sequences for all subtypes (D) as described under “Experimental Procedures.” The data were converted to copy the number/ng of total RNA (C). D, transcript variants of mouse FBXL10. JmjC, jumonji C domain; CxxC, ZF-CXXC domain; PHD, plant homeodomain; NoLS, nucleolar localization signal; LRR, leucine-rich repeat (top). The exons and introns of mouse Fbxl10 are shown in the middle panel. The first exon of each variant is indicated as V1, V2, and V3. Coding region is indicated as filled column and noncoding region is indicated as open column. Primers for qPCR (#1, #2 for FBXL10-1-specific; #3, #4 for FBXL10-2-specific, and #5, #6 for Fbxl10 all subtypes common sequence) are indicated by arrows (middle panel). Internal control plasmid for qPCR is indicated in the bottom panel. Internal sequences (nucleotides 1622–1651) of FBXL10-1 are replaced by FBXL10-2-specific nucleotides (nucleotides 20–49). E, Fbxl10 is expressed in white adipose tissue (WAT) of mice and down-regulated in diet-induced obese mice. Matched littermates were fed with a normal (NCD) or HFD for 8 weeks, and gene expressions of FBXL10-1 and FBXL10-2 in white adipose tissue were analyzed with qPCR (n = 6 mice per group). Ppib serves as the internal control for qPCR. Error bars represent ± S.E. (*, p < 0.05; ***, p < 0.001). F, ORO staining of 3T3-L1 preadipocytes stably expressing FBXL10-1, FBXL10-2, or empty vector was performed on day 8 of differentiation. G, transcriptional changes of adipogenic genes in FBXL10-1-transduced preadipocytes during adipogenesis. Cells were harvested at the indicated times of differentiation, and transcriptional analyses were carried out using a microarray. H, mRNA levels of Pparg1 and Pparg2 in 3T3-L1 preadipocytes stably expressing FBXL10-1 or empty vector at indicated time points during differentiation were measured by qPCR The mRNA values are depicted relative to mRNA in empty 3T3-L1 preadipocytes at day 0 of differentiation, which was arbitrarily defined as 1. Data represent mean ± S.E. of three technical replicates (error bars are too tiny to see). I, PPARγ and RING1B protein levels in FBXL10-overexpressing 3T3-L1 preadipocytes. Whole cell lysates from 3T3-L1 preadipocytes stably expressing FBXL10-1, FBXL10-2, or empty vector at indicated hours after induction for differentiation were subjected to immunoblot (IB) analysis. J, 3T3-L1 cells were reverse-transfected at a density of 3 × 105 cells/6-well plates with siRNAs for Fbxl10 (si-Fbxl10 #1 and #2) (20 nm) or the siRNA negative control (si-Ctr) using Lipofectamine RNAi MAX (Invitrogen). Knockdown efficiency of Fbxl10 in each 3T3-L1 preadipocyte was determined by qPCR. Data are presented as relative percentage of control siRNA-transfected cells (top panel). Cells were induced to differentiate with dexamethasone (DEX), DEX plus insulin (DI), or MDI and were stained with ORO at day 8 of differentiation (bottom panel).
Chromatin Immunoprecipitation (ChIP)
The chromatin immunoprecipitation (ChIP) assays were performed as described previously (8, 9, 37) with the following modifications. For ChIP using anti-RING1B, cells were cross-linked with 1.5 mm ethylene glycol bis(succinimidylsuccinate) (Thermo Fisher Scientific) for 30 min at followed by second cross-linking by addition of 1% formaldehyde for 10 min. For ChIP using anti-H2Aub, anti-H3K27me3, and anti-V5 antibodies, cells were cross-linked with 1% formaldehyde for 10 min. For both cross-linking conditions, fixations were stopped by adding 0.2 m glycine. After cross-linking, nuclear pellets were prepared and resuspended in 2 ml of lysis buffer (25 mm Tris-HCl (pH 8.0), 3 mm EDTA, 0.2% SDS, 0.9% Triton X-100, and 133 mm NaCl), and chromatin DNA was sonicated to generate a 0.5-kb average fragment (for anti-H2Aub, anti-H3K27me3, and anti-V5-tag) to a 2-kb average fragment (for anti-RING1B). ChIP was performed using anti-RING1B (clone D22F2; Cell Signaling Technology), anti-H3K27me3 (catalog no. 07449, Millipore), anti-V5 (R960-25; Invitrogen), anti-H2Aub (catalog no. 05–678, clone E6C5, Millipore), or control IgG. Prewashed magnetic Dynabeads® (Invitrogen) were incubated with each antibody in 500 μl of buffer A (PBS containing 0.01% Tween 20) for 1 h by wheel rotating at room temperature. Subsequently, sonicated cross-linked nuclear lysates (350 μg in 200 μl buffer A) were added and incubated overnight at 4 °C by wheel rotating. For H2Aub ChIP, anti-H2Aub was incubated overnight at 4 °C by wheel rotating; subsequently, anti-mouse IgM antibody preincubated with Dynabeads® was added and then incubated for 3 h. DNA was purified using QIAquick PCR purification kit (Qiagen, Valencia, CA), and the concentration was measured by Qubit® double-stranded DNA high sensitivity assay kit (Invitrogen).
qPCR, ChIP-qPCR, ChIP-seq, and Transcriptome Microarray Analysis
These procedures were described previously (8, 9, 41, 42). ChIP sequencing was done with an Ilumina/Solexa sequencer. qPCR was carried out in 384-well plates using ABI PRISM 7900HT sequence detection system (Applied Biosystems/Invitrogen). All reactions were done in triplicate. All primer sequences used in this article are listed in Tables 1 and 2. For transcriptome analysis of 3T3-L1 preadipocytes, GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA) was used as described previously.
TABLE 1.
Primers for qPCR
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Cdk1 | 5′-AAACTGGCTGATTTCGGCC-3′ | 5′-AGCAACACTTCTGGAGATCGGT-3′ |
| Cdk2 | 5′-AATTCATGGATGCCTCTGCTCT-3′ | 5′-CACGGTGAGAATGGCAGAA-3′ |
| Cdk4 | 5′-CCTGCCGGTTGAGACCAT-3′ | 5′-AGAAAATCCAGGCCGCTTAG-3′ |
| Cullin1 | 5′-CCCTGGTGATGTCAGCATTCA-3′ | 5′-GGCCATTTTGGTAACCGCA-3′ |
| Cyclin A2 (Ccna2) | 5′-TGTGAAGATGCCCTGGCTTT-3′ | 5′-CATGTGGTGATTCAAAACTGCC-3′ |
| Cyclin B1 (Ccnb1) | 5′-TTAGGCTGCTTCAGGAGACCAT-3′ | 5′-TACACCGACCAGCTGTAGCATC-3′ |
| Cyclin D1 (Ccnd1) | 5′-GCCGAGAAGTTGTGCATCTACA-3′ | 5′-TGTTCACCAGAAGCAGTTCCATT-3′ |
| Cyclin E1 (Ccne1) | 5′-TCTCCTCACTGGAGTTGATGCA-3′ | 5′-AACGGAACCATCCATTTGACA-3′ |
| FBXL10-1 (Kdm2b) | 5′-ATGGAGGCAGAGAAAGAC-3′ | 5′-CACGTCCGACAAGTCTTCGTT-3′ |
| FBXL10-2 (Kdm2b) | 5′-GCCGAGGACGACGACTATGAA-3′ | 5′-CAAAAGTGGCACTCTCCACA-3′ |
| FBXL10-all subtypes | 5′-TTGCTCAAGGAGCAGAAGATGAAC-3′ | 5′-GAGGCTCGCTTGCGTCCA-3′ |
| p15Ink4b (Cdkn2b) | 5′-GATCCCAACGCCCTGAA-3′ | 5′-GGCGCAGTTGGGTTCTG-3′ |
| p16Ink4a (Cdkn2a) | 5′-GCTCTGGCTTTCGTGAACATG-3′ | 5′-CGTGAACGTTGCCCATCAT-3′ |
| p21Cip1 (Cdkn1a) | 5′-TCTCAGGGCCGAAAACG-3′ | 5′-AATCTGCGCTTGGAGTGATAGA-3′ |
| p27Kip1 (Cdkn1b) | 5′-TGGACCAAATGCCTGACTCGT-3′ | 5′-GCCCTTTTGTTTTGCGAAGAAG-3′ |
| Pparg1 | 5′-CTGCGTAACTGACAGCCTAAC-3′ | 5′-ACTTGGTCACTCTCCGTCCT-3′ |
| Pparg2 | 5′-CTGGGAGATTCTCCTGTTGACC-3′ | 5′-CCTTGCAGCAACATCAGGAA-3′ |
| Ppib | 5′-GCAAAGTTCTAGAGGGCATGG-3′ | 5′-CAGCTGCTTAGAGGGATGAGG-3′ |
| Ring1b (Rnf2) | 5′-GTATGAAGCGCATCAGGAAAGG-3′ | 5′-CGCTGTAATCTGTTCATGGCCT-3′ |
| Skp1a | 5′-CTCCTCCTCCTGAGGATGATG-3′ | 5′-GGCCACAGTCTTGCATGTGAC-3′ |
TABLE 2.
