Background: Skeletal muscle atrophy is a condition of cellular stress leading to muscle mass loss, and involving the up-regulation of specific genes, known as atrogenes.
Results: The expression of the histone deacetylase 6 (HDAC6) is up-regulated by FoxO3 during skeletal muscle atrophy and its inactivation protects against atrophy.
Conclusion: HDAC6 is an atrogene.
Significance: HDAC6 acts downstream of FoxO3a in stress response.
Keywords: FOXO, Histone Deacetylase 6 (HDAC6), Muscle Atrophy, Proteasome, Skeletal Muscle, Ubiquitin, MAFbx
Abstract
Skeletal muscle atrophy is a severe condition of muscle mass loss. Muscle atrophy is caused by a down-regulation of protein synthesis and by an increase of protein breakdown due to the ubiquitin-proteasome system and autophagy activation. Up-regulation of specific genes, such as the muscle-specific E3 ubiquitin ligase MAFbx, by FoxO transcription factors is essential to initiate muscle protein ubiquitination and degradation during atrophy. HDAC6 is a particular HDAC, which is functionally related to the ubiquitin proteasome system via its ubiquitin binding domain. We show that HDAC6 is up-regulated during muscle atrophy. HDAC6 activation is dependent on the transcription factor FoxO3a, and the inactivation of HDAC6 in mice protects against muscle wasting. HDAC6 is able to interact with MAFbx, a key ubiquitin ligase involved in muscle atrophy. Our findings demonstrate the implication of HDAC6 in skeletal muscle wasting and identify HDAC6 as a new downstream target of FoxO3a in stress response. This work provides new insights in skeletal muscle atrophy development and opens interesting perspectives on HDAC6 as a valuable marker of muscle atrophy and a potential target for pharmacological treatments.
Introduction
Skeletal muscle atrophy results in a massive muscle mass loss and it is characterized by a decrease in the size of pre-existing muscle fibers. Muscle atrophy can occur as a result of simple inactivity and aging or can be associated with various pathologies such as myopathies, hyperthyroidism, AIDS, or cancer (1). Throughout the past 30 years, the prevention of muscle wasting has been a major issue both due to muscle atrophy being associated with multiple pathologies and to its tendency to severely impair the daily life of afflicted patients (2). Among the effectors of skeletal muscle atrophy, massive protein degradation by the ubiquitin-proteasome pathway and autophagy (3) play a pivotal role. Significant advances have been made in the understanding of muscle wasting through the identification of specific factors involved in the process and with the identification of common pathways shared by all types of muscle atrophy. The genes encoding such factors are referred to as atrogenes. The first to be identified were the E3 ubiquitin ligases MAFbx/Atrogin-1 and MuRF1. The expression of these two genes is up-regulated in all models of muscle atrophy, and their inactivation significantly reduces muscles wasting (4, 5).
The up-regulation of these atrogenes during muscle atrophy depends on the activation of specific transcription factors of the Forkhead box O (FoxO)3 family (6, 7) (their expression being activated during muscle atrophy and their activity being controlled by the Akt kinase). In normal conditions, phosphorylation of such factors by Akt prevents their translocation in the nucleus. In pro-atrophic conditions, Akt activity decreases, thus reducing FoxO phosphorylation and allowing its entry into the nucleus to activate its target genes (7). The muscle-specific transcription factor Myogenin also regulates MAFbx and MuRF1 expression: upon denervation, Myogenin expression is strongly up-regulated and participates in the activation of the two ubiquitin ligases (8).
Histone deacetylases (HDACs) are central regulators of gene expression. HDAC1 regulates the expression of the acetylcholine receptor in response to neural factors (9), whereas HDAC4 and HDAC9 together with HDAC1 and HDAC3 participate in the regulation of AChR expression by electrical activity via the control of myogenin expression (10, 11).
Recently, it was shown that HDACs are also involved in the regulation of atrogenes via Myogenin and FoxO transcription factors. FoxO3 association with the acetyltransferase p300 and subsequent acetylation induces its cytoplasmic translocation and degradation. Conversely, FoxO3 deacetylation is associated with transcriptional activation (12). Myogenin activation also involves HDAC4. HDAC4 is required for Myogenin activation and therefore participates in the activation of MAFbx during muscle atrophy induced by denervation (8).
The present study is based on our observation that the expression of another histone deacetylase, HDAC6, is up-regulated during skeletal muscle atrophy both in mouse and human. HDAC6 is a peculiar HDAC: mostly cytoplasmic, it deacetylates Tubulin, Cortactin and Hsp90 (rather than histones) and it contains an ubiquitin binding motif (13). In addition, HDAC6 interacts with components of the ubiquitin proteasome pathway (14) and has been shown to regulate ubiquitin-dependent cellular processes (15, 16). Altogether, these evidences suggested that HDAC6 could be involved in regulation or degradation of muscle atrophy effectors. In this work we show that HDAC6 expression is up-regulated during muscle atrophy and that its regulation is FoxO3-dependent. HDAC6 interacts with MAFbx and participates to its activity. More interestingly, HDAC6 inactivation in mouse reduces muscle wasting. We thus demonstrate that HDAC6 is a new FoxO3-dependent player in muscle atrophy.
MATERIALS AND METHODS
Patients Biopsies
Patient muscle samples were analyzed from surgical biopsies of the deltoïd muscle primarily performed for diagnosis purpose. All the patients signed a informed consent for research.
Plasmids
C2C12 cells and muscle fibers were transfected with Myc-tagged HDAC6, GST-tagged HDAC6, HA-tagged HDAC6 or HA-tagged HDAC6 deletion mutants (14), HA-tagged MyoD, and Flag-tagged MAFbx plasmids.
The HDAC6-shRNA plasmid was constructed by cloning double stranded oligonucleotides (Eurogentec, Belgium), into the BamH1/HindIII sites of the pRNAT-H1.1/Neo vector (Genescript Corp.) expressing the cytoplasmic GFP protein.
The sequences of the oligonucleotides were as follows: HDAC6-shRNA V1 gatcccGTGGGGTACAGCACACTTCttcaagagaGAAGTGTGCTGTACCCCACttttttccaaa HDAC6-shRNA V2, gatcccAGCTGGTTGCTCAGCTTCttcaagagaGAAGCTGAGCCAACCAGCTttttttccaaa with the shRNA sequences in antisense and sense orientations underlined. As a control an shRNA directed against luciferase was used (17).
