Abstract
The discovery that somatic cells can be induced into a pluripotent state by the expression of reprogramming factors has enormous potential for therapeutics and human disease modeling. With regard to aging and rejuvenation, the reprogramming process resets an aged, somatic cell to a more youthful state, elongating telomeres, rearranging the mitochondrial network, reducing oxidative stress, restoring pluripotency, and making numerous other alterations. The extent to which induced pluripotent stem cell (iPSC)s mime embryonic stem cells is controversial, however, as iPSCs have been shown to harbor an epigenetic memory characteristic of their tissue of origin which may impact their differentiation potential. Furthermore, there are contentious data regarding the extent to which telomeres are elongated, telomerase activity is reconstituted, and mitochondria are reorganized in iPSCs. Although several groups have reported that reprogramming efficiency declines with age and is inhibited by genes upregulated with age, others have successfully generated iPSCs from senescent and centenarian cells. Mixed findings have also been published regarding whether somatic cells generated from iPSCs are subject to premature senescence. Defects such as these would hinder the clinical application of iPSCs, and as such, more comprehensive testing of iPSCs and their potential aging signature should be conducted.
Keywords: aging, differentiation, epigenetic, induced pluripotent stem, reprogramming, telomeres
Introduction
It was discovered early on that somatic cells could be reset to a pluripotent state through somatic cell nuclear transfer (Gurdon, 1962; Tada et al., 1997; Hochedlinger & Jaenisch, 2002; Wilmut et al., 2007) and cell fusion (Tada et al., 2001). A landmark experiment in the cell reprogramming field was performed by Takahashi and Yamanaka, demonstrating that adult somatic cells could be restored to pluripotency through the exogenous expression of four transcription factors: Oct4, Sox2, Klf4, and c-Myc. These induced pluripotent stem cells (iPSCs) expressed markers exclusive to embryonic stem cells (ESCs), mimed their morphology and growth properties, and could differentiate into all three germ layers (Takahashi & Yamanaka, 2006).
Since their initial discovery, multiple methods of reprogramming have been generated. Adult somatic cells have been successfully induced into pluripotency using viral vectors (Zhou & Freed, 2009), nonintegrating episomes (Yu et al., 2009), and minicircle vectors (Jia et al., 2010). Pluripotency can also be induced by the use of reprogramming proteins, either by direct addition of purified protein (Zhou et al., 2009) or with extracts from cells stably expressing reprogramming factors (Kim et al., 2009). More recently, effective reprogramming was achieved using synthetic mRNA (Warren et al., 2010), a technique our group has used to derive iPSCs from disease and healthy donors. More comprehensive listings of successfully employed methods have been reviewed elsewhere (González et al., 2011).
With regard to aging and age-related disease, iPSCs represent enormous therapeutic potential. Reprogramming adult, somatic cells allows for the generation of patient-specific models that have already been used to generate a wealth of information regarding disease pathogenesis, drug testing, and drug discovery (Bellin et al., 2012). It was previously proposed that the ability to reprogram a cell to a youthful state without affecting the differentiation program may be an effective strategy for rejuvenating an aged organism (Rando & Chang, 2012). In order for such a method to be viable, reprogramming would have to reset the aging clock, clearing the damage that accrues with age and restoring a cell to a youthful state. This would require multiple types of restoration, as somatic cells accumulate nuclear and mitochondrial mutations as well as damaged macromolecules with age. Furthermore, aging cells are characterized by distinct changes in the epigenome, telomere shortening, increased oxidative stress, and numerous other alterations (Kirkwood, 2005; Haigis & Yankner, 2010; Johnson et al., 2012). Such restoration is not impossible, however, as evinced by the fertilization process, where an aged sperm and egg fuse to form a zygote devoid of aging damage or any evidence of the age of the parental cells (Rando & Chang, 2012).