Primers for ChIP-qPCR
| Amplified region | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Ppib (+4.0 kb) | 5′-CTCACCCCAACTAGTCTAATCC-3′ | 5′-GTGACACACAGTGACTAACTTCC-3′ |
| Cdk1 (TSS) | 5′-CGAGTGCCAGCAGTTTCAAA-3 | 5′-TCTACGTGCAATCGGATTGG-3 |
| Pparg1 (+0.2 kb) | 5′-CTAGGTGACTGGGTCTCCGCT-3′ | 5′-AGTCGAGACAGGGTTAGGCTGT-3′ |
| Pparg1 (+19 kb) | 5′-ACCTGTAGAGGGCAATAGTAG-3′ | 5′-TGATCTGAGTGGGCTCCTG-3′ |
| Pparg2 (TSS) | 5′-CTGGGAGATTCTCCTGTTGACC-3′ | 5′-CCTTGCAGCAACATCAGGAA-3′ |
| Hoxc8 (+5.7kb) | 5′-CGTCGTCCCATTACCGAAAT-3′ | 5′-TTTCGGAAGCCACTTTGATTG-3′ |
Computational Data Analysis
For ChIP-seq data processing, all bound DNA fragments were mapped to UCSC build mm9 (NCBI Build 37) assembly of the mouse genome by the mapping program ELAND based on the 5′-side 36-bp sequences. To identify ChIP-seq-enriched regions, regions of ChIP-seq that were enriched over the background were identified with SICER (43, 44) using the default values for the parameters: window size, 200 bp; gap size, 400; E-value threshold, 100. To annotate binding sites of V5-FBXL10 and RING1B, the closest genes and the intragenic binding sites were assigned to those genes. The closest gene is defined as follows: if binding sites of V5-FBXL10 or RING1B lie between gene A and B, and gene A is closer, V5-FBXL10 or RING1B assigns to gene A. The distance from the gene is not counted. Even if two genes are very far away from each other and the binding sites and gene A is very far, this rule is still applied. For gene ontology analysis, we used DAVID bioinformatics resources (david.abcc.ncifcrf.gov) (45).
Data Access
Microarray and ChIP-Seq data were deposited in the Gene Expression Omnibus (GEO) database with accession numbers GSE64154 and GSE64312, respectively.
Animal Experiments
All animal work was conducted according to the institutional guidelines. C57BL/6J mice (10 weeks of age) were purchased from Charles River and housed in a temperature-controlled (23 °C) facility with a 12-h light/dark cycle (09:00 to 21:00). Mice were fed with a normal chow diet (CLEA Japan) or a high fat diet (HFD) ad libitum for 8 weeks and then sacrificed for the total RNA extraction from epididymal white adipose tissues.
Statistical Analysis
All data are presented as mean ± S.E. Student's t test was performed for the comparison of two groups. A value of p < 0.05 was considered significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
FBXL10 Isoform Expression during 3T3-L1 Differentiation
To investigate further the epigenetic cues that mediate adipogenesis, we examined expression of histone modification enzyme genes in 3T3-L1 preadipocytes that were treated for 8 days with a hormone mixture containing MDI to induce differentiation. In a companion culture, we also induced differentiation with MDI in the presence of exogenous Wnt3a to inhibit adipogenesis (8, 46). Gene expression analyses by oligonucleotide microarray revealed that Fbxl10 shows a unique regulation pattern; its expression increased up to 4-fold over basal levels, peaked at 48 h, and then rapidly declined (Fig. 1A). This induction was blunted by Wnt3a treatment that keeps preadipocytes in a quiescent state (Fig. 1A). FBXL10 is a JmjC domain containing histone demethylase and contains several domains in addition to the NH2-terminal JmjC domain. These include a CXXC zinc finger domain, a PHD finger, the signature F-box, and eight leucine-rich repeats (Fig. 1, B and D). The mouse Fbxl10 gene encodes three isoforms due to the use of alternative promoters and splice sites (Fig. 1, B and D). Isoforms 1 and 3 (designated FBXL10-1 and -3, respectively) both contain all of the identified domains above, but they are encoded from transcripts that have unique exons at their 5′ end and differ in their 5′-UTR and amino-terminal protein sequences (Fig. 1, B and D). Isoform 2 (designated FBXL10-2) lacks the JmjC domain and has a distinct amino terminus when compared with isoforms 1 and 3 (Fig. 1, B and D). qPCR analyses revealed that about 80% of Fbxl10 mRNA in 3T3-L1 preadipocytes corresponds to isoform 2 (Fig. 1, C and D). Fig. 1C shows isoform 2 levels increase during first 48 h of differentiation but abruptly decrease afterward. Isoform 1 mRNA levels were constantly low during adipogenesis, and the estimated isoform 3 mRNA level, which was calculated by subtracting isoforms 1 and 2 from all Fbxl10 subtypes mRNA levels, was also low. These data indicate that isoform 2 is the predominant Fbxl10 isoform and is induced during the early phase of differentiation of 3T3-L1 preadipocytes.
Fbxl10 Levels Are Elevated in Mouse Models of Obesity
To see whether Fbxl10 isoforms show altered expression in the mouse model of obesity, we examined adipose tissue from C57BL/6J mouse fed a high fat diet. We randomized 6-week-old C57BL/6J littermates and fed them either normal or HFD for 8 weeks, extracted RNA from epididymal fat depots, and examined the expression of Fbxl10 by qPCR. Similar to 3T3-L1 cells, isoform 2 (FBXL10-2) was the predominant isoform expressed in white adipose tissue, and it was induced by more than 20-fold in HFD-fed mice relative to normal chow diet-fed mice (Fig. 1E). These data suggest that FBXL10 signals may be germane to fat differentiation and possibly pathophysiology in vivo as well.