Cell Culture
The mouse skeletal muscle cell line C2C12 was cultured in proliferation medium (DMEM, Invitrogen) supplemented with 15% of fetal bovine serum (Perbio). 5.105 C2C12 were plated on 35 mm wells, and transfected with 2 μg of total plasmid DNA. Cells were transfected with jet prime reagent (Ozyme) according to the manufacturer's recommendations.
Protein extracts were produced using standard lysis buffer (25 mm Tris-HCL, 50 mm NaCl, 0.5% deoxycholic acid, 2% Nonidet P-40, 0.2% SDS) supplemented with a protease inhibitor mixture (Complete, Roche). Protein extracts were analyzed by classic Western blotting procedures using anti HDAC6 (7612; Cell Signaling) and anti α-tubulin (Sigma-Aldrich). For protein half-life measurement, cells were treated for the indicated times by adding 50 μg/ml of cycloheximide (Sigma-Aldrich) into the proliferation medium, to inhibit protein synthesis. Protein extracts were analyzed by classic Western blotting procedures using anti HA (Covance) and anti α-tubulin (Sigma-Aldrich) antibodies.
Mutant Mice
Muscle specific triple Foxo KO mice were generated using the triple floxed mice, described in Paik J. H. (18), and the Human Skeletal Actin-cre mice that specifically express the cre recombinase in skeletal muscles (19).
Electroporation and Denervation
4–5-week-old Swiss Webster mice were anesthetized with 100 μl of 0.05% xylazine, 1.7% ketamine in NaCl 0.9%. 5 μg of HDAC6-shRNA vector or control luciferase-shRNA vector were injected transcutaneously into the tibialis anterior (TA) muscle using a sterile 1 ml syringe with a 27-gauge needle. Caliper electrode plates (Q-biogen, France) were immediately applied on each side of the muscle, and a series of 8 electrical pulses (2 Hz, 20 ms each) was delivered with a standard square-wave electroporator (ECM 830, Q-biogen). Electrical contact was ensured by shaving the leg of the animal and by conductive gel application.
Two days later, atrophy was induced in the left TA muscle of the previously electroporated animals, by sciatic nerve transection under anesthesia. The TA of the right leg of the same animal was left innevated, and used as a control. Alternatively, denervation was performed 7 days before electroporation in the curative experiments.
For the electroporated muscles, fourteen days after denervation (21 days for the curative experiments), TA muscles were dissected, weighted, fixed 1 h in 4% paraformaldehyde at 4 °C, washed briefly in PBS, and incubated 30 min in 0.1 m glycine in PBS at room temperature. The effectiveness of plasmid expression was monitored under fluorescence microscope by the presence of GFP expressing fibers. The muscles were then incubated 1h at 4 °C in 30% sucrose, embedded in Cryomatrix resin (Thermo Shandon) and frozen in liquid nitrogen before conservation at −20 °C. 25-μm thick transversal cryosections of the muscles were then realized, and immediately mounted on glass slides in a fluorescence medium (Dako, Denmark). Images were acquired on a Timelapse fluorescence microscope (Zeiss) with a Coolsnap digital camera (Nikon), and the surface measurement of the GFP-expressing fibers was performed with the Image J software.
Quantitative RT-PCR Analysis on Muscles
Total RNA was isolated from mice muscle tissues or from human muscle biopsies using Trizol (Invitrogen). RNA was analyzed by quantitative real-time PCR using SYBR Green (Roche). The data were normalized to mouse-GAPDH or to human-hypoxanthine-guanine phosphoribosyltransferase mRNA levels. Primers sequences were as follows: mGAPDH: 5′-GGTCACCAGGGCTGCCATTTG-3′, reverse 5′-TTCCAGAGGGGCCATCCACAG-3′; mHDAC6: forward: 5′-AAGTGGAAGAAGCCGTGCTA-3′, reverse 5′-CTCCAGGTGACACATGATGC-3′; mMAFbx: forward 5′-CAGACCTGCATGTGCTCAGT-3′, reverse 5′-CCAGGAGAGAATGTGGCAGT-3′. hHPRT: forward 5′-ACGAGCCCTCAGGCGAACCT-3′, reverse 5′-CACTAATCACGACGCCAGGGCT-3′; hHDAC6: forward 5′-ATGGCCATCATTAGGCCTCC-3′, reverse 5′-CGGATGCGGTGTTTCTGTTG-3′.
Chromatin Immunoprecipitation Assay
Mouse TA muscles were incubated for 15 min on ice in buffer RBI (100 mm KCl, 5 mm MgCl2, 5 mm EDTA, 5 mm Na pyrophosphate, pH 6.8, and protease inhibitors) then transferred to a 2% formaldehyde solution for 15 min at 37 °C. Reactions were stopped by addition of glycine at 0.125 m final and incubation at 25 °C for 5 min. After a PBS wash, muscles were crushed in lysis buffer A (10 mm Tris-HCl, pH 7.9, 85 mm KCl, 0.5% Nonidet P-40, and protease inhibitors). The mixture was homogenized by 20 strokes of Dounce followed by a 5-min spin at 5,000 rpm at 4 °C. The pellet was resuspended in lysis buffer B (50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% SDS, and protease inhibitors) incubated on ice for 10 min followed by a 5-min centrifugation at 5,000 rpm at 4 °C. The supernatant was sonicated in a Bioruptor (Diagenode) for 15 min (medium; 30 s/30 s cycles). The size of chromatin fragment was checked to be in the range of 300–500-bp length on an agarose gel after de-cross-linking. ChIP was performed using the EZ Magna ChIP kit (Millipore) with 200 μg of chromatin and 20 μg of antibody.
The antibody used was FoxO3 (Santa Cruz Biotechnology). QPCR values were calculated by extrapolation from a standard curve of input DNA dilutions. Enrichment values of the Foxo binding region (region A) were normalized to IgG signals and shown as the fold difference relative to a negative genomic region located between the 23rd and the 24th exon of the HDAC6 gene (region B).