There are currently conflicting data regarding the ability of reprogramming to fully rejuvenate an aged somatic cell and the extent to which iPSCs mime ESCs. Moreover, contentious data exist suggesting that cells derived from iPSCs may be subject to premature senescence. This review highlights recent data relevant to these controversies and discusses the conclusions that can be currently drawn.
Does reprogramming reset the aging clock?
Epigenetic memory
Epigenetic modifications such as histone acetylation and DNA methylation play a paramount role in regulating gene expression and exhibit unique changes during aging and age-related disease (Fraga et al., 2007; Johnson et al., 2012). Modifications to epigenetic machinery can directly impact longevity (Lin et al., 2005) and health (Klein et al., 2011) as well as prevent differentiation of stem cells into somatic tissues (Bröske et al., 2009), highlighting the importance of a well-functioning epigenome. Emerging studies suggest that iPSCs may harbor a higher number of genetic and epigenetic abnormalities than both ESCs and the somatic cells that they originate from (Pera, 2011). Moreover, there are mixed data regarding the epigenetic memory of iPSCs and whether this memory affects the differentiation potential of reprogrammed cells (Fig. 1).
Figure 1.
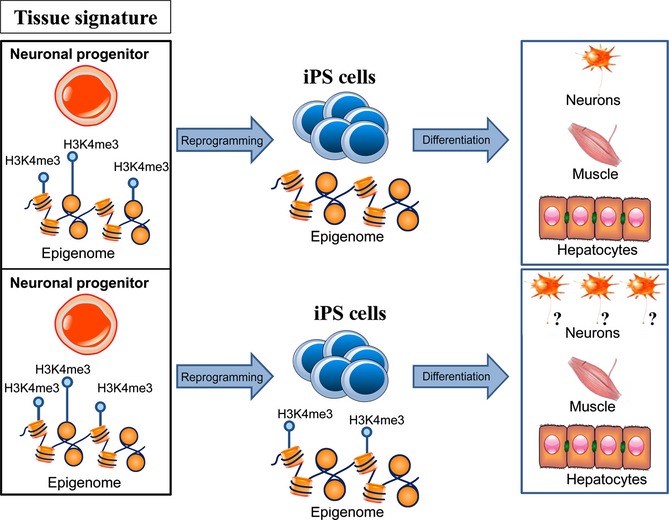
Epigenetic memory and reprogramming. There are controversial data regarding the epigenetic memory of induced pluripotent stem cell (iPSC)s and whether or not this memory affects the differentiation potential of reprogrammed cells. iPS cells have been reported to feature incomplete epigenetic reprogramming compared to ESCs, retaining residual methylation signatures characteristic of their tissue of origin that favor differentiation into lineages related to the donor cell.
It was recently shown that low-passage iPSCs can feature incomplete epigenetic reprogramming compared to ESCs, retaining residual DNA methylation signatures that are characteristic of their tissue of origin and favor differentiation into lineages related to the donor cell (Fig. 1). iPSCs derived from mouse neural progenitors, for example, contained methylomic signatures at loci important for hematopoietic differentiation, resulting in a decreased propensity for differentiating into hematopoietic cell types. Treatment with chromatin-modifying compounds reduced DNA methylation at these loci and increased the blood-forming potential of the low-passage iPSCs, suggesting that the effects of these epigenetic marks can be attenuated via pharmaceutical intervention (Kim et al., 2010). Conflicting data exist regarding the retention of these methylation signatures with passage number. Some iPSC clones derived from human neonatal keratinocytes and umbilical cord blood cells were documented to maintain tissue-specific methylation memory at high passage numbers (Kim et al., 2011), while iPSCs derived from mouse myogenic cells, fibroblasts, and hematopoietic cells reportedly lost their epigenetic memory with continued passage in culture (Polo et al., 2010). More recently, genetically matched human iPSC clones from dermal fibroblasts and bone marrow stromal cells of the same donor were generated and differentiated into osteogenic and chondrogenic lineages. The authors found that, although the iPSCs exhibited an epigenetic memory characteristic of the donor tissue used, the clones varied in their differentiation propensity. Moreover, no correlation was found between the cell type of origin and the propensity of an iPSC clone to differentiate into bone and cartilage (Nasu et al., 2013). Further work is required to determine whether this epigenetic memory affects the pluripotency of iPSCs and whether this influence declines with passage number or varies with the donor tissue or iPSC line used.