FBXL10 Inhibits Adipocyte Differentiation
To examine whether FBXL10 has a function in regulating adipocyte differentiation, we examined the effects of expression of FBXL10-1 and -2 isoforms by retroviral transduction during 3T3-L1 preadipocyte differentiation using a retroviral vector with a very weak long terminal repeat (LTR) promoter. Following transduction, 3T3-L1 preadipocyte were treated with the MDI mixture to induce differentiation. In this experiment, FBXL10 blocked the morphological changes associated with adipogenesis, most notably through a marked decrease in accumulation of lipids as stained by ORO (Fig. 1F). This morphological effect was accompanied by a profound decline in the induction of the key adipogenic transcription factors Cebpa and Pparg together with a PPARγ target gene Cd36 in FBXL10-transduced 3T3-L1 preadipocytes (Fig. 1G). Additional gene expression analysis by qPCR using isoform-specific primers for Pparg showed both Pparg1 and Pparg2 mRNA levels were suppressed in FBXL10-expressing cells, which was associated with reduced protein levels (Fig. 1, H and I). However, Cebpb and Cebpd, two critical adipogenic genes whose expressions are induced in the early phase of adipocyte differentiation (∼24 h after induction), were induced normally suggesting that FBXL10 acts on or upstream of Pparg and Cebpa in a transcriptional cascade but independent of Cebpb and Cebpd (Fig. 1G). These results demonstrate that the ectopic expression of FBXL10 inhibits adipogenesis, and this inhibition does not require its JmjC catalytic domain suggesting that FBXL10 repression of adipogenesis is not mediated by its demethylation activity. To complement the sufficiency results, we undertook necessity tests by predicting that a decrease in FBXL10 expression would enhance differentiation of 3T3-L1 preadipocytes (Fig. 1J). Because the MDI mixture is very efficient in converting 3T3-L1 preadipocytes into mature adipocytes, we reasoned that it would be difficult to assess an increase in adipogenesis using MDI. Therefore, we also examined induction with dexamethasone only or dexamethasone plus insulin to induce differentiation, either of which is much more inefficient by themselves for inducing differentiation than the complete MDI mixture. A control siRNA or two independent siRNA oligonucleotide sequences that target Fbxl10 were separately transfected into 3T3-L1 cells, which were then treated with dexamethasone, dexamethasone plus insulin, or MDI and cultured for 8 days. Treatment with either Fbxl10 siRNA decreased Fbxl10 mRNA levels (Fig. 1J, top); however, the difference in lipid accumulation was very subtle between control and Fbxl10 knockdown 3T3-L1 cells (Fig. 1J, bottom). In agreement with this, the level of Pparg mRNA was not elevated in 3T3-L1 cells transfected with two independent siRNAs that target Fbxl10 compared with those transfected with a control siRNA as determined by microarray analysis (data not shown). This suggests that induction of Fbxl10 mRNA expression itself may be to fine-tune some physiological responses during adipocyte differentiation and may not be actively suppressing adipogenesis (see under “Discussion”).
FBXL10 Transduction Results in G1 Arrest by Blocking Cyclin-dependent Entry into S Phase during MCE
Because MCE is an important step that takes place at the early stage during differentiation of 3T3-L1 preadipocytes to adipocytes (3), we examined the effect of ectopic expression of FBXL10 on MCE of preadipocytes. The cell proliferation before reaching confluency did not differ between empty and FBXL10 vector-transduced preadipocytes (Fig. 2A). After reaching confluence when cells were induced for differentiation by MDI, the number of control 3T3-L1 preadipocytes was increased ∼4-fold; however, FBXL10 consistently reduced this by 2-fold indicating that MCE was incomplete (Fig. 2B). Bromodeoxyuridine (BrdU) pulse labeling followed by flow cytometry analysis revealed that the percentage of S phase cells at 48 h of induction was dramatically reduced when constructs corresponding to the two naturally occurring FBXL10 isoforms 1 and 2 were transduced into preadipocytes relative to those of control preadipocytes (Fig. 2, C and D). This analysis also revealed that the percentage of M phase cells was reduced at 48 h after induction relative to control (Fig. 2E). These data indicate that MCE of 3T3-L1 preadipocytes transduced with FBXL10 was incomplete under conditions where control 3T3-L1 preadipocytes underwent two full rounds of MCE. Consistently, analysis of representative cell cycle-regulatory protein genes by oligonucleotide microarray and qPCR analyses revealed that the expression of Cdk2 and cyclin E1 genes, whose gene products mediate G1 to S transition, cyclin A2 and B1 genes, which mediate entry of G2 to M phase, and Cdk1 gene, whose product mediates both G1/S and G2/M transitions, were reduced by 40–80% at 48 h in FBXL10-transduced 3T3-L1 preadipocytes as compared with control cells (Fig. 2F). Altogether, these data demonstrate that FBXL10 transduction leads to G1/S and also G2/M cell cycle arrest during MCE.
FIGURE 2.
Effects of FBXL10 expression on cell growth and mitotic clonal expansion of 3T3-L1 preadipocytes. A and B, effect of FBXL10 expression on cell growth of 3T3 L1 cells (A) and mitotic clonal expansion (B). 3T3-L1 preadipocyte overexpressions of FBXL10-1 (left panel) or FBXL10-2 (right panel) were plated at densities of 5 × 104 cells in 6-well plates, and cell numbers were counted every 24 h. To examine the effect of FBXL10 expression on mitotic clonal expansion, 2 days post-confluent transduced 3T3-L1 preadipocytes were induced for differentiation, and cell numbers were counted every 12 h. C and D, fluorescence-activated cell sorting analysis (FACS) performed with 3T3-L1 preadipocytes overexpressing FBXL10-1 (C) or FBXL10-2 (D) in comparison with those overexpressing empty vector as described under “Experimental Procedures.” Purple indicates cells at G0/G1 phase; green indicates cells at S phase; blue indicates cells at G2/M phase (C and D, left panels). The ratios of the cells at S phase of FBXL10-1 (C, right panel) or FBXL10-2 (D, right panel)-overexpressed 3T3-L1 preadipocytes during differentiation are shown. Data represent ± S.E. of three technical replicates. **, p < 0.01 compared with control. E, cell cycle analysis using FACS with 3T3-L1 preadipocytes overexpressing FBXL10-1 (left panel) or FBXL10-2 (right panel) at day 2 of differentiation, as described in B and C, showing the ratio of the cells at G2/M phase. Data represent mean ± S.E. of three technical replicates. *, p < 0.05 compared with control. F, expression of genes involved in the cell cycle in FBXL10-1-transduced 3T3-L1 preadipocytes. At 48 h of induction, cells were harvested for isolation of total RNA and used for reverse transcription and qPCR. Mouse cyclophilin mRNA was used as the invariant control. The mRNA values are depicted relative to mRNA in empty vector transduced 3T3-L1 preadipocytes, which are arbitrarily defined as 1. Each bar represents mean ± S.E. of three technical replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with control.
F-box and LRRs Domain-dependent Inhibition of Adipogenesis by FBXL10
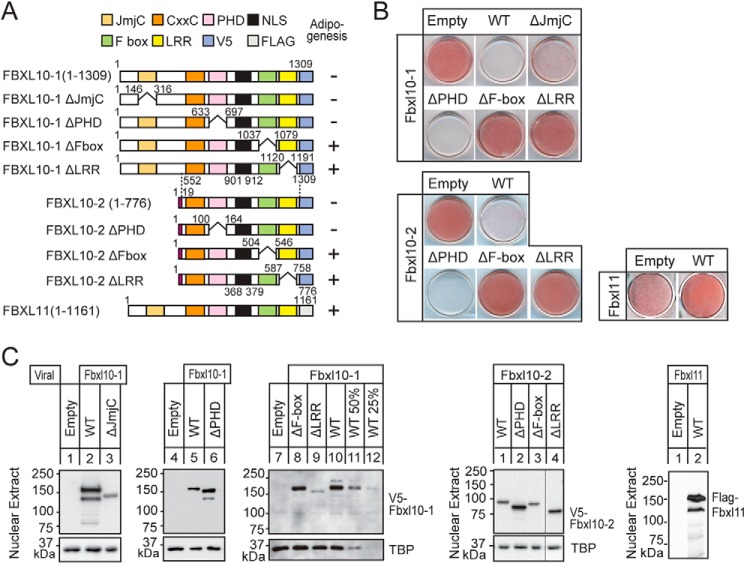
Because the inhibition of adipogenesis did not require the JmjC domain, the mechanism is independent of its demethylase activity. To investigate the domains of FBXL10 required for the inhibition of adipogenesis, we expressed a series of FBXL10 mutants in 3T3-L1 preadipocytes by retroviral transduction, and we compared their ability to influence adipogenesis following treatment with the MDI differentiation mixture. Engineered mutants lacking the JmjC or the PHD finger domains still resulted in the inhibition of adipogenesis, whereas the loss of either the F-box or LRRs resulted in no inhibition of adipogenesis by both FBXL10-1 and -2 (Fig. 3, A and B). In a companion experiment, we also examined the effects of overexpression of FBXL11 (also referred to as KDM2A or JHDM1A), which is the closest related paralog of FBXL10/KDM2B (Fig. 3, A and B). Transduction of FBXL11 into 3T3-L1 preadipocytes did not show any inhibitory effect on adipogenesis. Protein levels of the ΔF-box and WT-FBXL10 were comparable for both isoforms 1 and 2, so their inability to influence adipogenesis by ΔF-box FBXL10-1 and -2 could not be secondary to effects on protein expression. For loss of LRRs, although the protein levels of ΔLRRs and WT FBXL10-2 were comparable, that of ΔLRRs of FBXL10-1 was ∼50% of WT-FBXL10, so we cannot rule out the possibility that LRR deletion reduced protein stability and resulted in the loss of the inhibitory effect of FBXL10 on adipogenesis (Fig. 3C). These data show that the repression of adipogenesis by FBXL10-1 and -2 requires both F-box and LRRs domains and confirms that it is independent of JmjC. In agreement with this, FBXL10-2, the naturally occurring JmjC domain lacking variant isoform, is the most abundant FBXL10 isoform in both 3T3-L1 adipocytes and white adipose tissues of mice.