The primers used were 5′-TGGTGGAAAGAAGTGTGCTG-3′ and 5′-GTCTCTCTCACCAGCCCTTG-3′ for region A. 5′-TGACTGAGCCTTGGTCTCCTT-3′ and 5′-CTCAAGGCAGATCCCAAAAG-3′ for region B.
Immunoprecipitation and GST-pulldown
C2C12 cells were rinsed in cold PBS and lysed with IP buffer (50 mm Tris pH 7.4, 150 mm NaCl, 150 mm EDTA, 1% Nonidet P-40, protease and phosphatase inhibitors). 1 mg of lysate was immunoprecipitated with 30 μl of protein G or A, covalently conjugated with the antibodies overnight at 4 °C. Overexpressed proteins were immunoprecipitated and detected with anti-FLAG (F1804, Sigma Aldrich), anti-HA (Sigma-Aldrich) and anti-Myc (9E10; Covance). GST-pull down was performed as described in Lemercier et al. (20), except that cell extracts overexpressing atrogin were used instead of reticulocyte lysates.
Electrophoretic Mobility Shift Assay (EMSA)
Protein extracts were prepared as previously described (37).
DNA probes were prepared by polynucleotide kinase (NEB) labeling with 32P ATP-gamma (NEG/BLU502A) and purified on Illustra ProbeQuant G50 micro-columns (GE Healthcare). Sequence of oligonucleotides (Eurogentec) used were as follows: FoxO3-HDAC6-forward: 5′-GCGGAGGTAAAGAAGAAAGG-3′; FoxO3-HDAC6-reverse: 5′-CCTTTCTTCTTTACCTCCGC-3′; FoxO3mut-HDAC6-forward: 5′-GCGGAGGTCACGAAGAAAGG-3′; FoxO3mut-HDAC6-reverse: 5′-CCTTTCTTCGTGACCTCCGC-3′; NFKB-forward: 5′-GCAAACAGAAATTCCACCAGCCTC-3′; NFKB-reverse: 5′GAGGCTGGTGGAATTTCTGTTTGC03′.
EMSA was performed by incubating optimal (experimentally defined) amounts of extracts (5 μg) and probe (up to 5000 cpm) at 30 °C for 45 min in a 20 μl final reaction volume containing 50 mm KCl, 5 mm MgCl2, 0,1 μg BSA, 0,05% Nonidet P-40, and 500 ng of poly(dI-dC)-poly(dI-dC) (Sigma). 1 μl of glycerol was added to the reaction before loading on a 4,5% acrylamide/bis (50/1 ratio) gel in TNE (6.7 mm Tris HCl, pH 7.9, 3.3 mm NaOAc, 1 mm EDTA). A bromphenol blue marker was run in parallel to monitor the migration. Electrophoresis was done at 25 mA, 50 V until Bromphenol Blue was at 3/4 of the gel. Gels were then autoradiographed.
RESULTS
Muscle Atrophy Triggers HDAC6 Expression
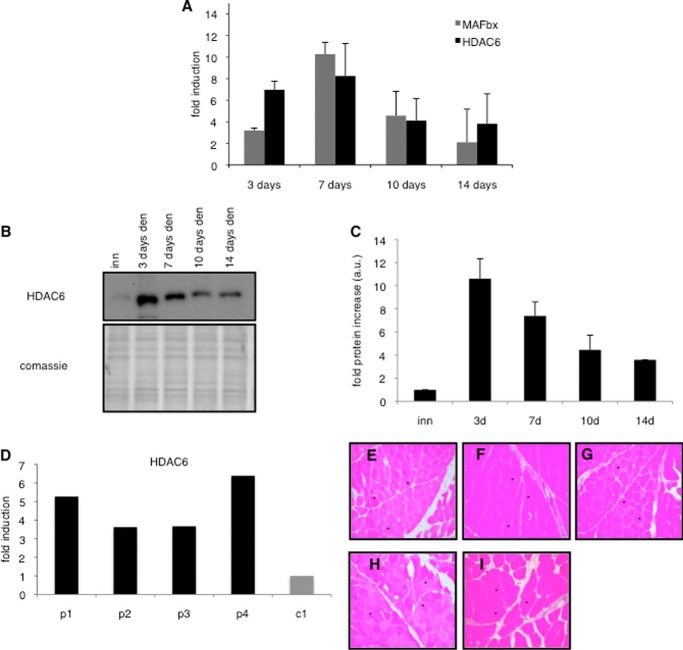
Muscle denervation was used to induce muscle atrophy in mice to identify new HDACs up-regulated during this process. As shown in Fig. 1A, denervation-induced atrophy induced an increase of HDAC6 expression. This increase was concomitant with the up-regulation of MAFbx expression, a classic marker of muscle atrophy (5). Even 14-days after denervation, HDAC6 mRNA levels remained 4-fold higher than in innervated muscles (Fig. 1A). HDAC6 up-regulation was also visible at the protein level (Fig. 1, B and C).
FIGURE 1.
A, up-regulation of MAFbx (gray bars) and HDAC6 (black bars) in adult mice TA muscle induced by 3, 7, 10, 14 days of denervation. Quantitative PCR (QPCR) analysis was performed on 3 mice for each time point. Results are represented as fold induction compared with innervated. Error bars represent S.E. B, Western blot of extracts from adult mice TA muscle after 3, 7, 10, 14 days of denervation. Upper panel: HDAC6 blot. Lower panel: Coomassie staining as a total protein loading control. The experiment was repeated three times on 3 independent sets of mice. One representative experiment is shown. C, quantification of the fold increases of HDAC6 protein compared with the innervated control. Quantifications of 3 different Western blots are shown. Data are mean of three independent experiments. Error bars represent S.E. D, relative expression levels of HDAC6 were measured by QPCR in muscles biopsies of patients with muscle atrophy (patient 1 = p1, patient 2 = p2, patient 3 = p3, patient 4 = p4) or control muscles (c1). Results are normalized to the levels of HPRT expression. E–H, hematoxylin eosin staining of deltoid muscle cryosections performed on patients with muscle atrophy (p1, p2, p3, p4) and (I) control muscles (c1). Magnification ×200.