Telomere length
Unlike stem cells, somatic cells have a limited division capacity and senesce in vitro. The inability to further replicate is termed replicative senescence and can be induced by a plethora of factors, such as short and uncapped telomeres, oxidative stress, and mitochondrial DNA damage (Fig. 2) (Chen et al., 2007). The enzyme telomerase, which maintains telomere length and long-term self-renewal potential of stem cells, is strongly expressed in ESCs and is inactive in most somatic cells. As such, telomere length gradually decreases with every cell division in a typical somatic cell, eventually resulting in replicative senescence (Harley et al., 1990). The lifespan of normal human fibroblasts can be extended in vitro by exogenous introduction of plasmids expressing the catalytic subunit of telomerase hTERT, resulting in an increased telomerase activity (Bodnar et al., 1998). Donor cells that are difficult to reprogram can be more efficiently induced into the pluripotent state by adding hTERT and SV40 large T antigen to the reprogramming factors originally used by Yamanaka’s laboratory (Park et al., 2008). Furthermore, telomerase-deficient mice exhibit a sharp reduction in reprogramming efficiency that can be restored by the reintroduction of telomerase (Marion et al., 2009), highlighting the imperative role telomerase plays in iPS reprogramming.
Figure 2.

Aspects of aging and reprogramming. There are currently conflicting data regarding the ability of reprogramming to fully rejuvenate an aged somatic cell and reverse age-related changes such as DNA damage, shortened telomeres, and dysfunctional mitochondria. Moreover, contentious data exist suggesting that cells derived from induced pluripotent stem cells may be subject to premature senescence.
Through the use of factor-based reprogramming, Yu et al. reported that human iPSCs display levels of telomerase activity characteristic of ESCs (Yu et al., 2007). In a recent report using mouse iPSCs, it was found that, at passage eight, iPSCs have shorter telomeres than ESCs, yet longer telomeres than the embryonic fibroblasts they were derived from. It was only after subsequent cell divisions and further passaging that telomeres were restored to the lengths found in ESCs (Marion et al., 2009). In six of seven iPS cell lines derived from human skin fibroblasts, telomeres were substantially elongated compared to their parental cells and had telomere lengths comparable to ESCs at passage five (Suhr et al., 2009) (Table S1). At passage 25, several iPSC clones derived from human neonatal foreskin fibroblasts had significantly longer telomeres than their parental donors (Yehezkel et al., 2011). iPSCs reprogrammed from senescent and centenarian human cells exhibited telomere lengths that were identical to or greater than the lengths observed in ESCs and did not shorten with increased passage number (Lapasset et al., 2011) (Table S1).
A study conducted by Vaziri et al. compared telomerase activity and telomere length in iPSCs to hESCs. The authors reported that five well-studied human iPS cell lines had significantly shorter telomeres than three commonly used ES cell lines as well as reduced levels of telomerase activity. Six iPS cell clones were then generated from the ESC-derived cell line EN13 and, after further culturing, telomeres in five of the six lines shortened to lengths comparable to those observed in the widely used iPS cell lines. One clone, however, expressed high levels of telomerase and displayed a progressive increase in telomere length for over 60 days, procuring a length equivalent to that of the parental ES cell line (Vaziri et al., 2010). Mathew et al. also reported the variability in telomere length and telomerase expression in human iPSC clones. Only clones with the highest level of telomerase expression were observed to have telomere lengths comparable to ESCs (Mathew et al., 2010). In iPSCs derived from patients with dyskeratosis congenita, a disease characterized by shortened telomeres and defects in telomerase function, reprogramming was capable of restoring telomere length in some iPSCs (Agarwal et al., 2010), but not others (Batista et al., 2011) (see Table S1 for a detailed overview of telomere length data). For those iPSCs where telomere length was elongated, telomere length was observed to increase with further passaging (Agarwal et al., 2010).