FIGURE 3.
F-box and LRR-dependent inhibition of adipogenesis by FBXL10. A, schematic representations of wild-type FBXL10-1 and -2 isoforms and their different mutants and FBXL11. B, 3T3-L1 preadipocytes overexpressing V5-tagged wild-type FBXL10-1, -2, their mutants, and FLAG-tagged FBXL11 were induced for differentiation with MDI mixture as described, and ORO was performed at day 8. C, expressions of transduced proteins were assessed by immunoblot analysis with anti-V5 or anti-FLAG. In lanes 9–12, sequentially diluted lysates from 3T3-L1 preadipocytes expressing full-length FBXL10-1 are shown as the standard to estimate the protein level of ΔLRR-FBXL10-1. Equal loading of the proteins were confirmed by the detection of nuclear protein TATA-binding protein (TBP).
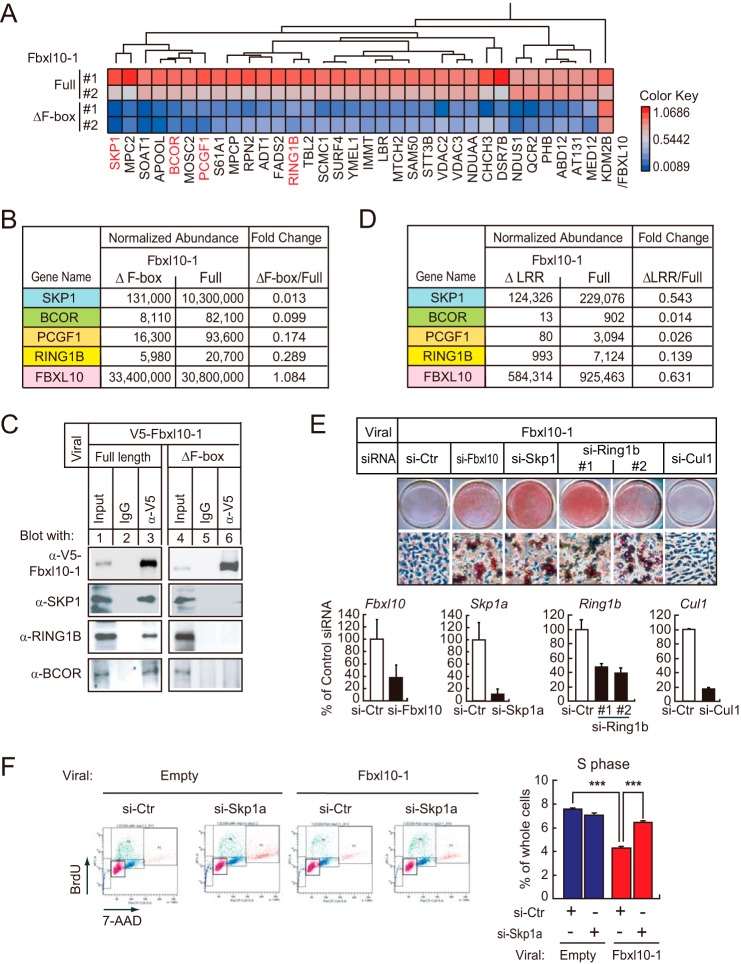
Identification of F-box and LRR-dependent PRC1 Complex
To investigate the mechanism for repression through the F-box and LRRs domains, we performed a comprehensive proteomics analysis to characterize possible proteins that might interact with FBXL10 through these domains. 3T3-L1 preadipocytes were transduced with retroviruses expressing full-length FBXL10-1 and its mutants lacking either F-box (ΔF-box) or LRRs (ΔLRRs) tagged with the V5 epitope, and we compared the profiles of interacting proteins following immunoprecipitation with the V5 antibody by mass spectrometry. The analysis between the ΔF-box mutant and full-length FBXL10-1 yielded several peptide sequences that were present in the full-length FBXL10-1 immunoprecipitate and absent in the ΔF-box interacting sample (Fig. 4A). Proteins corresponding to these peptides include SKP1 (47), BCOR (27, 28), PcG Ring finger protein PCGF1, and E3 ubiquitin-protein ligase RING1B (Fig. 4B) (48). These proteins are part of the PRC1 repression complex suggesting that FBXL10 interacts with PRC1 through its F-box domain, and this conclusion is consistent with recent reports that identified FBXL10 as part of a noncanonical PRC1 complex in several different cell lines, including ES cells (26–29, 49).
FIGURE 4.
FBXL10/KDM2B forms a noncanonical PRC1 complex in 3T3-L1 preadipocytes containing RING1B and SKP1 via F-box and LRRs. A–D, FBXL10 in 3T3-L1 associates with a variant PRC1 complex containing RING1B, BCOR/BCORL1, PCGF1, and SKP1. 3T3-L1 preadipocytes overexpressing either V5-tagged full-length FBXL10 (Full) or F-box deletion mutant (ΔF-box) were immunoprecipitated by anti-V5 and then subjected to mass spectrometry as described under “Experimental Procedures.” A, heat map representing identified protein whose abundance was 3-fold higher in full-length FBXL10 relative to that in ΔF-box (n = 2). B and D, abundance of peptides co-immunoprecipitated was normalized by ProgenesisTM software (Nonlinear Dynamics, Newcastle, UK) using those of the FBXL10 peptide peaks (LAGLDITDVSLR(2+), ASSLQTSPGSSSHLS(3+), IKESEGVVNDELPNC(3+)) as a control. B, normalized abundance of SKP1, BCOR, PCGF1, or RING1B co-immunoprecipitated with either full-length or ΔF-box FBXL10. C, immunoblot analysis verifying that FBXL10 interacts with RING1B, SKP1, and BCOR. Full-length or ΔF-box FBXL10-1 was immunoprecipitated by anti-V5 or mouse IgG cross-linked with Dynabeads protein G. Aliquots of protein were subjected to SDS-PAGE followed by immunoblot analysis using anti-V5, anti-SKP1 (#2156), anti-BCOR (SAB4502272), or anti-RING1B (#5694). D, identified proteins (SKP1, BCOR, PCGF1, and RING1B) that interact with FBXL10 dependently of LRRs domain of FBXL10. 3T3-L1 preadipocytes overexpressing V5-tagged full-length FBXL10 (Full) and LRR deletion mutant (ΔLRR) were immunoprecipitated with anti-V5 and then subjected to mass spectrometry and the abundance of peptides were normalized. E, 3T3-L1 preadipocyte-transduced FBXL10-1 was transfected with siRNA specifically targeting Fbxl10, Skp1, Ring1b, or Cul1 or control siRNA. Cells were induced to differentiate with MDI and were stained with ORO at day 8 of differentiation (top panel). Knockdown efficiency of Fbxl10, Skp1, Ring1b, and Cul1 in 3T3-L1 preadipocytes overexpressing FBXL10-1 at day 0 of differentiation was determined by qPCR. Data were presented as relative percentage of control siRNA-transfected cells (bottom panel). F, cell cycle analysis using FACS performed with FBXL10-1 overexpressed 3T3-L1 preadipocytes in comparison with those overexpressing empty vector transfected with control siRNA or siRNA targeting Skp1 at 48 h of induction by MDI mixture as described in the legends to Fig. 2, C and D (left panel). The ratio of the cells at S phase of cells at 48 h after induction are shown (right panel). Data represent S.E. of three technical replicates. ***, p < 0.001 compared with control.