HDAC6 up-regulation was next evaluated in human skeletal muscle in different conditions of muscle atrophy. Muscle biopsies of patients were analyzed to evaluate HDAC6 expression. Patients were selected on the basis of their muscle histology showing no other abnormality than muscle fiber atrophy (Fig. 1, E–H). Among the four patients selected, three had muscle atrophy associated to muscle disuse, and one had muscle atrophy due to peripheral neuropathy associated with type II diabetes. As a control, a patient with fibromyalgia devoid of any histological abnormality in the muscle biopsy was used (Fig. 1I). QPCR analysis of the biopsies revealed that HDAC6 expression was up-regulated in the four atrophied patients (p1, p2, p3, and p4) compared with the control (c1) (Fig. 1D).
HDAC6 Inactivation Reduces Muscle Atrophy Induced by Denervation
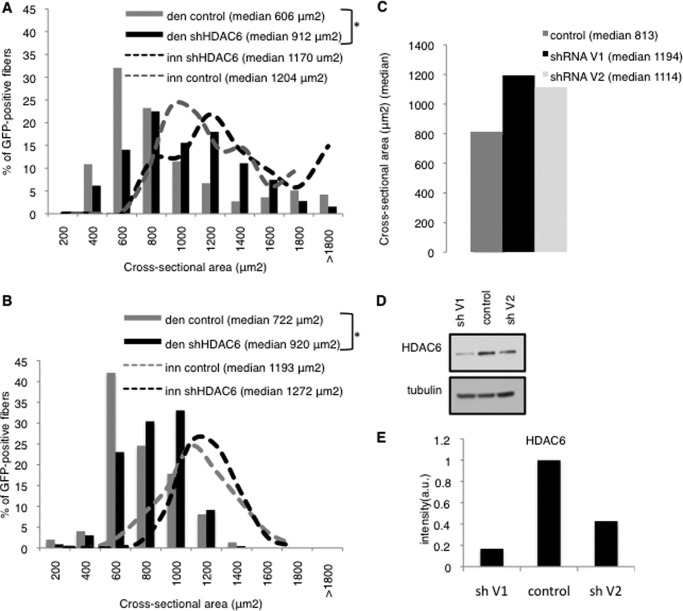
To evaluate if HDAC6 had a role in muscle atrophy, we tested whether inactivation of HDAC6 could prevent muscle wasting. To address this question, HDAC6 expression was specifically down regulated in muscle fibers by electroporation of shRNA expression vectors directed against HDAC6. This technique allows the introduction of plasmid vectors in muscle fibers without affecting the surrounding mononucleated cells (21). The shRNAs vectors contained a GFP reporter gene to allow visualization of the transfected fibers. The HDAC6 or the control shRNA plasmids were electroporated bilaterally into tibialis anterior (TA) of adult mice. Denervation by sciatic nerve section was performed 2 days after on the left TA muscle. 15 days later, the muscles were collected and cryo-sectioned, and the cross-sectional area of the transfected fibers was measured. As shown in Fig. 2A, the HDAC6-shRNA electroporated fibers were significantly less sensitive to atrophy than the control electroporated fibers.
FIGURE 2.
A and B, muscles were electroporated either 2 days before (A) or 7 days (B) after denervation. 14 days later, cryosections were preformed and the cross sectional area of electroporated fibers was measured. Frequency histograms show the distribution of cross-sectional areas of muscle fibers of TA either innervated or 15 days denervated and electroporated with shHDAC6-GFP or control shRNA. As a control an shRNA against the luciferase was used. The median cross sectional area value is indicated for each condition. n = 5 mice for each condition. The asterisks indicate statistically significant changes at p < 0.05 (1 asterisk), calculated by Mann-Whitney's U test. den: 14 days denervated muscle; inn: innervated muscle. C, histogram shows the median values of the cross-sectional areas of TA fibers, electroporated with two different HDAC6 shRNA expressing vector (V1 and V2). D, Western blot showing the efficiency of V1 and V2 HDAC6 shRNA to reduce HDAC6 levels in C2C12. The experiment was repeated 3 times. One representative experiment is shown. E, quantification of panel D by Image J software. HDAC6 levels were normalized to tubulin levels.
To determine if HDAC6 inhibition could also be beneficial after the onset of muscle atrophy, TA muscles were denervated to induce muscle atrophy, and 1 week later they were electroporated with the HDAC6 shRNA expression vector or the control vector. Two weeks after the electroporation, muscle wasting was evaluated as described above. Depletion of HDAC6 resulted in a significant attenuation of the atrophy phenotype, as shown in Fig. 2B, suggesting that HDAC6 is not only involved in the outbreak of skeletal muscle atrophy, but also in maintaining the rate of muscle wasting. To determine if the efficiency of the shRNA to repress HDAC6 expression correlated with its ability to limit muscle atrophy, a second HDAC6 shRNA was designed and electroporated in TA muscles. Muscle atrophy was evaluated in the electroporated muscles 2 weeks after denervation (Fig. 2C). In parallel, the effect of the shRNAs on HDAC6 levels was evaluated by QPCR and Western blot 24h after transfection in C2C12 cells. The shRNA that induced the stronger reduction in HDAC6 levels was also the more potent to reduce muscle atrophy (Fig. 2, D and E).
