Background: FcRn controls the serum persistence of antibodies.
Results: A panel of novel Fc mutations reveals sites controlling pH dependence and FcRn affinity.
Conclusion: FcRn affinity thresholds determine IgG recycling efficiency.
Significance: Knowledge of the relationship between FcRn binding and serum persistence can aid in designing better therapeutic antibodies.
Keywords: Antibody Engineering, Fc Receptor, Monoclonal Antibody, Phage Display, Pharmacokinetics, FcRn
Abstract
The Fc domain of IgG has been the target of multiple mutational studies aimed at altering the pH-dependent IgG/FcRn interaction to modulate IgG pharmacokinetics. These studies have yielded antibody variants with disparate pharmacokinetic characteristics, ranging from extended in vivo half-life to those exhibiting extremely rapid clearance. To better understand pH-dependent binding parameters that govern these outcomes and limit FcRn-mediated half-life extension, we generated a panel of novel Fc variants with high affinity binding at acidic pH that vary in pH 7.4 affinities and assessed pharmacokinetic outcomes. Pharmacokinetic studies in human FcRn transgenic mice and cynomolgus monkeys showed that multiple variants with increased FcRn affinities at acidic pH exhibited extended serum half-lives relative to the parental IgG. Importantly, the results reveal an underappreciated affinity threshold of neutral pH binding that determines IgG recycling efficiency. Variants with pH 7.4 FcRn affinities below this threshold recycle efficiently and can exhibit increased serum persistence. Increasing neutral pH FcRn affinity beyond this threshold reduced serum persistence by offsetting the benefits of increased pH 6.0 binding. Ultra-high affinity binding to FcRn at both acidic and neutral pH leads to rapid serum clearance.
Introduction
pH-dependent FcRn binding is considered crucial to both the transfer of maternal IgG to the neonate via the placenta and homeostasis of IgG and serum albumin (SA)3 in humans. The generally accepted model of FcRn-mediated IgG homeostasis states that IgG, upon internalization via pinocytosis, binds to FcRn in the acidic endosome and is salvaged via trafficking to the cell surface (pH 7.4), where it is returned to circulation, thereby maintaining high serum levels of IgG (1–7). It is proposed that specific histidine residues on the IgG Fc and SA facilitate pH-dependent binding by toggling between positively charged and neutral states in different pH environments (5, 8–11). In the acidic endosome, positively charged histidine side chains can interact favorably with negatively charged side chains of FcRn; however, this affinity is greatly diminished at the neutral cell surface where IgG and SA are released into circulation. This hypothesis is supported by numerous genetic and mutagenesis studies as well as recent crystallographic data (12, 13).
The mechanism of FcRn-mediated IgG recycling has been exploited to engineer therapeutic antibodies and Fc-fused biologics with altered pharmacokinetic (PK) properties, such as longer serum persistence (14, 15). The guiding engineering principle for generating antibodies with longer half-lives has been to increase binding affinity to FcRn at acidic pH while maintaining minimal binding at neutral pH (11). Despite the observation that mutations generating increased FcRn affinity at acidic pH generally produce concurrent neutral pH affinity increases (16), multiple variants have been described that exhibit higher serum half-life than endogenous IgG in transgenic human FcRn (hFcRn) mice and primates (16–20), including the M252Y/S254T/T256E mutation (referred to as the YTE mutation hereafter) (11, 21, 22) capable of extending antibody serum half-lives by more than 90 days in humans (23). However, some engineered variants with similarly improved FcRn binding characteristics do not exhibit expected increases in in vivo half-life (24–26). Furthermore, IgG with greatly increased FcRn binding at both pH 6.0 and pH 7.4 can exhibit very fast clearance and can enhance the degradation of endogenous IgG (11, 27). These examples highlight the complex relationship between FcRn affinity, pH dependence, and in vivo clearance as well as the engineering challenges inherent in this system.
In this study, we explore the potential limits of FcRn-mediated half-life extension to better understand the binding parameters that govern IgG recycling. Our work focuses on engineering the FcRn binding of an Fc variant that shows ultra-high binding affinity at both pH 6.0 and pH 7.4 and exhibits rapid serum clearance. We targeted His-435 and adjacent residues in the CH3 domain because mutations in this region have been shown to drastically influence the pH-dependent capability of IgG to bind FcRn and have yielded variants with either increased or decreased serum half-lives (11, 17, 20, 27). By pH binding selection, we isolated a panel of variants that maintain high affinity binding at pH 6.0 but vary in pH 7.4 affinities. Many of these variants extend serum half-life similarly to YTE in hFcRn mice and in cynomolgus monkeys. Our results reveal an underappreciated affinity threshold at neutral pH that governs IgG PK outcomes of affinity-improved FcRn-binding variants. Once this binding threshold is satisfied, serum clearance decreases with increased pH 6.0 FcRn binding. Understanding the parameters that govern pH-dependent binding and IgG recycling will aid in the design of Fc containing biologics and expand their application.
EXPERIMENTAL PROCEDURES
Reagents and Figures
All chemicals were of analytical grade. Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs. Oligonucleotides were purchased from Integrated DNA Technologies. An anti-CD20 antibody (HB20-3) (28) and the anti-RSV antibody motavizumab (29) were used as backbones for characterization of Fc variants. Recombinant human and cynomolgus FcRn were expressed and purified as described previously (11). Antibody positions are listed according to the Kabat EU numbering convention (30). Images for Fig. 1 were generated using the YTE Fc-FcRn-SA complex structure (Protein Data Bank entry 4N0U) (12) and PyMOL (Schrödinger, LLC). Sequence Logo (31) figures were generated using Weblogo (32).
FIGURE 1.
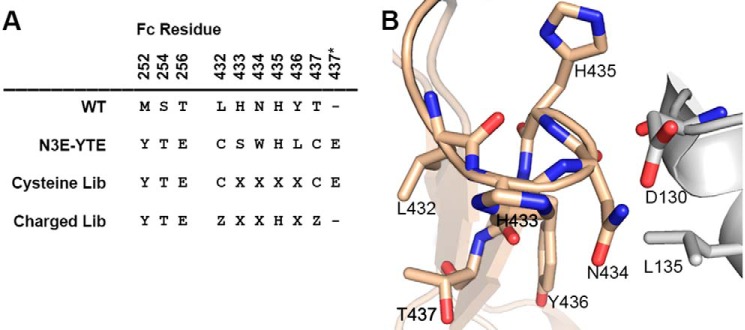
Library design and the His-435 loop. A, alignment of N3E-YTE mutations with corresponding wild-type IgG1 residues. Kabat EU numbering (30) is used; 437* designates an amino acid insertion after amino acid 437. The design of cysteine and charged libraries is shown (where X represents any amino acid and Z represents H, D, E, or Q). B, the His-435 loop (residues 432–437) of human IgG (wheat) is shown in complex with interacting residues on FcRn (gray; Protein Data Bank code 4N0U).