F-box-dependent interactions of RING1B and SKP1 with FBXL10-1 in 3T3-L1 preadipocytes were further validated by co-immunoprecipitation and immunoblot analyses (Fig. 4C). The interaction between FBXL10-1 and PCGF1 was not clearly detected by this method (data not shown). Additional proteomic analysis comparing V5-tagged ΔLRR- and full-length FBXL10-1-interacting proteins also identified the same series of proteins (i.e. SKP1, BCOR, PCGF1, and RING1B) bound to FBXL10 in an LRR-dependent manner (Fig. 4D). These data demonstrated that F-box and LRRs are both required for interaction with PRC1.
Knockdown of Either SKP1 or RING1B Reversed the Inhibition of FBXL10 on Adipogenesis
SKP1 showed the highest number of peptides that matched in the co-immunoprecipitates of FBXL10, which were reduced in ΔF-box mutant and ΔLRR. A direct interaction between SKP1 and FBXL10 was also recently reported (50). If SKP1 is the responsible molecule for the F-box and LRR-mediated repression of adipogenesis, knockdown of SKP1 would rescue FBXL10-induced suppression of adipogenesis. Accordingly, we transfected siRNA designed to inactivate SKP1 into FBXL10-1-transduced preadipocytes, and we examined adipocyte differentiation. Retroviral transduction of FBXL10 inhibited the ability of the MDI mixture to induce differentiation, and inactivation of FBXL10 by siRNA transfection restored adipocyte differentiation as demonstrated by Oil Red O staining (Fig. 4E). Consistent with our hypothesis, specific knockdown of SKP1 reversed the inhibition of FBXL10 on adipogenesis (Fig. 4E). Similar results were also obtained by knockdown of RING1B (Fig. 4E) indicating that SKP1 and RING1B mediate the inhibitory effect of overexpressed FBXL10 on adipogenesis through the formation of a protein complex. This recovery of adipogenesis was associated with the recovery of G1 to S phase entry at the second round of MCE (Fig. 4F). Although SKP1 is a component of SCF ubiquitin ligase complex composed of SKP1, CUL1, and F-box proteins and contributes to cell cycle control through the ubiquitin proteasome pathway (47, 51), knockdown of CUL1 did not rescue the inhibition of adipogenesis by FBXL10 (Fig. 4E); in agreement with this, CUL1 was not identified in our proteomics analyses (Fig. 4A). Taken together, these data show that forced expression of FBXL10 inhibits 3T3-L1 preadipocyte differentiation through interaction with a noncanonical PRC1, at least in part, by regulating the second MCE.
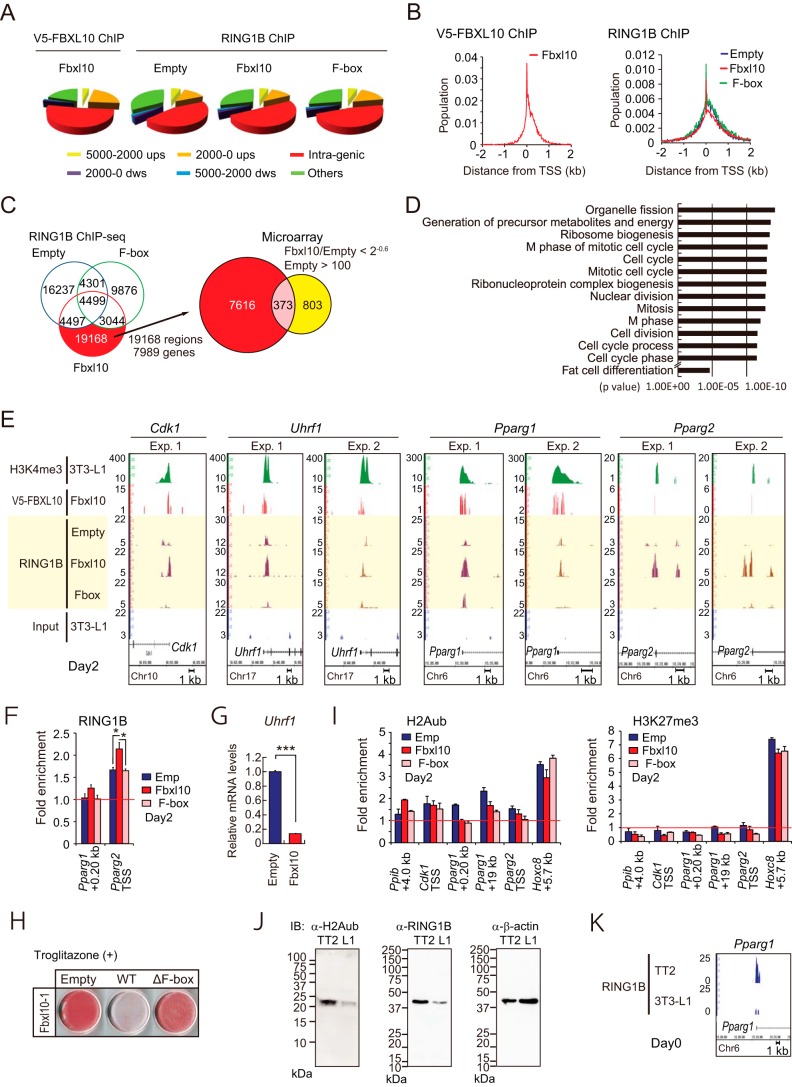
Genomic Localization of FBXL10 in 3T3-L1 Preadipocytes
To identify direct targets of FBXL10 and its associated noncanonical PRC1 complex, we performed ChIP-seq and global gene expression analysis. 3T3-L1 preadipocytes transduced with the retroviral vector for V5-tagged FBXL10 or its ΔF-box mutant were treated with the MDI mixture for 2 days, and ChIP-seq was conducted using anti-V5 tag or anti-RING1B antibodies. ChIP-seq peak calling by MACS showed V5-FBXL10 localized mostly on proximal promoter (22%) or intragenic regions (52%) (Fig. 5A, left panel). Peak calling analysis by SICER program for the RING1B-ChIP-seq identified 29,475, 31,111, and 21,720 regions as significant binding sites of RING1B in empty, FBXL10-1, and ΔF-box mutant vector-transduced 3T3-L1 preadipocytes, respectively. This analysis also revealed that RING1B localized mostly on the intragenic region (45%), intergenic region (33%), and proximal promoter (13%) (Fig. 5A, right panel). The sequence tag densities of both V5-FBXL10 and RING1B were mostly concentrated within the proximal regions of TSS (Fig. 5B).
FIGURE 5.