HDAC6 Is a Target of Foxo3
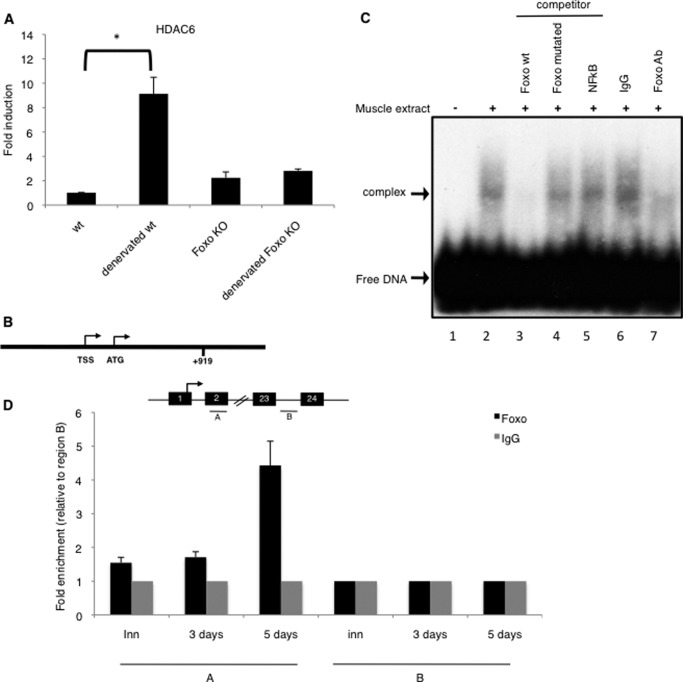
FoxO transcription factors are key regulators of cellular stress-response and in the atrophy process, they drive the up-regulation of atrogenes and of autophagy genes. The involvement of FoxO in the activation of HDAC6 during atrophy was evaluated. For this purpose, HDAC6 expression was measured in muscle-specific FoxOs triple knock out mice. Interestingly, after 5 days of denervation, HDAC6 expression was still not up-regulated in the FoxOs triple KO mice compared with the wild type, indicating a FoxOs dependent regulation of HDAC6 (Fig. 3A). Consistently, a potential FoxO3 binding site is present in the HDAC6 regulatory region (GTAAAGA), at position +919 (Fig. 3B). The ability of FoxO3 to bind this site was first evaluated by electrophoretic mobility shift assay (Fig. 3C). Incubation of 5 days denervated-muscle extracts with an oligonucleotide containing the FoxO3 binding site allowed visualization of a retarded complex. An excess of non-labeled probe prevented the formation of the complex, but an oligonucleotide containing a mutated FoxO binding site did not, neither did an NFkB binding sequence. Addition of an anti-FoxO3 antibody markedly reduced the formation of the complex. Altogether, these results indicate that FoxO3 binds a sequence present in the HDAC6 promoter.
FIGURE 3.
A, up-regulation of HDAC6 in FoxO triple KO and wild type mice TA muscles after 5 days of denervation. QPCR analysis was performed on 3 mice for each condition. Results are presented as fold induction compared with innervated. Error bars represent S.E. The asterisk indicates statistically significant changes at p < 0.01 (p = 0.004). B, consensus sequence for FoxO3 (GTAAAGA) at position +919 in the HDAC6 gene region. C, muscle extracts from denervated TA muscles were tested for binding to double stranded 32P-labeled oligonucleotides containing a FoxO3 binding site by EMSA. Arrows indicate the FoxO3-DNA complex and the free DNA. Lane 1: free DNA alone; all other lanes contain the protein extract and, as indicated at the top of the figure, no oligonucleotide (positive control; lane 2), a competitor oligonucleotide (FoxO wt, FoxO mutant or NFkB; lanes 3–5) or an antibody (IgG or anti-FoxO3; lanes 6–7). D and E, localization of FoxO3 protein at the HDAC6 gene. Chromatin immunoprecipitation (ChIP) analysis were performed in wild type mice TAs muscles, either innervated, or 3 or 5 days denervated. Two indicated regions (A and B) were tested for QPCR amplification. QPCR values were calculated by extrapolation from a standard curve of Inputs DNA dilutions. Enrichment values were normalized to IgG signals and shown as the fold difference relative to region B. Each ChIP-QPCR histogram indicates the mean of triplicate results, error bars indicate S.E. A: FoxO binding site region at +919, B: negative region at +15074.
The ability of FoxO3 to bind the binding site at +919 was next evaluated by chromatin immunoprecipitation (ChIP) on innervated and denervated muscle extracts. ChIP analysis revealed that denervation stimulated the recruitment of FoxO3 in the region containing the GTAAAGA binding site (Fig. 3D).
HDAC6 Interacts with MAFbx
MAFbx/Atrogin-1 was among the first atrogenes identified. It belongs to a SCF E3-ubiquitin ligase complex that contributes to muscle protein degradation via the ubiquitin-proteasome system (5).
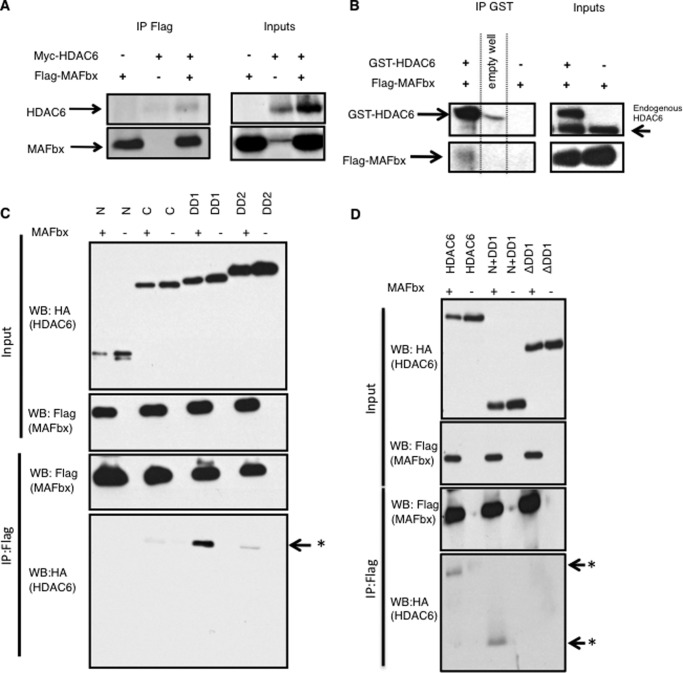
Given the concomitant activation of HDAC6 and MAFbx during atrophy (Fig. 1A) (7), and given the ability of HDAC6 to bind ubiquitinated proteins and to transport them along microtubules to allow their degradation (15), we hypothesized that HDAC6 and MAFbx could work together during muscle wasting. In the absence of suitable MAFbx antibodies, possible interactions between HDAC6 and MAFbx were first investigated by co-immunoprecipitation in C2C12 cells transfected with Flag-MAFbx and Myc-HDAC6 expression vectors. Western blot analysis showed that MAFbx coimmunoprecipitated with HDAC6 (Fig. 4A). To verify that MAFbx and HDAC6 also interacted in muscle fibers, a GST-pulldown experiment was performed in tibialis anterior muscles electroporated with GST-HDAC6 and Flag-MAFbx expression vectors and showed that HDAC6 and MAFbx also interact in muscle fibers (Fig. 4B).
FIGURE 4.