Construction of Phage Libraries
DNA encoding IgG1 Fc (including YTE mutations in CH2) (11) starting from Glu-216 (Kabat numbering) to the Fc C terminus was cloned as a gene-3 fusion into a phagemid vector. The cysteine loop library was constructed using a degenerate primer that randomized at positions 433, 434, 435, and 436 with the NNS codon (Fig. 1A). Positions 432 and 437 encoded cysteines, and an additional codon encoding glutamic acid was inserted following T437C. For the charged loop library, a PCR primer was designed with a fixed histidine at position 435 while randomizing positions 433, 434, and 436 (via an NNS codon). At positions 432 and 437, an STE codon was placed in the primer, encoding histidine, glutamic acid, aspartic acid, or glutamine. The Fc gene generated with the designed primers was cloned into a phagemid vector by restriction cloning.
pH Binding Screening of Phage Library
Phage libraries were panned against biotinylated, recombinant FcRn essentially as described (11). Briefly, 2 μg/ml biotinylated, recombinant human and mouse FcRn were mixed and added to Maxisorp neutravidin immunoplate (Pierce). The plate was then blocked with 3% milk for 1 h at room temperature and then washed with 20 mm MES buffer (pH 6.0) before adding phage. Library phage (6 × 1012 pfu) prepared in 20 mm MES buffer, pH 6.0, plus 0.05% Tween 20 and 3% milk was added to FcRn-coated wells. Following 2 h of incubation at 37 °C, the wells were washed 20 times with 20 mm MES buffer, pH 6.0, plus 0.05% Tween 20 and 0.3 m NaCl. The bound phage were eluted by 100 μl of PBS, pH 7.4, for 30 min at 37 °C. To discourage pH 7.4 binding, eluted phage were added to wells freshly coated with FcRn at pH 7.4 for 1 h at 37 °C. Unbound phage were collected and used to infect TG1 cells. The pH binding screen to each library was performed four times with the pH 7.4 binding depletion step at the end of each round of screening.
Phage ELISA
Maxisorb neutravadin plates were coated with 2 μg/ml biotinylated hFcRn at 4 °C overnight. Plates were then washed three times with 100 mm sodium phosphate buffer and blocked with 3% milk. pH 6.0 and pH 7.4 binding ELISAs were performed in phosphate buffers of the appropriate pH. 109 phage were used for the respective ELISA, and phage incubation was performed at room temperature for 1 h. Following binding, plates were washed three times with pH 6.0 or pH 7.4 sodium phosphate buffer with 0.2% Tween 20. Anti-M13 HRP (1:5000) was then added and incubated 30 min before washing and the addition of 3,3′,5,5′-tetramethylbenzidine substrates. Absorbance at 450 nm was measured after development with 3,3′,5,5′-tetramethylbenzidine substrate (KPL) according to the manufacturer's directions.
Surface Plasmon Resonance (SPR) Measurements
The interaction of recombinant hFcRn with immobilized antibody variants was monitored by SPR with a ProteOnTM XPR36 (Bio-Rad) (33). Antibodies were first coupled to a GLC sensor chip using a ProteOnTM amine coupling kit (Bio-Rad) according to the manufacturer's instructions. Excess reactive groups were blocked with a 6-min injection of 1 m ethanolamine. Antibodies were immobilized at a surface density between ∼1000 and 5000 reference units for equilibrium measurements and <500 response units for kinetic binding experiments. FcRn was used at concentrations ranging from 0.45 nm to 3 μm at a flow rate of 25 μl/min for steady-state measurements and 75 μl/min for kinetic measurements. One channel was always left unmodified to provide a blank reference surface. Dilutions and binding experiments were carried out at 25 °C in phosphate-buffered saline (PBS), pH 6.0 or pH 7.4, containing 0.05% Tween 20. Antibody surfaces were regenerated with a 15-s injection of 5 mm HCl. Binding affinities were determined by using the ProteOnTM Manager software (Bio-Rad). Dissociation constants (KD values) were determined by fitting the corresponding binding isotherms for steady-state data or by fitting the kinetics for association and dissociation employing a 1:1 Langmuir mass transfer model. Equilibrium KD values greater than 10,000 nm are reported in tables as “>10,000” because the highest concentration of FcRn evaluated was 3 μm. Representative sensograms and kinetic data for all variants listed in Table 2 are included in supplemental Table 1.
TABLE 2.
Human FcRn binding to various motavizumab IgGs and hFcRn transgenic mouse PK data
| Motavizumab construct | pH 6 Hu FcRn KDa | pH 7.4 Hu FcRn KDa | Clearance | β-Phase t½ | AUCinf |
|---|---|---|---|---|---|
| nm | nm | ml/day/kg | days | μg·day/kg | |
| Wild type | 2140 | >10,000 | 60.1 | 0.73 | 42 |
| YTE | 314 | >10,000 | 23.3 | 3.28 | 108 |
| N3E-YTE | 3 | 25 | 373.0 | 0.78 | 7 |
| N3-YTE | 13 | 383 | 61.9 | 0.54 | 40 |
| N3E | 41 | 860 | 33.3 | 2.16 | 75 |
| Y31-YTE | 84 | 1160 | 23.8 | 3.65 | 105 |
| Y3-YTE | 53 | 2680 | 26.4 | 2.07 | 95 |
| Y12-YTE | 86 | 5040 | 26.4 | 2.75 | 94 |
| Y12 | 254 | >10,000 | 32.3 | 2.44 | 71 |
| Y31 | 491 | >10,000 | 37.5 | 2.99 | 67 |
a Steady-state affinity measurements carried out by ProteOn as described under “Experimental Procedures.” Values in italic type were determined via kinetic model (Langmuir with mass transfer).
In Vivo PK in hFcRn Transgenic Mice
hFcRn transgenic mice used in this study are the F1 cross of murine FcRn-deficient B6.129X1-Fcgrttm1Dcr/DcrJ and hFcRn cDNA transgenic line B6.Cg-Fcgrttm1Dcr Tg (CAG-FCGRT) 276 Dcr/DcrJ (7, 34). Sex-matched (6–16-week-old) hFcRn mice were given a bolus intravenous dose of 2.5 mg/kg antibody on day 0. Eight mice were used per antibody, with two groups of mice (A group or B group) bled at alternate time points. Blood samples were obtained from the retro-orbital plexus using capillary pipettes at different time points throughout the 2–3-week studies. A quantitative ELISA was used to monitor the serum concentrations of the tested antibodies. Briefly, 96-well plates were coated with 2 μg/ml AffiPure goat anti-human F(ab′)2 fragment-specific antibody (Jackson Immunoresearch). Plates were blocked with 3% BSA in PBS for 1 h and then incubated with appropriately diluted serum samples (1:200 for earlier time points and 1:50 or 1:100 for later time points). Goat anti-human Fd-specific HRP-conjugated antibody (Southern Biotechnology Associates) was used to detect the human antibody (dilution 1:10,000). Absorbance at 450 nm was measured after development with 3,3′,5,5′-tetramethylbenzidine substrate (KPL) according to the manufacturer's directions. Standard curves were generated for each antibody variant diluted into 1:100 prebleed mouse serum (taken at day −3). The linear portions of standard curves generated in Prism (GraphPad Software) were then used to quantify human IgG in the serum samples.