FBXL10 represses Cdk1, Uhrf1, Pparg1, and Pparg2 expression via F-box-dependent recruitment of RING1B. A and B, genome-wide distributions of binding sites of V5-FBXL10 in 3T3-L1 preadipocytes transduced with V5-FBXL10-1 (A) or those of RING1B in 3T3-L1 preadipocytes transduced with empty, wild type, or ΔF-box FBXL10 (B) as determined by ChIP-seq. ups, upstream; dws, downstream. C and D, Ring1B ChIP-seq and transcriptional microarray analysis performed in 3T3-L1 preadipocytes stably expressing wild-type (FBXL10) or F-box deletion (ΔF-box) FBXL10-1 or empty vector (Empty) at day 2 of differentiation. C, Venn diagram showing number of RING1B-binding sites identified with SICER in transduced 3T3-L1 preadipocytes (left panel). Full-length FBXL10-1-induced RING1B-binding sites (19168 sites) were annotated to 7,989 genes. The number of RING1B-binding genes depending on V5-tagged wild-type FBXL10-1 expression and the number of genes suppressing more than 2−0.6 in FBXL10-1 transduced cells are shown in the Venn diagram in the right panel. All transcripts with average difference call below 100 in empty 3T3-L1 preadipocytes were excluded. D, histogram displaying fold enrichment values for GO term analysis of the genes that are down-regulated more than 2−0.6 in FBXL10-1-overexpressing 3T3-L1 preadipocytes relative to those of control cells. E, ChIP-seq profiles for H3K4me3, V5-FBXL10, and RING1B on Cdk1, Uhrf1, Pparg1, and Pparg2 genomic regions. 3T3-L1 preadipocytes stably expressing wild-type (FBXL10) or F-box deletion mutant (ΔF-box) FBXL10-1, or empty vector (Empty) at day 2 of differentiation were subjected to ChIP-seq analysis. ChIP-seq using anti-RING1B antibody was performed twice (Exp. 1 and Exp. 2) and presented with ChIP-seq profiles for H3K4me3 and V5-FBXL10. Scale bars indicate 1 kb. F, ChIP-qPCR analysis of RING2B on the Pparg1 and Pparg2 genes using set of primers that amplifies TSS (as listed in Table 2) in 3T3-L1 preadipocytes at day 2. Data are normalized to precipitated DNA (fold enrichment). Error bars represent ± S.E. of three technical replicates. *, p < 0.05 compared with control. G, qPCR of Uhfr1 in 3T3-L1 preadipocytes stably expressing FBXL10-1 or empty vector. The mRNA values are depicted relative to mRNA in the control group (empty vector), which is arbitrarily defined as 1. Data represent ± S.E. of three technical replicates. ***, p < 0.001 compared with control. H, 3T3-L1 preadipocytes overexpressing wild type or ΔF-box FBXL10 were induced for differentiation with MDI and 1 μm troglitazone, and ORO was performed at day 8. I, ChIP-qPCR analysis of H2Aub (left panel) and H3K27me3 (right panel) on the Ppib, Cdk1, Pparg1, Pparg2, and HoxC8 genes using set of primers as listed in Table 2 in 3T3-L1 preadipocytes at day 2. Data are normalized to precipitated DNA (fold enrichment). Error bars represent ± S.E. of three technical replicates. J, immunoblot (IB) analysis of H2AK119ub (left panel) and RING1B (middle panel) in 3T3-L1 preadipocytes and TT2 ES cells using either anti-H2Aub (catalog no. 8240, Cell Signaling Technology) or anti-H3K27me3 (catalog no. 07449, Millipore) antibody. Equal loading of the proteins was confirmed by the detection of β-actin (ACTB; A5441) (right panel). K, ChIP-seq profiles of RING1B on PPARg1 genomic region in TT2 ES cells or 3T3-L1 preadipocytes (day 0). Scale bars indicate 1 kb.
Because inhibition of adipogenesis was dependent on the FBXL10 F-box domain, we sought to identify RING1B target genes whose recruitment was dependent on the expression of FBXL10 wild-type but not ΔF-box mutant. The Venn diagram in Fig. 5C, left panel, shows 19,168 RING1B binding regions (annotated to 7,989 genes) that met the above criteria (shown in red). A global gene expression analysis in empty and FBXL10 vector-transduced 3T3-L1 preadipocytes at 48 h of induction demonstrated that 1,176 genes were down-regulated by more than 2−0.6-fold by FBXL10 (Fig. 5C, right panel, Venn diagram). A gene ontology enrichment analysis revealed that many of these genes were associated with GO terms related to mitotic cell cycle-related genes (p < 10−11) (Fig. 5D). Of these, 373 genes overlapped with RING1B-binding sites (Fig. 5C, right panel, Venn diagram, and supplemental Table 1). These genes include cyclin-dependent kinase 1 (Cdk1), ubiquitin-like, containing PHD and RING finger domains, 1 (Uhrf1), and peroxisome proliferator activated receptor γ (Pparg) (supplemental Table 1). Cdk1 gene product is pivotal for the mammalian cell cycle that regulates both S phase and M phase (52, 53); Uhrf1 (also referred to as Icbp90 or Np95) gene product is reported to regulate G1/S transition (54, 55), and the Pparg1 and -2 gene product is a master regulator for adipogenesis. ChIP-seq and ChIP-qPCR analyses around these genes confirmed recruitment of RING1B to the TSS sites of the above genes (Fig. 5, E and F). Recruitment of RING1B at these genes in FBXL10-transduced 3T3-L1 preadipocytes was associated with reduction in expression of their mRNAs (Figs. 1H, 2F, and 5G). These data further indicate that Pparg1 and -2 genes are direct targets of RING1B recruited by forced expression of FBXL10 wild-type but not the ΔF-box mutant, which barely associated with RING1B (Fig. 4, B and C). These data suggest that Pparg1 and Pparg2 gene expressions were directly suppressed by FBXL10-RING1B containing variant PRC1. Consistent with this notion, the inhibitory effect of FBXL10 on adipogenesis was not rescued by agonistic activation of PPARγ by synthetic ligand troglitazone (56) in FBXL10-transduced 3T3-L1 preadipocytes (Fig. 5H). Note that this inhibition was restored by inactivation of RING1B by siRNA transfection (Fig. 4E).
RING1B recruitment to target genes by FBXL10 is known to cause monoubiquitination of H2AK119, which in turn recruits PRC2 for H3K27me3 modification (26, 29, 30). To examine whether FBXL10-RING1B complex also leads to H2AK119ub or H3K27me3 to repress transcriptions of Cdk1, Uhrf1, and Pparg1 and -2, we performed ChIP-qPCR using anti-H2Aub and anti-H3K27me3 antibodies in retrovirally transduced 3T3-L1 preadipocytes. Although we detected appreciable levels of H2A monoubiquitination and H3K27me3 modifications at the HoxC8 gene, we could hardly detect H2Aub or H3K27me3 modifications across Cdk1 and Pparg1 and -2 genes (Fig. 5I). In addition, forced expression of FBXL10 did not affect these histone modification levels (Fig. 5I). The total cellular level of H2AK119ub was much lower in 3T3-L1 preadipocytes as compared with TT2 ES cells (Fig. 5J). In agreement with this, the cellular RING1B protein levels and the recruitment of RING1B to Pparg regions were much higher in TT2 ES cells, which are not committed to preadipocytes, compared with those in 3T3-L1 preadipocytes (Fig. 5, J and K).
Taken together, we concluded that FBXL10 recruits the noncanonical PRC1 complex to directly repress Cdk1, Uhrf1, and Pparg that may account for the FBXL10-mediated inhibition of adipogenesis. The transcriptional repression may not be mediated by H2Aub or H3K27me3.
DISCUSSION
Mitotic clonal expansion is a critical step that takes place in 3T3-L1 adipocyte differentiation. In this study, we showed that FBXL10/KDM2B regulates adipogenesis via the formation of noncanonical PRC1 by regulating MCE and also Pparg genes expression. In 3T3-L1 preadipocyte differentiation, Fbxl10 gene expression is induced by 4-fold during the early phase of differentiation (first 48 h), which corresponds to the two rounds of MCE, and Fbxl10 expression is abruptly declined at the timing of the completion of second MCE, reciprocally, Pparg1 and -2 gene expression abruptly increases at this time.
Noncanonical Polycomb Recessive Complex 1-mediated Inhibition of MCE and Adipogenesis
When FBXL10 was overexpressed in 3T3-L1 preadipocytes, MCE was arrested at G1/S (and also G2/M) after the first MCE, and the cells did not undergo terminal differentiation. This inhibition was independent of the demethylase activity of FBXL10 because the JmjC domain deletion mutant of FBXL10 still had the ability to inhibit adipogenesis. Also, the major isoform of Fbxl10 in both 3T3-L1 cells and adipose tissue in vivo lacks the JmjC domain, and its expression is induced dramatically in diet-induced obese mice (Fig. 1, C and E). Intriguingly, FBXL10-induced inhibition of adipogenesis was depending on the F-box domain. The best understood role of F-box proteins is as a substrate recognition component of an SCF ubiquitin ligase complex that contain SKP1, CUL1, ROC1, and one F-box protein. Some F-box proteins such as FBXW7 contribute to cell cycle control through the ubiquitin-proteasome pathway (51). FBXL10 formed a complex with SKP1; however, it did not interact with CUL1 in 3T3-L1 preadipocytes consistent with previous studies performed in three other cell lines (50). In agreement with this result, the knockdown of CUL1 did not restore FBXL10-induced inhibition of adipocyte differentiation. Thus, we conclude that the inhibition of adipogenesis by FBXL10 is not mediated by through interacting with an SCF complex.