A, indicated tagged proteins (Myc-HDAC6 and Flag-MAFbx) were expressed in C2C12 cells (input panel). Flag-MAFbx was then immunoprecipitated with an anti-Flag antibody and detected by Western blot with the same antibody (lower panels) and the co-immunoprecipitated Myc-HDAC6 was detected by Western blot using an anti-Myc antibody (upper panel). B, indicated tagged proteins expressing vectors (GST-HDAC6 and Flag-MAFbx) were electroporated in TA muscles (input panel). GST-HDAC6 was then immunoprecipitated with GST antibody conjugated Sepharose beads and detected with an anti-HDAC6 antibody (upper panels). The co-immunoprecipitated Flag-MAFbx was detected by Western blot using an anti-Flag (lower panels). C, indicated HA-tagged fragments of HDAC6 and Flag-MAFbx were expressed in C2C12 cells (input panel). Flag-MAFbx was then immunoprecipitated with an anti-Flag antibody and detected by Western blot with the same antibody. The coimmunoprecipitated HA-HDAC6 fragments were detected by Western blot using an anti-HA antibody (IP panel). Arrow asterisk, co-immunoprecipitated DD1. HDAC6 fragments are: N is the N-terminal non-catalytic domain, DD1 and DD2 respectively correspond to the catalytic domains 1 and 2 of HDAC6, and C corresponds to the ubiquitin-binding domain of HDAC6. D, indicated HA-tagged mutants of HDAC6 and Flag-MAFbx were expressed in C2C12 cells (input panel). Flag-MAFbx was immunoprecipitated with an anti-Flag antibody and detected by Western blot with the same antibody. Coimmunoprecipitated HA-HDAC6 mutants were detected by Western blot using an anti-HA antibody (IP panel). Arrow asterisk, co-immunoprecipitated N+DD1 and HDAC6. Arrow, absence of the co-immunoprecipitated ΔDD1. HDAC6 mutants are: HDAC6 is full-length HDAC6, N+DD1 is the N-terminal plus DD1, ΔDD1 is a full-length HDAC6 lacking the DD1.
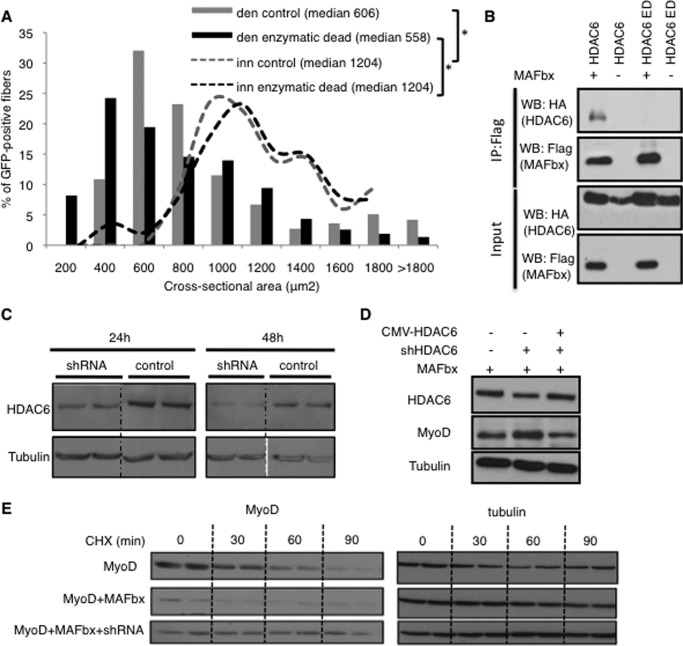
To map the site of interaction between HDAC6 and MAFbx, different HA-tagged fragments of HDAC6 were transfected into C2C12 cells and the interactions were tested by coimmunoprecipitation. As shown in Fig. 4C, MAFbx interacts with the first catalytic domain of HDAC6 (DD1). To confirm that this domain of HDAC6 mediated the interaction, coimmunoprecipitation experiments were performed with MAFbx and deletion mutants of HDAC6. As expected, the deletion of the DD1 domain abolished the interaction with MAFbx (Fig. 4D). The involvement of the DD1 domain in the interaction between HDAC6 and MAFbx raised the question of the involvement of HDAC6 deacetylase activity. The coimmunoprecipitation experiments were thus repeated with an enzymatic dead mutant of HDAC6 (22). No interaction with MAFbx could be detected (Fig. 5B). To evaluate if this mutant had a dominant negative action on HDAC6, it was electroporated into muscle. Evaluation of muscle atrophy 15 days after denervation revealed no significant difference compared with control muscles (Fig. 5A).
FIGURE 5.
A, frequency histograms showing the distribution of cross-sectional areas of muscle fibers of TA muscles either innervated or 14 days denervated and electroporated with the HDAC6 enzymatic dead mutant or control plasmids. 14 days later, cryosections were preformed, and the cross sectional area of electroporated fibers was measured. The median cross sectional area value is indicated for each condition. n = 5 mice for each condition. The asterisks indicate statistically significant changes at p < 0.05 (1 asterisk), calculated by Mann-Whitney's U test. den: 14 days denervated muscle; inn: innervated muscle. B, C2C12 were cotransfected with and Flag-tagged MAFbx and either pcDNA expression vector, HA-tagged full-length HDAC6 or HDAC6 enzymatic dead (ED). Interactions were evaluated by immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with an anti-Flag or anti-HA antibody. Total lysates used in immunoprecipitation are shown as input. C, HDAC6-shRNA is efficient in cultured cells. C2C12 myoblasts were transfected with either the HDAC6-shRNA or the control vector. 24 to 48 h after transfection, α-tubulin and HDAC6 were detected on protein extracts by Western blot. D, C2C12 cells were transfected either with a MAFbx expression vector, or with a MAFbx expression vector, and a HDAC6 shRNA vector, or with a MAFbx expression vector, a HDAC6 shRNA vector and a CMV-HDAC6 expression vector, to restore normal HDAC6 levels. 36 h after transfection, MyoD levels were evaluated by Western blot. E, C2C12 cells were cotransfected with vectors expressing HA-MyoD, MAFbx, and the HDAC6-shRNA as indicated. The cells were incubated with cycloheximide (CHX) 24 h after transfection, and total proteins harvested at the indicated times of treatment for analysis by Western blotting with anti-HA or anti α-tubulin.