In Vivo PK in Cynomolgus Monkey
A non-GLP PK study was carried out to determine antibody serum persistence. Twelve female cynomolgus monkeys were randomized and assigned to one of three study groups. Each animal received an intravenous dose of motavizumab, YTE-motavizumab, or Y31-YTE-motavizumab at 10 mg/kg. Blood samples were drawn prior to dosing on day 0; at 1, 4, and 12 h after dosing; and at 2, 3, 4, 8, 15, 22, 29, 42, 57, 71, and 85 days after dosing. The serum concentrations of antibodies were determined using the anti-motavizumab ELISA as described previously (21) with a separate standard curve generated for each variant tested.
PK Data Analysis
Data collected from hFcRn mice and the cynomolgus monkey PK studies were analyzed using non-compartmental analysis model 201 of Phoenix WinNonlin version 5.2 (Certara, Princeton, NJ). AUCinf (area under the curve to infinity) was calculated using the log-linear trapezoidal method. Terminal half-life (t½) was calculated using log-linear regression of the concentration data, including at least the last three sampling time points with measurable concentrations. Serum clearance was estimated as CL = dose/AUCinf. Descriptive statistics for major PK parameters were then calculated.
RESULTS
Engineering and Identification of pH-dependent, High Affinity FcRn-binding Fc Variants from a Phage Library
A high affinity FcRn-binding Fc variant exhibiting poor pH dependence, N3E-YTE, was identified by screening a phage library containing combinatorial mutations in the His-435 loop, including an amino acid insertion after residue 437. N3E-YTE also harbors the YTE mutation in CH2, which improves FcRn binding and serum half-life in experimental models and in humans (21, 23). Intriguingly, N3E-YTE was found to contain two cysteine residues at positions 432 and 437, forming the base of the His-435 loop. The N3E-YTE sequence and the His-435 loop of IgG1 are shown in Fig. 1. To engineer pH-dependent binding into this high affinity variant, a new Fc library was generated by randomizing the intervening sequences between L432C and T437C (cysteine library; Fig. 1A). This library was screened with a goal of identifying variants that retained high affinity binding at pH 6.0 yet exhibited low affinity at neutral pH. A pH 7.4 binding step following pH 6.0 binding selection was added to deplete variants with high neutral pH affinity to FcRn. A phage ELISA was also used to characterize binding of individual phage clones at both pH 6.0 and pH 7.4 after four rounds of panning. Several clones were identified that showed significantly reduced binding at pH 7.4 yet still bind strongly to FcRn at pH 6.0 (Fig. 2). Sequence analysis of clones showed that His-435 was unanimously selected, highlighting the key role of this residue in FcRn binding. Additionally, clones that showed improved pH binding dependence (e.g. C37-YTE, C56-YTE, C73-YTE, and C59-YTE) were highly enriched with positively charged amino acid arginine at position 433, 434, or 436 (Fig. 2).
FIGURE 2.
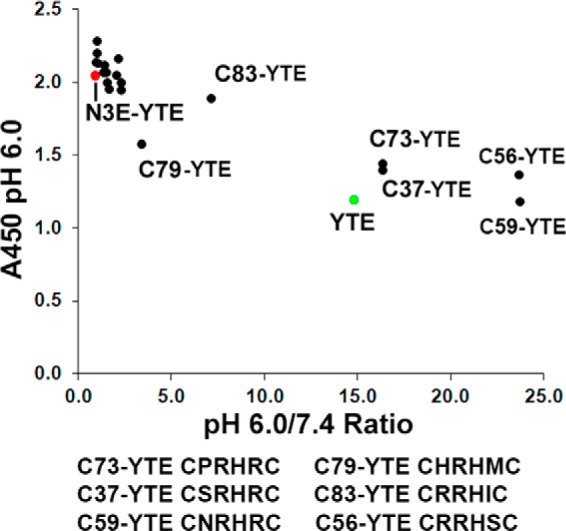
Phage ELISA results of variants identified from the cysteine library. Binding at pH 6.0 (A450) is plotted against the ratio of pH 6.0 over pH 7.4 binding. High pH 6.0 A450 values and a high pH 6.0/7.4 ratio suggest high affinity and pH-dependent binding, respectively. N3E-YTE (red) is a high affinity, pH-independent variant that shows a high A450 reading at pH 6.0 yet a low pH 6.0/7.4 binding ratio. Sequences (residues 432–437) are shown for selected clones. All variants contain the YTE mutation (M252Y/S254T/T256E).
Another library (charged library) was designed to replace the cysteine residues at positions 432 and 437 with charged residues, including histidine, and randomized amino acids at positions 433, 434, and 436 while leaving His-435 unchanged (Fig. 1A). We reasoned that charged amino acids at the base of the loop may introduce pH-dependent changes that could be additionally beneficial to pH-dependent FcRn binding. In this charged library, Glu, Asp, or His was introduced at positions 432 and 437. Glutamine was also encoded at these positions due to the use of degenerated oligonucleotides in library construction. The Glu insertion at position 437* was not included in the charged library (Fig. 1). The same pH-dependent selection scheme used to screen the cysteine library was used to screen the charged library. Following four rounds of panning, individual phage clones were sequenced, and their binding was screened via phage ELISA as described above. Unlike the cysteine library results, the majority of the clones identified from the charged library exhibited improved pH-dependent binding compared with N3E-YTE, demonstrated by the high ratio of pH 6.0 versus pH 7.4 binding in the phage ELISA (Fig. 3A). Furthermore, a consensus sequence pattern of 432ER(Y/F)HRQ437 was revealed in the clones (Fig. 3B). Residues with aromatic side chains were highly enriched at position 434, as were positively charged amino acids (especially arginine) at 433 and 436. Glu and Gln were the preferred amino acids at positions 432 and 437, respectively, in the pH-dependent binding clones.
FIGURE 3.