By contrast, our proteomics revealed that FBXL10 associated with SKP1, RING1B, and BCOR, and this required both the F-box and LRR domains. Other studies also showed that FBXL10, BCOR, and RING1B form a complex, which is recently referred to as a noncanonical PRC1 complex (26–28). Knockdown of either RING1B or SKP1 restored FBXL10-induced inhibition of adipogenesis, and in addition, this restoration of adipocyte differentiation was associated with the restoration of G1/S transition at the second MCE (Fig. 4, E and F). Thus, we conclude that forced expression of FBXL10 inhibited adipogenesis through the formation of noncanonical PRC1, at least in part, by arresting the cell cycle.
Noncanonical PRC1 Targets Pparg Genes in FBXL10-transduced Preadipocytes
We identified the adipogenic master regulator Pparg genes as a direct target of RING1B that may account for the inhibitory effect of FBXL10-PRC1 on adipogenesis. This finding is further supported by the observations that agonistic activation of PPARγ could not rescue the inhibitory effect of FBXL10 on adipogenesis (Fig. 5H). By contrast, knockdown of RING1B rescued this inhibition by FBXL10 indicating that the PRC1 complex directly represses Pparg gene transcription.
Our ChIP-seq analyses in 3T3-L1 preadipocytes demonstrated that the major binding sites of FBXL10 are intragenic. It has been demonstrated that FBXL10 preferentially binds to CpG islands (CGI) and co-localizes with RING1B on polycomb target genes in ES cells. Although some target genes are classified as non-CGI genes, they still contain CpG-rich sequences just below the strict threshold for classifying UCSC-annotated CpG islands (26, 29, 30). In agreement with this, our data showed that RING1B was recruited to TSS and intragenic sites within FBXL10 targets (i.e. Cdk1, Uhrf1, Pparg1, and -2) in WT-FBXL10-expressing 3T3-L1 preadipocytes but not in those overexpressing ΔF-box-FBXL10 mutant. Interestingly, this mutant does not associate with RING1B (Fig. 5E). These data support the previous reports that FBXL10 interacts with RING1B forming noncanonical PRC1 and targets to CpG-rich sequences in intragenic regions at the target loci.
It was previously reported that FBXL10 is highly expressed in ES cells and is down-regulated upon differentiation (19). In this context, FBXL10 enhances reprogramming, importantly, not through normal demethylation activity but through a noncanonical PRC1-mediated mechanism (31). Our data show that RING1B levels at the TSS of Pparg1 were pretty high in ES cells (Fig. 5K); by contrast, its recruitment in 3T3-L1 preadipocytes was hardly detectable. Because forced expression of FBXL10 in 3T3-L1 preadipocytes led to higher RING1B recruitment at the Pparg1 locus, which was accompanied by the inhibition of Pparg expression and adipocyte differentiation, we propose that FBXL10 may function to maintain pluripotency prior to the induction of adipogenesis. This notion is in agreement with the role of FBXL10 in maintaining pluripotency of ES/iPS cells (19, 31). These data also suggest that Pparg is a key target of FBXL10 and FBXL10-PRC1 to inhibit the commitment of ES cells to preadipocytes to maintain pluripotency.
We failed to detect H2Aub at this locus in transduced 3T3-L1 preadipocytes, although we cannot rule out the possibility that H2Aubs in 3T3-L1 preadipocytes were too low to be detected by commercially available current anti-H2Aub antibody. Recently, enzymatic activity-independent functions for histone modification enzymes have been reported where they can function as a scaffold for co-activator or repressor recruitment (57, 58). H2A ubiquitination plays an important role in PRC1-mediated silencing; however, how H2A ubiquitination influences repression is not fully understood. RING1B is associated with chromatin compaction at target loci independently of histone ubiquitination activity (58, 59). Therefore, other mechanisms, including chromatin compaction, are likely to mediate the FBXL10-PRC1-repressive capacity in 3T3-L1 preadipocytes.
Acknowledgments
We thank Dr. Toshio Kitamura for a retroviral packaging cell line and pMX plasmid and Drs. Nobuaki Yoshida and Manabu Ozawa for helpful advice. We also thank Aoi Uchida, Hiroko Kawasaki, and Akashi Taguchi-Izumi for technical assistance and other members of the Sakai laboratory for helpful discussions.
This work was supported by Grants-in-aid for Scientific Research (S) 22229009 and (B) 25291002, Grant-in-aid for Scientific Research on Innovative Areas 25126704, and a grant for Translational Systems Biology and Medicine Initiative from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

This article contains Table S1.
- MDI
- methylisobutylxanthine, dexamethasone, and insulin
- PPAR
- proliferator-activated receptor
- MCE
- mitotic clonal expansion
- HFD
- high fat diet
- qPCR
- quantitative real time PCR
- LRR
- leucine-rich repeat
- ORO
- Oil Red O
- TSS
- transcription start site
- 7-AAD
- 7-aminoactinomycin D
- PcG
- polycomb group
- BCOR
- BCL-6 co-repressor.
REFERENCES
- 1. Rosen E. D., MacDougald O. A. (2006) Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885–896 [DOI] [PubMed] [Google Scholar]
- 2. Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., Spiegelman B. M. (1999) Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3, 151–158 [DOI] [PubMed] [Google Scholar]
- 3. Tang Q. Q., Otto T. C., Lane M. D. (2003) Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacDougald O. A., Lane M. D. (1995) Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 64, 345–373 [DOI] [PubMed] [Google Scholar]
- 5. Rosen E. D., Spiegelman B. M. (2006) Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farmer S. R. (2006) Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okamura M., Inagaki T., Tanaka T., Sakai J. (2010) Role of histone methylation and demethylation in adipogenesis and obesity. Organogenesis 6, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okamura M., Kudo H., Wakabayashi K., Tanaka T., Nonaka A., Uchida A., Tsutsumi S., Sakakibara I., Naito M., Osborne T. F., Hamakubo T., Ito S., Aburatani H., Yanagisawa M., Kodama T., Sakai J. (2009) COUP-TFII acts downstream of Wnt/β-catenin signal to silence PPARγ gene expression and repress adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 5819–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N. S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., Kodama T., Aburatani H., Sakai J. (2009) The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell. Biol. 29, 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwartz Y. B., Pirrotta V. (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22 [DOI] [PubMed] [Google Scholar]
- 11. Simon J. A., Kingston R. E. (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 [DOI] [PubMed] [Google Scholar]
- 12. Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J., 3rd, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Müller J., Verrijzer P. (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150–158 [DOI] [PubMed] [Google Scholar]
- 14. Simon J. A., Lange C. A. (2008) Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647, 21–29 [DOI] [PubMed] [Google Scholar]
- 15. Cao R., Tsukada Y., Zhang Y. (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854 [DOI] [PubMed] [Google Scholar]
- 16. Francis N. J., Follmer N. E., Simon M. D., Aghia G., Butler J. D. (2009) Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell 137, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King I. F., Emmons R. B., Francis N. J., Wild B., Müller J., Kingston R. E., Wu C. T. (2005) Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 25, 6578–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon J. A., Kingston R. E. (2013) Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang G., He J., Zhang Y. (2012) Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat. Cell Biol. 14, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He J., Kallin E. M., Tsukada Y., Zhang Y. (2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat. Struct. Mol. Biol. 15, 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Janzer A., Stamm K., Becker A., Zimmer A., Buettner R., Kirfel J. (2012) The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology, and the metabolome of fibroblasts. J. Biol. Chem. 287, 30984–30992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. (2007) JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450, 309–313 [DOI] [PubMed] [Google Scholar]