HDAC6 Is Required for MAFbx-dependent MyoD Degradation
MyoD is the best-characterized substrate of MAFbx, and it was previously shown the half-life of MyoD was significantly reduced by MAFbx overexpression in C2C12 muscle cells (23). Since the HDAC6 shRNA was efficient in C2C12 cells (Fig. 5C), we sought to determine if HDAC6 was involved in this process. C2C12 cells were transfected with a MAFbx expression vector and the HDAC6 shRNA. As shown in Fig. 5D, the presence of HDAC6 shRNA significantly increased MyoD levels. The rescue of normal HDAC6 expression by HDAC6 overexpression abrogated this effect.
To further evaluate the involvement of HDAC6 in MyoD degradation by MAFbx, the effect of HDAC6 and MAFbx on MyoD half-life was measured. Cycloheximide was thus added at this time, and the levels of MyoD were monitored afterward in Western blots at different time points (Fig. 5E). As expected, the half-life of MyoD was significantly reduced by MAFbx overexpression. The knock down of HDAC6 abolished the effect of MAFbx and even conferred to MyoD a longer half-life than in the absence of MAFbx transfection. This probably results from the inhibition of MyoD degradation by endogenous MAFbx. Altogether these results demonstrate that HDAC6 is required for MAFbx-dependent MyoD degradation.
DISCUSSION
Our results demonstrate that HDAC6 expression is up-regulated during muscle atrophy and that it interacts with MAFbx in muscle cells. Furthermore, HDAC6 depletion in vivo protects against muscle atrophy. The transcriptional activation of HDAC6 during atrophy is FoxOs dependent, and FoxO3 directly binds to the HDAC6 regulatory region.
The activation of HDAC6 expression in murine and human models of muscle atrophy suggests that HDAC6 could be considered as a new atrogene. A transcriptome analysis of muscle wasting due to limb immobilization, that includes HDAC6 among the genes still up-regulated after 7 days of atrophy (24), corroborates our hypothesis.
FoxO transcription factors are known to promote the activation of cellular stress response (25). HDAC6 is also involved in stress response, but its regulation remains largely unknown. The fact that FoxO3a mediates HDAC6 activation places HDAC6 downstream of this factor in the cascade of events leading to stress response.
The proposed role of HDAC6 in the ubiquitin-proteasome system is to regulate the fate of ubiquitinated proteins accumulated under stress conditions by addressing them to various ubiquitin-dependent degradation structures (proteasome or aggresomes). The interaction between HDAC6 and MAFbx and their co-regulation by FoxO transcription factors indicates that these two actors of muscle atrophy work together. Accordingly, our results show that HDAC6 is required for MAFbx-dependent degradation of MyoD. The described function of HDAC6 in the transport of ubiquitinated proteins along microtubules (15) suggests that HDAC6 could convey the proteins ubiquitinated by MAFbx to degradation.
The experiments performed above do not allow determining if the interaction between MAFbx and HDAC6 is direct. Since MAFbx belongs to a multiprotein complex, it is possible that HDAC6 is recruited to the complex via another subunit.
Interestingly, the valosin-containing protein (p97-VCP) was recently shown to be involved in muscle atrophy (26). VCP expression is up-regulated during muscle atrophy and a VCP dominant negative mutant can reduce muscle atrophy (26). Since HDAC6 is a partner of VCP for the ubiquitination of substrates targeted to the proteasome (27), we hypothesize that HDAC6 could cooperate with VCP to favor muscle protein degradation.
The fact, that HDAC6 and MAFbx are both enzymes, raises the question of possible mutual modifications. We could imagine that MAFbx function is regulated by acetylation. However, using anti-acetyl lysine antibodies we could not detect any variation of MAFbx acetylation in muscle cells expressing the HDAC6 shRNA (data not shown). The fact that an enzymatic dead mutant of HDAC6 does not interact with MAFbx indicates that the enzymatic activity of HDAC6 could be required for the interaction with MAFbx. The action of HDAC6 on microtubules might be required to bring HDAC6 and MAFbx together. Alternatively, it is possible that the mutation affects the structure of the DD1 domain in such a way that the interaction with MAFbx is abolished.
It is also conceivable that MAFbx could ubiquitinate HDAC6, but again, anti-ubiquitin antibodies did not reveal any ubiquitination of HDAC6 in muscle cells, even with MAFbx overexpression (data not shown).
Muscle atrophy is associated with most neuromuscular diseases of neural or muscle origin, with aging, muscle inactivity due to loss of motor innervation or immobilization, as well as various pathologies such as type 2 diabetes, AIDS. Cancer cachexia is a major cause of muscle wasting, and negatively interferes with chemotherapies and is associated with 20% of cancer mortality (1).
These evidences make muscle atrophy a major public health problem as current treatments are limited and not efficient in curing or preventing this disorder, that seriously impact quality of life and responsiveness to therapies. The fact that HDAC6 is activated during atrophic conditions and that it potentially cooperates with MAFbx indicates that HDAC6 could be an interesting pharmacological target to treat muscle wasting. In addition, HDAC6 inactivation in muscle already undergoing atrophy efficiently reduced the evolution of muscle wasting. HDAC6 is therefore the first atrogene for which both prophylactic and curative interest has been demonstrated.
HDAC6 is an unconventional histone deacetylase, whose activity goes well beyond deacetylating histones and is involved in many cellular processes. Through the deacetylation of many cytoplasmic substrates such as Tubulin, Cortactin, and Hsp90, HDAC6 is able to control cell migration, aggresomes formation, autophagy and stress response (15, 28–30). The multiple partners-mode of action implies a multi-faceted involvement in vivo as well. Therefore, HDAC6 can be considered as a potential therapeutic target for many human diseases. Several anti-cancer treatments targeting HDAC6 have been proposed, given its capacity to control cancer cells and metastasis migration (31–33). As a result of its ability to degrade protein aggregates, HDAC6 has also been proposed to be a relevant target for neurodegenerative diseases (34–36). In this work, we provide evidence that targeting HDAC6 could also be relevant to prevent skeletal muscle atrophy.
Acknowledgments
We thank Valérie Risson as well as the imaging platform (PLATIM) and animal facility (PBES) of the UMS 3444 for technical assistance.