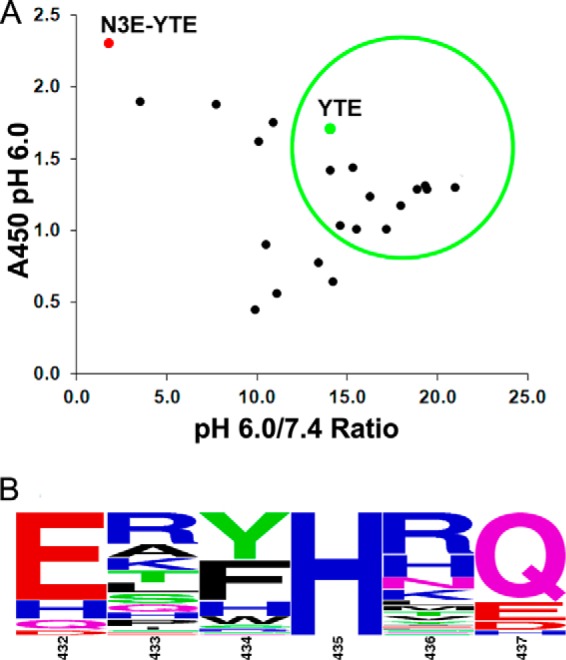
Sequence analysis and phage ELISA results of variants identified from the charged library. A, phage ELISA data are presented as shown in Fig. 2. The majority of variants identified from the charged library show high affinity and pH-dependent binding. Clones displayed in the green circle were converted to full-length IgG and reassessed for FcRn binding (see Table 1). B, sequence logo representation of a total of 68 phage clones identified after four rounds of phage panning. The relative letter size represents the frequency of the amino acid occurring at the position. Positions 433, 434, 436, and 437 are randomized in the library. Amino acids Glu, Asp, His, or Gln are encoded in positions 432 and 437. His-435 was not mutated in the library.
SPR Binding Characterization of FcRn-binding Variants as IgG
Variants of N3E-YTE and a panel of variants from the cysteine and charged libraries were initially generated as IgGs in the HB20.3 (anti-CD20) background (28) for further characterization. SPR was used to measure binding of purified antibodies to FcRn at both pH 6.0 and pH 7.4 (Table 1).
TABLE 1.
Human FcRn binding to various anti-CD20 IgGs including cysteine loop and charged loop library variants
| Variant | Sequence (432–437)a | pH 6.0 KDb | pH 7.4 KDb | Notes |
|---|---|---|---|---|
| nm | nm | |||
| Wild type | LHNHYT | 880 | >10,000 | Anti-CD20 IgG1 (28) |
| YTE | LHNHYT | 215 | 5750 | (M252Y, S254T, T256E) (11) |
| N3E-YTE | CSWHLCEc | 4 | 41 | Original construct identified from phage panning (contains glutamine insertion after residue 437) |
| N3-YTE | CSWHLC | 24 | 353 | Glutamine insertion removed from N3E-YTE |
| N3E | CSWHLCE | 49 | 1260 | YTE removed from N3E-YTE |
| CwtC-YTE | CHNHYC | 367 | 7700 | L432C, T437C, YTE |
| C83-YTE | CRRHICE | 160 | 6650 | Cysteine loop library |
| C37-YTE | CSRHRCE | 225 | 10,000 | Cysteine loop library |
| C56-YTE | CRRHSCE | 364 | >10,000 | Cysteine loop library |
| C59-YTE | CNRHRCE | 737 | >10,000 | Cysteine loop library |
| Y3-YTE | ERYHTQ | 36 | 2640 | Charged loop library |
| Y31-YTE | ERFHRQ | 57 | 4730 | Charged loop library |
| Y12-YTE | EAWHRQ | 34 | 6970 | Charged loop library |
| Y83-YTE | ERFHSQ | 120 | >10,000 | Charged loop library |
| Y53-YTE | ERFHTQ | 68 | 5460 | Charged loop library |
| Y43-YTE | EPHHRQ | 152 | >10,000 | Charged loop library |
| Y34-YTE | HTHHRQ | 187 | >10,000 | Charged loop library |
| Y39-YTE | EPWHYQ | 25 | 947 | Charged loop library |
| Y37-YTE | HRFHLQ | 179 | 1520 | Charged loop library |
| Y38-YTE | EQFHRQ | 152 | >10,000 | Charged loop library |
a Residue numbering is according to Kabat et al. (30).
b Steady-state affinity measurements carried out by ProteOn as described under “Experimental Procedures.” Values in italic type were determined via kinetic model (Langmuir with mass transfer).
c Boldface type indicates insertion residue 437*.
To determine which mutations conferred the extreme FcRn affinity increases and poor pH dependence to N3E-YTE, N3-YTE (lacking the Glu insertion after position 437) and N3E (lacking YTE) were generated. N3-YTE had a slight pH 6.0 decrease and a significant affinity decrease at 7.4 relative to N3E-YTE, indicating that the glutamate insertion does play a role in reducing pH dependence (Table 1). N3E had significantly reduced affinity at both pH 6.0 and pH 7.4, suggesting that the combination of N3E with the YTE mutations in CH2 is responsible for the ultra-high affinity of N3E-YTE. Additionally, the L423C/T437C mutation (CwtC-YTE) was generated. This variant did not improve FcRn binding compared with YTE, indicating that the cysteines alone do not confer affinity increases in N3E-YTE (Table 1).
Variants from the charged library, such as Y3-YTE, Y12-YTE, and Y31-YTE, show ∼3–6-fold binding affinity improvements over YTE at pH 6.0. The pH 7.4 binding of all tested variants identified in either library was significantly reduced compared with N3E-YTE. Many variants exhibit neutral pH FcRn affinity similarly to YTE IgG (>5 μm). Cysteine loop library variants showing pH dependence in phage ELISA maintained pH-dependent binding as full-length IgGs as well; however, these variants did not exhibit improved pH 6.0 affinity to FcRn compared with YTE.
As previous studies have shown that variable domains can influence Fc and FcRn interaction (24, 35, 36), the FcRn binding of a few high affinity, pH-dependent clones was tested in an additional IgG1 background, motavizumab (Table 2) (29). Clones Y12-YTE, Y31-YTE, and Y3-YTE were generated in themotavizumab background, and FcRn binding was examined. The pH 6.0 binding of these clones, as well as of YTE, N3E, N3-YTE, and N3E-YTE in motavizumab, is consistent with binding in the anti-CD20 background. Interestingly, binding at pH 7.4 varied in some mutants upon switching variable domains, with either increased binding (Y31-YTE) or decreased binding (YTE) to FcRn observed. Additionally, clones Y12 and Y31 (lacking the YTE mutation) exhibited reduced FcRn binding at both pH 6.0 and pH 7.4, suggesting that the contribution of YTE to binding is not sensitive to pH.
SPR Binding Characterization of Y31-YTE Variants
The role that highly enriched hydrophobic aromatic residues at the 434 position play in FcRn affinity was interrogated by mutating N434F to all 19 other amino acids in the Y31-YTE motavizumab background (Table 3). SPR binding to FcRn shows that aromatic amino acids (including histidine) confer large affinity increases to FcRn at both acidic and neutral pH. Neither hydrophobic non-aromatic residues nor polar amino acids at the 434-position had improved binding, and charged amino acids were detrimental to FcRn binding.
TABLE 3.