When telomere chromatin is transcribed, long noncoding RNA transcripts referred to as telomeric-repeat-containing RNA (TERRAs) are generated. TERRA levels are positively correlated with telomere length and may act as regulators of telomerase expression (Schoeftner & Blasco, 2008). In mouse iPSCs, TERRAs were observed to be increased compared to differentiated cells, yet lower compared to ESCs at passage eight (Marion et al., 2009). At passage 25, human iPSCs had higher TERRA levels than their parental source, yet these levels were found to vary from clone to clone (Yehezkel et al., 2011).
These results indicate that, although telomerase activity is clearly reconstituted during the reprogramming process, there is a significant variability in telomere length among various iPS cell lines. This length is sensitive to passage number and can even vary among cell lines derived from the same tissue or parental cell type. This is not unique, however, as it was recently shown that substantial variability in telomere length and telomerase expression exists for ESCs as well. For both iPSCs and ESCs, telomere length was found to be highly correlated with proliferation efficiency and developmental pluripotency (Huang et al., 2011). As such, caution should be made when drawing comparisons for telomere length, ensuring that passage number is controlled for and that appropriate controls (e.g., numerous ES cell lines and donor tissue) are used.
Mitochondrial alterations and oxidative stress
Aging in somatic cells is accompanied by mitochondrial dysfunction and oxidative stress (Fig. 2) (Passos et al., 2007; Moiseeva et al., 2009). Compared to somatic cells, ESCs have less mitochondrial mass, decreased ATP levels, reduced reactive oxygen species (ROS), and more active repair mechanisms that mitigate mitochondrial DNA damage (Cho et al., 2006; Saretzki et al., 2008). Mitochondria in ESCs have also been reported to be sparser, have underdeveloped or condensed cristae, and rely more on anaerobic glycolysis for energy (Prigione et al., 2010; Suhr et al., 2010).
It was recently shown that the reprogramming process reorganizes mitochondria, causing them to adopt the immature morphology, distribution, and DNA content characteristic of the pluripotent state. Like ESCs, these human iPSCs also displayed reduced levels of oxidative stress, decreased amounts of intracellular ATP, and increased production of lactate. Expression of nuclear factors involved in mitochondrial biogenesis was also comparable (Prigione et al., 2010). Age does not appear to limit the ability of reprogramming to mediate these changes, as iPSCs with chromosomal anomalies from an 84-year-old woman exhibited restructured mitochondria and decreased levels of oxidative stress and DNA damage. These changes were similar to those observed for iPSCs generated from young donors (Prigione et al., 2011) (see Table S1 for details). In senescent and centenarian human fibroblasts, the reprogramming process reset the expression of genes associated with mitochondrial metabolism and oxidative stress. Mitochondrial morphology and distribution were also both comparable to ESCs (Lapasset et al., 2011). In iPSC clones derived from human dermal fibroblasts, mitochondrial biogenesis and ROS stress defense mechanisms were analogous to those observed in ESCs (Armstrong et al., 2010). Resetting of metabolic signature and restructuring of mitochondria have also been observed by others (Suhr et al., 2010). Varum et al. obtained more mixed results (Fig. 2), finding that human iPSCs exhibit a metabolic signature that, while not completely identical to ESCs, clusters more closely with ESCs than with differentiated cells. It was also observed that the mitochondrial morphology in iPSCs is more similar to ESCs than somatic cells, yet still distinct (Varum et al., 2011). Although different reports have been published regarding the extent of rejuvenation, these data clearly demonstrate that the reprogramming process resets mitochondria and related metabolic and stress mechanisms to a more youthful state (Fig. 2). Further studies are required to illuminate the extent of this rejuvenation, however, and why varied results have been obtained.