- 23. Ge R., Wang Z., Zeng Q., Xu X., Olumi A. F. (2011) F-box protein 10, an NF-κB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 18, 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konuma T., Nakamura S., Miyagi S., Negishi M., Chiba T., Oguro H., Yuan J., Mochizuki-Kashio M., Ichikawa H., Miyoshi H., Vidal M., Iwama A. (2011) Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp. Hematol. 39, 697–709.e695 [DOI] [PubMed] [Google Scholar]
- 25. Fukuda T., Tokunaga A., Sakamoto R., Yoshida N. (2011) Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol. Cell. Neurosci. 46, 614–624 [DOI] [PubMed] [Google Scholar]
- 26. Farcas A. M., Blackledge N. P., Sudbery I., Long H. K., McGouran J. F., Rose N. R., Lee S., Sims D., Cerase A., Sheahan T. W., Koseki H., Brockdorff N., Ponting C. P., Kessler B. M., Klose R. J. (2012) KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 1, e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sánchez C., Sánchez I., Demmers J. A., Rodriguez P., Strouboulis J., Vidal M. (2007) Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell. Proteomics 6, 820–834 [DOI] [PubMed] [Google Scholar]
- 28. Gearhart M. D., Corcoran C. M., Wamstad J. A., Bardwell V. J. (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu X., Johansen J. V., Helin K. (2013) Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 49, 1134–1146 [DOI] [PubMed] [Google Scholar]
- 30. Blackledge N. P., Farcas A. M., Kondo T., King H. W., McGouran J. F., Hanssen L. L., Ito S., Cooper S., Kondo K., Koseki Y., Ishikura T., Long H. K., Sheahan T. W., Brockdorff N., Kessler B. M., Koseki H., Klose R. J. (2014) Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He J., Shen L., Wan M., Taranova O., Wu H., Zhang Y. (2013) Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 15, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsui T., Leung D., Miyashita H., Maksakova I. A., Miyachi H., Kimura H., Tachibana M., Lorincz M. C., Shinkai Y. (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 [DOI] [PubMed] [Google Scholar]
- 33. Watanabe-Okochi N., Kitaura J., Ono R., Harada H., Harada Y., Komeno Y., Nakajima H., Nosaka T., Inaba T., Kitamura T. (2008) AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood 111, 4297–4308 [DOI] [PubMed] [Google Scholar]
- 34. Kitagawa J., Hara T., Tsurumi H., Kanemura N., Kasahara S., Shimizu M., Moriwaki H. (2010) Cell cycle-dependent priming action of granulocyte colony-stimulating factor (G-CSF) enhances in vitro apoptosis induction by cytarabine and etoposide in leukemia cell lines. J. Clin. Exp. Hematopathol. 50, 99–105 [DOI] [PubMed] [Google Scholar]
- 35. Ohguchi H., Tanaka T., Uchida A., Magoori K., Kudo H., Kim I., Daigo K., Sakakibara I., Okamura M., Harigae H., Sasaki T., Osborne T. F., Gonzalez F. J., Hamakubo T., Kodama T., Sakai J. (2008) Hepatocyte nuclear factor 4α contributes to thyroid hormone homeostasis by cooperatively regulating the type 1 iodothyronine deiodinase gene with GATA4 and Kruppel-like transcription factor 9. Mol. Cell. Biol. 28, 3917–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakai J., Duncan E. A., Rawson R. B., Hua X., Brown M. S., Goldstein J. L. (1996) Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 37. Iguchi H., Ikeda Y., Okamura M., Tanaka T., Urashima Y., Ohguchi H., Takayasu S., Kojima N., Iwasaki S., Ohashi R., Jiang S., Hasegawa G., Ioka R. X., Magoori K., Sumi K., Maejima T., Uchida A., Naito M., Osborne T. F., Yanagisawa M., Yamamoto T. T., Kodama T., Sakai J. (2005) SOX6 attenuates glucose-stimulated insulin secretion by repressing PDX1 transcriptional activity and is down-regulated in hyperinsulinemic obese mice. J. Biol. Chem. 280, 37669–37680 [DOI] [PubMed] [Google Scholar]
- 38. Iguchi H., Urashima Y., Inagaki Y., Ikeda Y., Okamura M., Tanaka T., Uchida A., Yamamoto T. T., Kodama T., Sakai J. (2007) SOX6 suppresses cyclin D1 promoter activity by interacting with β-catenin and histone deacetylase 1, and its down-regulation induces pancreatic beta-cell proliferation. J. Biol. Chem. 282, 19052–19061 [DOI] [PubMed] [Google Scholar]
- 39. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 40. Daigo K., Kawamura T., Ohta Y., Ohashi R., Katayose S., Tanaka T., Aburatani H., Naito M., Kodama T., Ihara S., Hamakubo T. (2011) Proteomic analysis of native hepatocyte nuclear factor-4α (HNF4α) isoforms, phosphorylation status, and interactive cofactors. J. Biol. Chem. 286, 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanaka T., Tahara-Hanaoka S., Nabekura T., Ikeda K., Jiang S., Tsutsumi S., Inagaki T., Magoori K., Higurashi T., Takahashi H., Tachibana K., Tsurutani Y., Raza S., Anai M., Minami T., Wada Y., Yokote K., Doi T., Hamakubo T., Auwerx J., Gonzalez F. J., Nakajima A., Aburatani H., Naito M., Shibuya A., Kodama T., Sakai J. (2014) PPARβ/δ activation of CD300a controls intestinal immunity. Sci. Rep. 4, 5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mimura I., Nangaku M., Kanki Y., Tsutsumi S., Inoue T., Kohro T., Yamamoto S., Fujita T., Shimamura T., Suehiro J., Taguchi A., Kobayashi M., Tanimura K., Inagaki T., Tanaka T., Hamakubo T., Sakai J., Aburatani H., Kodama T., Wada Y. (2012) Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell. Biol. 32, 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zang C., Schones D. E., Zeng C., Cui K., Zhao K., Peng W. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoang S. A., Xu X., Bekiranov S. (2011) Quantification of histone modification ChIP-seq enrichment for data mining and machine learning applications. BMC Res. Notes 4, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 46. Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., MacDougald O. A. (2000) Inhibition of adipogenesis by Wnt signaling. Science 289, 950–953 [DOI] [PubMed] [Google Scholar]
- 47. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 48. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 49. Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., Reinberg D. (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koyama-Nasu R., David G., Tanese N. (2007) The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat. Cell Biol. 9, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 51. Nakayama K. I., Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 52. Santamaría D., Barrière C., Cerqueira A., Hunt S., Tardy C., Newton K., Cáceres J. F., Dubus P., Malumbres M., Barbacid M. (2007) Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815 [DOI] [PubMed] [Google Scholar]
- 53. Hochegger H., Dejsuphong D., Sonoda E., Saberi A., Rajendra E., Kirk J., Hunt T., Takeda S. (2007) An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J. Cell Biol. 178, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arima Y., Hirota T., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H. (2004) Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells 9, 131–142 [DOI] [PubMed] [Google Scholar]
- 55. Bonapace I. M., Latella L., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P. P. (2002) Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 157, 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kliewer S. A., Willson T. M. (1998) The nuclear receptor PPARγ–bigger than fat. Curr. Opin. Genet. Dev. 8, 576–581 [DOI] [PubMed] [Google Scholar]
- 57. Bittencourt D., Wu D. Y., Jeong K. W., Gerke D. S., Herviou L., Ianculescu I., Chodankar R., Siegmund K. D., Stallcup M. R. (2012) G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U.S.A. 109, 19673–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eskeland R., Leeb M., Grimes G. R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A. I., Wutz A., Bickmore W. A. (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Endoh M., Endo T. A., Endoh T., Isono K., Sharif J., Ohara O., Toyoda T., Ito T., Eskeland R., Bickmore W. A., Vidal M., Bernstein B. E., Koseki H. (2012) Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 8, e1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]