Human FcRn binding to Y31-YTE motavizumab (432–437 sequence: ERFHRQ) variants with amino acid substitutions at the 434-position
| Amino acid at 434 position | pH 6.0 KDa | pH 7.4 KDa |
|---|---|---|
| nm | nm | |
| Phe (Y31-YTE) | 84 | 1160 |
| Tyr | 48 | 1350 |
| Trp | 41 | 2120 |
| Leu | 6050 | >10,000 |
| Ile | 2750 | >10,000 |
| Val | 1390 | >10,000 |
| Met | 1740 | >10,000 |
| Ala | 390 | 2640 |
| Ser | 328 | 4540 |
| Thr | 858 | >10,000 |
| Gln | 369 | 4920 |
| Asn | 673 | 6920 |
| Asp | 4850 | >10,000 |
| Glu | >10,000 | >10,000 |
| His | 67 | 1440 |
| Lys | 1900 | >10,000 |
| Arg | 1860 | >10,000 |
| Gly | 200 | 2290 |
| Cys | 1680 | >10,000 |
| Prob |
a Steady-state affinity measurements carried out by ProteOn as described under Experimental Procedures. Values in italics were determined via kinetic model (Langmuir with mass transfer).
b Y31-YTE 434P had no expression.
Serum Clearance in hFcRn Mice
Serum clearance of several motavizumab variants harboring the engineered Fcs were tested in hFcRn transgenic mice. In these experiments, mice were dosed with 2.5 mg/kg antibody, and serum antibody concentrations were determined in an ELISA. Table 2 summarizes the relevant PK parameters as well as the sequences and FcRn binding properties of the tested antibodies. Serum clearance of representative variants is plotted in Fig. 4A. As expected, the YTE mutation reduced the serum clearance by ∼2.5-fold compared with motavizumab. N3E-YTE showed rapid clearance of 373 ml/day/kg, approximately 6 times faster than motavizumab, confirming that high affinity binding at pH 7.4 can be detrimental to the antibody recycling. All engineered variants tested except for N3-YTE and N3E-YTE exhibited 1.6–2.5-fold reduced serum clearance compared with motavizumab, despite notably increased pH 7.4 binding in many variants compared with YTE or the parental IgG. For instance, N3E exhibits substantial binding at pH 7.4 (KD of 860 nm) yet has a clearance significantly less than motavizumab (Fig. 4A). However, N3-YTE with further increased neutral pH binding (KD of 383 nm) did not confer reduced clearance, indicating that the neutral pH binding is beginning to detrimentally affect serum persistence. As expected, the ultra-high affinity binding at both acidic and neutral pH in variant N3E-YTE led to over 6- and 16-fold faster serum clearance relative to motavizumab and YTE-motavizumab, respectively. Fig. 4B further illustrates the relationships between binding and clearance.
FIGURE 4.
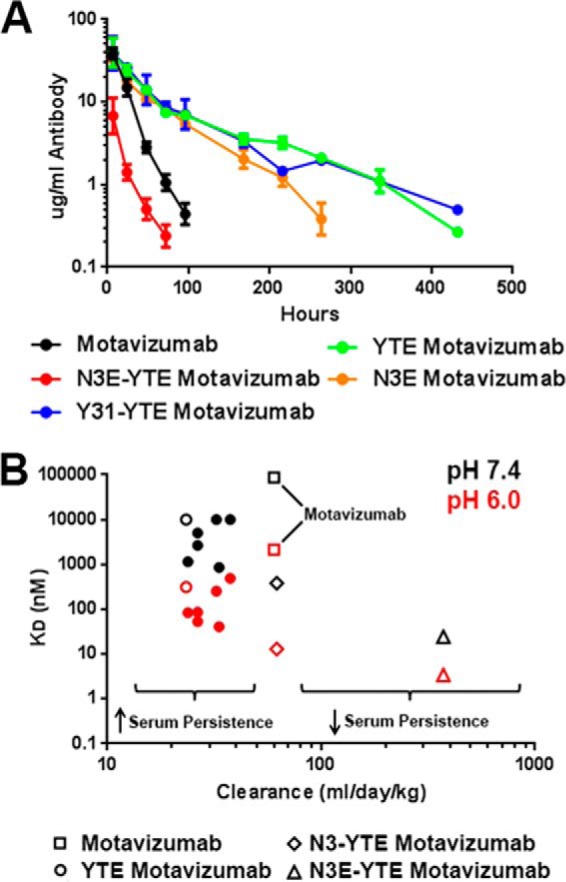
Pharmacokinetic properties of motavizumab variants in hFcRn transgenic mice. A, clearance curves for wild-type (black), YTE (green), Y31-YTE (blue), N3E (orange), and N3E-YTE (red) versions of motavizumab in hFcRn transgenic mice. B, clearance of all antibody variants described in Table 2 plotted against pH 6.0 (red) and pH 7.4 (black) hFcRn binding affinity. YTE (open circles), motavizumab (open squares), N3-YTE (open diamonds), and N3E-YTE (open triangles) are highlighted. A previously reported value of 88 μm is used for wild-type IgG1, pH 7.4, affinity (38) because motavizumab, pH 7.4, binding could not be quantified. Values of 10 μm were used for plotting YTE, Y12, and Y31, pH 7.4, KD. High FcRn affinity at acidic pH generally corresponds with decreased clearance with low pH 7.4 binding (less than 860 nm). FcRn affinity higher than 860 nm at pH 7.4 results in faster clearance (as with N3-YTE and N3E-YTE). Error bars, S.E.
PK in Cynomolgus Monkeys
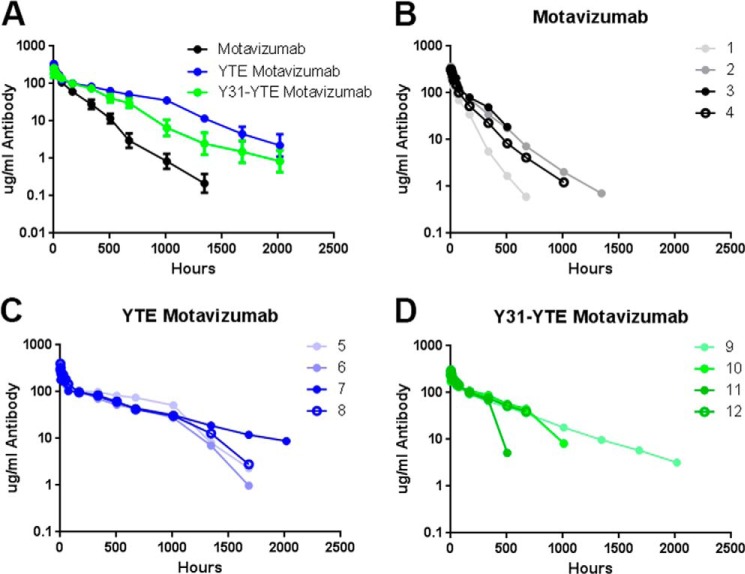
The PK of Y31-YTE, which exhibits slow clearance in hFcRn mice, was further tested in cynomolgus monkeys together with motavizumab and YTE-motavizumab. Fig. 5 shows the average serum concentration-time profiles as well as the profiles of individual animals within each group. In agreement with a previous study (21), YTE-motavizumab had extended serum half-life versus motavizumab in cynomolgus monkeys. Y31-YTE also had improved PK properties versus motavizumab; however, three animals in the Y31-YTE group exhibited abrupt drop-offs in serum concentrations near the 500 h time point (Fig. 5D), possibly indicating that anti-drug antibodies were clearing the variant. Similar decreases in the motavizumab and YTE-motavizumab group were not seen or were less profound during this period. Cynomolgus FcRn binding affinities and PK parameters are summarized in Table 4.