Effect of donor age on reprogramming efficiency
It is currently possible to generate iPSCs from human centenarian cells (Lapasset et al., 2011; Yagi et al., 2012) that can differentiate into all three germ layers (Yagi et al., 2012). The reprogramming process causes senescent and centenarian cells to acquire a more youthful signature after reprogramming, resetting telomere lengths and gene expression profiles to those observed in ESCs (Lapasset et al., 2011). Although these data demonstrate that old age does not abrogate the ability to reprogram somatic cells, there are conflicting data on whether donor age impacts reprogramming efficiency.
Using human tissue, Boulting and colleagues (Boulting et al., 2011) characterized 16 iPSCs from seven individuals of different ages. The authors found that, although the lines had variable expression of early pluripotency markers, all 16 lines passed stringent tests for differentiation capacity (Boulting et al., 2011). In mice, dermal skin fibroblasts procured from old mice (121 weeks old) had shorter telomeres than those obtained from young mice (22 weeks old). Despite the difference in telomere length, iPSCs could be derived from both old and young donors and the resultant iPSCs had equally elongated telomeres (Marion et al., 2009). iPSCs generated from bone marrow of old mice (23 months old) were more difficult to produce than those generated from the bone marrow of young mice (2 months old), requiring twice as much time to reprogram (Cheng et al., 2011). In iPSCs sourced from 14-month-, 6-month-, and 6-week-old mice, reprogramming efficiency declined with age. Unlike those from younger mice, iPSCs from 14-month-old mice exhibited unstable pluripotency, regressing after expansion in culture and exhibiting faint staining of the pluripotency marker alkaline phosphatase. A gradual loss of expression in the ESC marker Nanog was also observed in elderly mice. This was ameliorable, however, as inhibition of BMP and TGF-ß signaling helped to stimulate self-renewal as well as to stabilize Nanog expression (Wang et al., 2011).
A few age-related genes have been identified that play inhibitory roles in the reprogramming process. Using mouse and human fibroblasts, Li et al. reported that reprogramming efficiency declined with age. Knockdown of the Ink4/Arf locus, which encodes for tumor suppressors and is upregulated with age, was sufficient to rescue the age-dependent defect in reprogramming efficiency (Li et al., 2009). Similarly, expression of lamin A, a protein that maintains nuclear structure integrity, increases with age in somatic cells and is expressed at lower levels in undifferentiated cells. Using human fibroblasts, an inverse correlation was observed between reprogramming efficiency and the expression level of lamin A. Like the Ink4/Arf locus, knockdown of lamin A using short hairpin RNA accelerated the rate of iPSC induction. Furthermore, overexpression of lamin A severely hindered reprogramming (Zuo et al., 2012). Interestingly, mutations in lamin A/C cause defects in the nuclear envelope and underlie Werner syndrome and Hutchinson Gilford progeria, two diseases of accelerated aging. Recently, iPSCs were generated from patients suffering from these disorders. Compared to their donor fibroblasts, these iPSCs had normal nuclear membrane morphology, suggesting that the reprogramming process could rejuvenate nuclear defects (Ho et al., 2011).
Although additional age comparisons are necessary, these results suggest that mammalian aging may decrease reprogramming efficiency (for an overview of donor age of the generated iPSC lines, see Table S1, and for factors used for reprogramming, see Table S2). Old age does not prevent successful reprogramming, however, as these studies demonstrate that somatic cells of any age – even those that are senescent – can be coaxed into a more youthful, pluripotent state. Moreover, the loss in efficiency can be mitigated via inhibition of specific signaling pathways and genes. Hence, old age is unlikely to nullify the rejuvenative potential of iPSCs.
Do cells derived from iPSCs age prematurely?