This work was supported by the Association Française contre les Myopathies (AFM), the Centre National de la Recherche Scientifique (CNRS), the Ministere de l'Education Nationale, de la Recherche et de la Technologie, and the Institut National du Cancer (INCA).
- FoxO
- Forkhead box O
- HDAC
- histone deacetylase
- TA
- tibialis anterior.
REFERENCES
- 1. Penet M. F., Gadiya M. M., Krishnamachary B., Nimmagadda S., Pomper M. G., Artemov D., Bhujwalla Z. M. (2011) Metabolic signatures imaged in cancer-induced cachexia. Cancer Res. 71, 6948–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Inui A. (2002) Cancer anorexia-cachexia syndrome: current issues in research and management. CA: A Cancer Journal for Clinicians 52, 72–91 [DOI] [PubMed] [Google Scholar]
- 3. Mitch W. E., Goldberg A. L. (1996) Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. NE J. Med. 335, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 4. Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U.S.A. 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 6. Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., Glass D. J. (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14, 395–403 [DOI] [PubMed] [Google Scholar]
- 7. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moresi V., Williams A. H., Meadows E., Flynn J. M., Potthoff M. J., McAnally J., Shelton J. M., Backs J., Klein W. H., Richardson J. A., Bassel-Duby R., Olson E. N. (2010) Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravel-Chapuis A., Vandromme M., Thomas J. L., Schaeffer L. (2007) Postsynaptic chromatin is under neural control at the neuromuscular junction. EMBO J. 26, 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang H., Macpherson P., Marvin M., Meadows E., Klein W. H., Yang X. J., Goldman D. (2009) A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol. Biol. Cell 20, 1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Méjat A., Ramond F., Bassel-Duby R., Khochbin S., Olson E. N., Schaeffer L. (2005) Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nature Neuroscience 8, 313–321 [DOI] [PubMed] [Google Scholar]
- 12. Bertaggia E., Coletto L., Sandri M. (2012) Posttranslational modifications control FoxO3 activity during denervation. Am. J. Physiol. 302, C587–C596 [DOI] [PubMed] [Google Scholar]
- 13. Hook S. S., Orian A., Cowley S. M., Eisenman R. N. (2002) Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. U.S.A. 99, 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seigneurin-Berny D., Verdel A., Curtet S., Lemercier C., Garin J., Rousseaux S., Khochbin S. (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21, 8035–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 16. Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. (2007) Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178, 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alais S., Simoes S., Baas D., Lehmann S., Raposo G., Darlix J. L., Leblanc P. (2008) Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles. Biology of the Cell / under the auspices of the European Cell Biology Organization 100, 603–615 [DOI] [PubMed] [Google Scholar]
- 18. Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Risson V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., Freyssenet D., Tanti J. F., Le-Marchand-Brustel Y., Ferrier B., Conjard-Duplany A., Romanino K., Bauché S., Hantaï D., Mueller M., Kozma S. C., Thomas G., Rüegg M. A., Ferry A., Pende M., Bigard X., Koulmann N., Schaeffer L., Gangloff Y. G. (2009) Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 187, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemercier C., Verdel A., Galloo B., Curtet S., Brocard M. P., Khochbin S. (2000) mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275, 15594–15599 [DOI] [PubMed] [Google Scholar]
- 21. Lagirand-Cantaloube J., Cornille K., Csibi A., Batonnet-Pichon S., Leibovitch M. P., Leibovitch S. A. (2009) Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PloS one 4, e4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwon S., Zhang Y., Matthias P. (2007) The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. (2005) Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 280, 2847–2856 [DOI] [PubMed] [Google Scholar]
- 24. Kim J. W., Kwon O. Y., Kim M. H. (2007) Differentially expressed genes and morphological changes during lengthened immobilization in rat soleus muscle. Differentiation; Research in Biological Diversity 75, 147–157 [DOI] [PubMed] [Google Scholar]
- 25. Sandri M. (2012) FOXOphagy path to inducing stress resistance and cell survival. Nature Cell Biology 14, 786–788 [DOI] [PubMed] [Google Scholar]
- 26. Piccirillo R., Goldberg A. L. (2012) The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J. 31, 3334–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boyault C., Gilquin B., Zhang Y., Rybin V., Garman E., Meyer-Klaucke W., Matthias P., Müller C. W., Khochbin S. (2006) HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 25, 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S. H., Garrido C., Yao T. P., Vourc'h C., Matthias P., Khochbin S. (2007) HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 21, 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao Y. S., Hubbert C. C., Yao T. P. (2010) The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. J. Biol. Chem. 285, 11219–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 31. Kanno K., Kanno S., Nitta H., Uesugi N., Sugai T., Masuda T., Wakabayashi G., Maesawa C. (2012) Overexpression of histone deacetylase 6 contributes to accelerated migration and invasion activity of hepatocellular carcinoma cells. Oncology Reports 28, 867–873 [DOI] [PubMed] [Google Scholar]
- 32. Lafarga V., Aymerich I., Tapia O., Mayor F., Jr., Penela P. (2012) A novel GRK2/HDAC6 interaction modulates cell spreading and motility. EMBO J. 31, 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zuo Q., Wu W., Li X., Zhao L., Chen W. (2012) HDAC6 and SIRT2 promote bladder cancer cell migration and invasion by targeting cortactin. Oncology Reports 27, 819–824 [DOI] [PubMed] [Google Scholar]
- 34. d'Ydewalle C., Krishnan J., Chiheb D. M., Van Damme P., Irobi J., Kozikowski A. P., Vanden Berghe P., Timmerman V., Robberecht W., Van Den Bosch L. (2011) HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nature Med. 17, 968–974 [DOI] [PubMed] [Google Scholar]
- 35. Gal J., Chen J., Barnett K. R., Yang L., Brumley E., Zhu H. (2013) HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J. Biol. Chem. 288, 15035–15045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taes I., Timmers M., Hersmus N., Bento-Abreu A., Van Den Bosch L., Van Damme P., Auwerx J., Robberecht W. (2013) Hdac6 deletion delays disease progression in the SOD1G93A mouse model of ALS. Human Mol. Genet. 22, 1783–1790 [DOI] [PubMed] [Google Scholar]
- 37. Schaeffer L., Duclert N., Huchet-Dymanus M., Changeux J. P. (1998) Implication of a multisubunit Ets related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 17, 3078–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]