FIGURE 5.
Individual clearance curves of motavizumab variants in cynomolgus monkeys. A, average ± S.E. (error bars) serum concentrations of the three motavizumab variants dosed. Data points below the limit of quantitation (0.1 μg/ml) were given values of 0.05 μg/ml to generate averaged serum concentration-time profiles. Serum IgG levels of each animals within the motavizumab group (1–4) (B), YTE-motavizumab-dosed group (5–8) (C), and Y31-YTE-motavizumab-dosed group (9–12) (D) are shown. Data points below the limit of quantitation are not shown. YTE and Y31-YTE show prolonged serum half-life compared with motavizumab.
TABLE 4.
Cynomolgus monkey FcRn binding to various motavizumab IgGs and PK data
| Motavizumab construct | pH 6 Cyno FcRn KDa | pH 7.4 Cyno FcRn KDa | Clearance | β-Phase t½ | AUCinf |
|---|---|---|---|---|---|
| nm | nm | ml/day/kg | days | μg·day/kg | |
| Wild type | 1920 | >10,000 | 8.5 | 5.8 | 1420 |
| YTE | 275 | 8000 | 2.9 | 11.6 | 3720 |
| N3E-YTE | 4 | 37 | NTb | NT | NT |
| Y31-YTE | 76 | 1190 | 4.6 | 10.6 | 2510 |
| Y3-YTE | 44 | 7700 | NT | NT | NT |
| Y12-YTE | 90 | >10,000 | NT | NT | NT |
a Values in italic type were determined via kinetic model (Langmuir with mass transfer).
b NT, variant not tested in cynomolgus monkeys.
DISCUSSION
Engineering the pH-dependent interaction between IgG and FcRn has been a central approach for modulating the PK of potential therapeutic antibodies. There are currently many examples of Fc variants with increased FcRn binding at acidic pH having improved in vivo half-lives (16–21). However, additional studies suggest that a more complex relationship exists between FcRn affinity increase and improved half-life (25, 26, 37). In this study, we sought to understand how increased neutral pH binding can limit in vivo half-life improvements and to better define at what point gains in serum persistence are hindered by excess FcRn affinity at the neutral cell surface.
We characterized a panel of variants with increased FcRn binding affinity at acidic pH and variable binding at pH 7.4. Our data show that high affinity FcRn binding at pH 6.0 can yield substantial serum half-life increases compared with parental IgG1 despite considerable FcRn binding at pH 7.4 (up to 860 nm). These findings are exemplified by Y31-YTE and N3E motavizumab, which have neutral pH KD values of 1160 and 860 nm, respectively (KD of 84 and 41 nm at pH 6.0) and exhibit 2–3-fold slower clearance compared with motavizumab in hFcRn mice. In cynomolgus monkeys, Y31-YTE motavizumab exhibits ∼2-fold lower clearance than the parental antibody. These data support a direct correlation between increased FcRn affinity at acidic pH and increased serum persistence until a point where neutral pH binding limits and eventually reverses these improvements. This neutral pH threshold is exemplified by N3-YTE motavizumab, which lacks any PK improvement yet has substantial FcRn affinity increases compared with motavizumab (pH 6.0 KD of 13 nm and pH 7.4 KD of 383 nm). Similarly to N3-YTE, IgG Fc variants having high neutral pH FcRn affinities (as low as ∼100 nm) have been described that exhibit serum persistence values near that of the parental IgG (16, 38). In these variants and with N3-YTE, PK improvements achieved by increased FcRn affinity at acidic pH are offset by hindered exocytosis at the neutral cell surface, yielding no net gain in serum persistence. Finally, variants characterized by ultra-high affinity FcRn binding at neutral pH (e.g. N3E-YTE) exhibit serum persistence much shorter than the parental IgG. The reduced half-life is probably driven by severely impaired exocytosis, which can in turn even impair endogenous IgG recycling, resulting in Abdeg antibodies (27).
The novel variants described in this paper have a wide range of FcRn affinities at pH 6.0 and 7.4. Analysis of the isolated mutations in the context of the recently solved YTE Fc-FcRn-HSA crystal structure (12) can aid in further explaining the individual affinity contributions of mutations in the variants described. The ultra-high affinity of N3E-YTE at both acidic and neutral pH is probably due to several contributing factors, including the YTE mutations, hydrophobic residues at positions 434 and 436, a glutamic acid insertion after residue 437, and the cysteines encoded at positions 432 and 437 (Fig. 1). Among these mutations, N434W is probably responsible for a significant portion of the affinity gain via introducing a new hydrophobic interaction with Leu-135 of FcRn (Fig. 1B). The N434W point mutation has been shown to confer increased binding affinity to FcRn at both acidic and neutral pH (16). Our results from substituting all 19 additional amino acids for N434F in Y31-YTE motavizumab show that only amino acids with aromatic side chains (Phe, Tyr, Trp, and His) at position 434 have significant FcRn affinity gains relative to YTE-motavizumab (Table 3), supporting the role of these mutations in the noted affinity gains. The disulfide bond between L432C and T437C, verified by mass spectrometry (data not shown), serves to stabilize the loop and orients the solvent-exposed hydrophobic residues responsible for the FcRn affinity increase; L432C/T437C alone, however (Table 1), has no effect on FcRn binding.
The cysteine library was designed to identify pH-dependent FcRn-binding variants by retaining the N3E-YTE cysteines at positions 432 and 437 but randomizing intervening sequences. His-435 was unanimously selected in round four variants, confirming the importance of this residue in FcRn binding (10). However, recent structural data suggest that although His-435 may have an important structural role, it does not interact directly with negatively charged side chains on FcRn (12), and His-310 is actually the critical histidine for imparting pH dependence. Additionally, cysteine library variants with improved pH dependence were enriched for arginine at positions 433, 434, and 436. Although these mutations (e.g. C37-YTE, C83-YTE, etc.) reintroduced pH dependence into L432C/T437C-containing variants, it was at the expense of pH 6.0 affinity because these variants showed no binding improvement over YTE (Table 1). It was only with the charged loop library that multiple pH-dependent, higher affinity Fcs were isolated. We speculate that the rigidity imposed by an L432C/T437C disulfide bond may be deleterious to pH dependence, whereas charged or polar residues at these positions (as in the charged loop library) can maintain loop flexibility and adopt a loop orientation favorable to pH-dependent binding.
Multiple pH-dependent, high affinity pH 6.0 binding variants were identified from the charged loop library. These variants had bulky hydrophobic residues at position 434 (like N3E-YTE) yet were still able to confer pH-dependent FcRn binding and significantly improved serum persistence. Modeling again suggests that hydrophobic residues at the 434-position can interact with Leu-135 on FcRn, explaining the affinity increase. Intriguingly, in the orientation observed in the Fc-FcRn complex (12), bulky side chains at position 434 will clash with the Asp-130 side chain on FcRn (Fig. 1B). To account for this bulky residue at 434, we propose that an alternate Asp-130 rotamer (which faces away from the 434 side chain and toward His-435) may be imposed, which positions Asp-130 within distance to interact with His-435, resulting an additional pH-dependent interaction. We suspect that this, in the context of L432E/T437Q mutations at the base of the loop, allows the charged loop library variants to exhibit pH dependence, whereas N3E-YTE (and to a lesser extent N3-YTE and the majority of cysteine loop library clones) does not.
In this study, the novel variant with the longest half-life in hFcRn mice, Y31-YTE, had almost 3-fold slower clearance than motavizumab and ∼5-fold longer half-life. These data put Y31-YTE among the top PK improved variants described so far (14). PK data in cynomolgus monkeys also showed improvements for Y31-YTE relative to motavizumab (Table 4), despite sharp drops in the Y31-YTE serum levels in three of four animals, indicative of anti-drug antibody-mediated clearance. PK modeling based on our data and those of others suggests that there is probably little room for significant half-life extension (beyond the 3–4-fold enhancement already achieved) by continued FcRn binding engineering. Although our results are consistent with many others in showing that altering FcRn affinity can be used to predictably change the PK outcomes of antibodies, many factors other than FcRn binding (e.g. antibody stability and variability in antigen distribution) also play a role in determining the ultimate PK outcome of an engineered antibody. In this regard, we do envision that extremely long half-lives of Fc-containing molecules could be achieved by combining FcRn binding engineering with FcRn-independent half-life-extending technologies, such as pI optimization (39) and pH-dependent antigen binding engineering (40, 41).
Acknowledgments
We thank Mun Mun Patniak for performing cyno PK quantification and Lu Shan for reviewing the manuscript.

This article contains supplemental Table 1.
- SA
- serum albumin
- PK
- pharmacokinetic
- hFcRn
- human FcRn
- SPR
- surface plasmon resonance
- YTE
- M252Y/S254T/T256E.
REFERENCES
- 1. Ghetie V., Hubbard J. G., Kim J. K., Tsen M. F., Lee Y., Ward E. S. (1996) Abnormally short serum half-lives of IgG in β2-microglobulin-deficient mice. Eur. J. Immunol. 26, 690–696 [DOI] [PubMed] [Google Scholar]
- 2. Ghetie V., Ward E. S. (2000) Multiple roles for the major histocompatibility complex class I-related receptor FcRn. Annu. Rev. Immunol. 18, 739–766 [DOI] [PubMed] [Google Scholar]
- 3. Israel E. J., Wilsker D. F., Hayes K. C., Schoenfeld D., Simister N. E. (1996) Increased clearance of IgG in mice that lack β2-microglobulin: possible protective role of FcRn. Immunology 89, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim J. K., Firan M., Radu C. G., Kim C. H., Ghetie V., Ward E. S. (1999) Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. Eur. J. Immunol. 29, 2819–2825 [DOI] [PubMed] [Google Scholar]
- 5. Martin W. L., West A. P., Jr., Gan L., Bjorkman P. J. (2001) Crystal structure at 2.8 angstrom of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol. Cell 7, 867–877 [DOI] [PubMed] [Google Scholar]
- 6. Roopenian D. C., Akilesh S. (2007) FcRn: the neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7, 715–725 [DOI] [PubMed] [Google Scholar]
- 7. Roopenian D. C., Christianson G. J., Sproule T. J., Brown A. C., Akilesh S., Jung N., Petkova S., Avanessian L., Choi E. Y., Shaffer D. J., Eden P. A., Anderson C. L. (2003) The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J. Immunol. 170, 3528–3533 [DOI] [PubMed] [Google Scholar]
- 8. Medesan C., Matesoi D., Radu C., Ghetie V., Ward E. S. (1997) Delineation of the amino acid residues involved in transcytosis and catabolism of mouse IgG1. J. Immunol. 158, 2211–2217 [PubMed] [Google Scholar]
- 9. Raghavan M., Bonagura V. R., Morrison S. L., Bjorkman P. J. (1995) Analysis of the pH dependence of the neonatal Fc receptor/immunoglobulin G interaction using antibody and receptor variants. Biochemistry 34, 14649–14657 [DOI] [PubMed] [Google Scholar]
- 10. Shields R. L., Namenuk A. K., Hong K., Meng Y. G., Rae J., Briggs J., Xie D., Lai J., Stadlen A., Li B., Fox J. A., Presta L. G. (2001) High resolution mapping of the binding site on human IgG1 for FcγRI, FcγRII, FcγRIII, and FcRn and design of IgG1 variants with improved binding to the FcγR. J. Biol. Chem. 276, 6591–6604 [DOI] [PubMed] [Google Scholar]
- 11. Dall'Acqua W. F., Woods R. M., Ward E. S., Palaszynski S. R., Patel N. K., Brewah Y. A., Wu H., Kiener P. A., Langermann S. (2002) Increasing the affinity of a human IgG1, for the neonatal Fc receptor: biological consequences. J. Immunol. 169, 5171–5180 [DOI] [PubMed] [Google Scholar]
- 12. Oganesyan V., Damschroder M. M., Cook K. E., Li Q., Gao C., Wu H., Dall'Acqua W. F. (2014) Structural insights into neonatal Fc receptor-based recycling mechanisms. J. Biol. Chem. 289, 7812–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt M. M., Townson S. A., Andreucci A. J., King B. M., Schirmer E. B., Murillo A. J., Dombrowski C., Tisdale A. W., Lowden P. A., Masci A. L., Kovalchin J. T., Erbe D. V., Wittrup K. D., Furfine E. S., Barnes T. M. (2013) Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface. Structure 21, 1966–1978 [DOI] [PubMed] [Google Scholar]
- 14. Presta L. G. (2008) Molecular engineering and design of therapeutic antibodies. Curr. Opin. Immunol. 20, 460–470 [DOI] [PubMed] [Google Scholar]
- 15. Strohl W. R. (2009) Optimization of Fc-mediated effector functions of monoclonal antibodies. Curr. Opin. Biotechnol. 20, 685–691 [DOI] [PubMed] [Google Scholar]
- 16. Yeung Y. A., Leabman M. K., Marvin J. S., Qiu J., Adams C. W., Lien S., Starovasnik M. A., Lowman H. B. (2009) Engineering human IgG1 affinity to human neonatal Fc receptor: Impact of affinity improvement on pharmacokinetics in primates. J. Immunol. 182, 7663–7671 [DOI] [PubMed] [Google Scholar]
- 17. Deng R., Loyet K. M., Lien S., Iyer S., DeForge L. E., Theil F. P., Lowman H. B., Fielder P. J., Prabhu S. (2010) Pharmacokinetics of humanized monoclonal anti-tumor necrosis factor-α antibody and its neonatal Fc receptor variants in mice and cynomolgus monkeys. Drug Metab. Dispos. 38, 600–605 [DOI] [PubMed] [Google Scholar]
- 18. Hinton P. R., Johlfs M. G., Xiong J. M., Hanestad K., Ong K. C., Bullock C., Keller S., Tang M. T., Tso J. Y., Vásquez M., Tsurushita N. (2004) Engineered human IgG antibodies with longer serum half-lives in primates. J. Biol. Chem. 279, 6213–6216 [DOI] [PubMed] [Google Scholar]
- 19. Yeung Y. A., Wu X., Reyes A. E., 2nd, Vernes J. M., Lien S., Lowe J., Maia M., Forrest W. F., Meng Y. G., Damico L. A., Ferrara N., Lowman H. B. (2010) A therapeutic anti-VEGF antibody with increased potency independent of pharmacokinetic half-life. Cancer Res. 70, 3269–3277 [DOI] [PubMed] [Google Scholar]
- 20. Zalevsky J., Chamberlain A. K., Horton H. M., Karki S., Leung I. W. L., Sproule T. J., Lazar G. A., Roopenian D. C., Desjarlais J. R. (2010) Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dall'Acqua W. F., Kiener P. A., Wu H. (2006) Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J. Biol. Chem. 281, 23514–23524 [DOI] [PubMed] [Google Scholar]
- 22. Oganesyan V., Damschroder M. M., Woods R. M., Cook K. E., Wu H., Dall'acqua W. F. (2009) Structural characterization of a human Fc fragment engineered for extended serum half-life. Mol. Immunol. 46, 1750–1755 [DOI] [PubMed] [Google Scholar]
- 23. Robbie G. J., Criste R., Dall'acqua W. F., Jensen K., Patel N. K. L., Losonsky G. A., Griffin M. P. (2013) A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob. Agents Chemother. 57, 6147–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Datta-Mannan A., Chow C. K., Dickinson C., Driver D., Lu J., Witcher D. R., Wroblewski V. J. (2012) FcRn affinity-pharmacokinetic relationship of five human IgG4 antibodies engineered for improved in vitro FcRn binding properties in cynomolgus monkeys. Drug Metab. Dispos. 40, 1545–1555 [DOI] [PubMed] [Google Scholar]
- 25. Datta-Mannan A., Witcher D. R., Tang Y., Watkins J., Jiang W., Wroblewski V. J. (2007) Humanized IgG(1) variants with differential binding properties to the neonatal Fc receptor: relationship to pharmacokinetics in mice and primates. Drug Metab. Dispos. 35, 86–94 [DOI] [PubMed] [Google Scholar]
- 26. Datta-Mannan A., Witcher D. R., Tang Y., Watkins J., Wroblewski V. J. (2007) Monoclonal antibody clearance: impact of modulating the interaction of IgG with the neonatal Fc receptor. J. Biol. Chem. 282, 1709–1717 [DOI] [PubMed] [Google Scholar]
- 27. Vaccaro C., Zhou J., Ober R. J., Ward E. S. (2005) Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat. Biotechnol. 23, 1283–1288 [DOI] [PubMed] [Google Scholar]
- 28. Tedder T. F., Uchida J., Hamaguchi Y., Poe J. C. (May 28, 2009) Cd20-specific antibodies and methods of employing same. United States Patent Application Publication, US2009/0136516 A1
- 29. Wu H., Pfarr D. S., Johnson S., Brewah Y. A., Woods R. M., Patel N. K., White W. I., Young J. F., Kiener P. A. (2007) Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol. 368, 652–665 [DOI] [PubMed] [Google Scholar]
- 30. Kabat E. A., Wu T. T., Perry H. M., Gottesman K. S., Foeller C. (1991) Sequences of Proteins of Immunological Interest, 5th Ed., NIH Publication 91-3442, National Institutes of Health, Bethesda, MD [Google Scholar]
- 31. Schneider T. D., Stephens R. M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bravman T., Bronner V., Lavie K., Notcovich A., Papalia G. A., Myszka D. G. (2006) Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 358, 281–288 [DOI] [PubMed] [Google Scholar]
- 34. Chaudhury C., Mehnaz S., Robinson J. M., Hayton W. L., Pearl D. K., Roopenian D. C., Anderson C. L. (2003) The major histocompatibility complex related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J. Exp. Med. 197, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki T., Ishii-Watabe A., Tada M., Kobayashi T., Kanayasu-Toyoda T., Kawanishi T., Yamaguchi T. (2010) Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J. Immunol. 184, 1968–1976 [DOI] [PubMed] [Google Scholar]
- 36. Wang W., Lu P., Fang Y., Hamuro L., Pittman T., Carr B., Hochman J., Prueksaritanont T. (2011) Monoclonal antibodies with identical Fc sequences can bind to FcRn differentially with pharmacokinetic consequences. Drug Metab. Dispos. 39, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 37. Gurbaxani B., Dela Cruz L. L., Chintalacharuvu K., Morrison S. L. (2006) Analysis of a family of antibodies with different half-lives in mice fails to find a correlation between affinity for FcRn and serum half-life. Mol. Immunol. 43, 1462–1473 [DOI] [PubMed] [Google Scholar]
- 38. Igawa T., Maeda A., Haraya K., Tachibana T., Iwayanagi Y., Mimoto F., Higuchi Y., Ishii S., Tamba S., Hironiwa N., Nagano K., Wakabayashi T., Tsunoda H., Hattori K. (2013) Engineered monoclonal antibody with novel antigen-sweeping activity in vivo. PLoS One 8, e63236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dahiyat B. I., Lazar G. A., Bernett M. J. (January 28, 2014) Antibodies with modified isoelectric points. United States Patent Publication, US8637641 B2
- 40. Igawa T., Ishii S., Tachibana T., Maeda A., Higuchi Y., Shimaoka S., Moriyama C., Watanabe T., Takubo R., Doi Y., Wakabayashi T., Hayasaka A., Kadono S., Miyazaki T., Haraya K., Sekimori Y., Kojima T., Nabuchi Y., Aso Y., Kawabe Y., Hattori K. (2010) Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat. Biotechnol. 28, 1203–1207 [DOI] [PubMed] [Google Scholar]
- 41. Igawa T., Tsunoda H., Tachibana T., Maeda A., Mimoto F., Moriyama C., Nanami M., Sekimori Y., Nabuchi Y., Aso Y., Hattori K. (2010) Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng. Des. Sel. 23, 385–392 [DOI] [PubMed] [Google Scholar]