Recent data have emerged suggesting that cells derived from iPSCs may exhibit signs of premature senescence (please see Fig. 2 for an overview of premature senescence in iPSCs). As with epigenetic memory and telomere length, these data are also controversial.
Suhr et al. reprogrammed human skin fibroblasts into iPSCs and then produced differentiated cell lines derived from three iPSC-teratoma explants. Although one line exhibited elongated telomeres, the other two displayed telomere lengths comparable to the input fibroblasts (Suhr et al., 2009). The same group examined the mitochondria of iPSCs generated from human fibroblasts as well as fibroblasts re-derived from iPSCs. The authors observed that the quality and function of mitochondrial complement of the re-derived fibroblasts was dramatically improved compared to the input fibroblasts (Suhr et al., 2010). Upon differentiation, the mitochondrial network and metabolic signature of both human ESCs and iPSCs changed to match features observed in fibroblasts. Expression of the antioxidant GPX1 was higher in fibroblasts differentiated from iPSCs, however, suggesting that iPS-derived somatic cells may differ with regard to their handling of ROS (Prigione et al., 2010). Feng et al. successfully differentiated human iPSCs into multiple cell types, although the efficiency was markedly lower than it was for ESCs. Moreover, the authors observed that, unlike cells derived from ESCs, somatic cells derived from iPSCs exhibited early senescence and possessed dramatic defects in expansion capability (Feng et al., 2010) (for an overview of all iPSC lines tested, see Table S1). This fate is not inexorable, however, as others have generated somatic cells from iPSCs that do not exhibit premature senescence (Gokoh et al., 2011).
Although it is too early to conclusively determine, issues of premature senescence may, like telomere length, vary considerably from line to line. Subsequent research drawing detailed comparisons between cell types derived from multiple ESC and iPSC lines will help resolve this contention.
Conclusions
It is quite clear that the reprogramming reverses many aspects of aging. Even iPSCs derived from senescent and centenarian cells exhibit a more youthful signature, displaying elongated telomeres and gene expression profiles comparable to ESCs (Lapasset et al., 2011). Metabolic signatures, mitochondrial networks, handling of ROS, telomerase expression, and other factors are all reset to a state characteristic of pluripotency (Suhr et al., 2009; Prigione et al., 2010, 2011). These data are controversial, however, as differential reports have been published regarding the extent to which reprogramming rejuvenates aged, somatic cells and whether iPSCs exhibit aging signatures (summarized in Table S1).
Telomere length, for example, has been observed to be shortened (Vaziri et al., 2010), similarly sized, or even elongated compared to ESCs (Lapasset et al., 2011). Considerable variation in telomere length as well as telomerase activity has also been reported for both iPSCs and ESCs, however, suggesting that telomere profiles may be unique to the stem cell line used (Huang et al., 2011). The same may be true for epigenetic memory, as Nasu et al. found no correlation between the differentiation potential of an iPSC clone and the type of progenitor cell used. Instead, the authors observed that differentiation propensity varied substantially from clone to clone (Nasu et al., 2013). Mitochondrial networks have been reported to be rearranged to a state indistinguishable from ESCs (Prigione et al., 2010, 2011) or to a mixed phenotype in between that of ESCs and somatic cells (Varum et al., 2011). Some groups have reprogrammed cells from an elderly organism with no comment on decreased reprogramming efficiency (Marion et al., 2009), while others have observed reprogramming efficiency to decline with age (Li et al., 2009). Similar discrepancies are noted regarding premature senescence, which has been observed in somatic cells derived from some iPSCs (Feng et al., 2010), but not others (Gokoh et al., 2011).
That variations in differentiation propensity are observed in iPSCs derived from the same donor (Nasu et al., 2013) indicates that the reprogramming process may not reset cells to a younger state in an invariable manner. Furthermore, many of the studies cited used different reprogramming protocols as well as donor cells from different cell types or species (summarized in Table S2). Conflicting reports regarding the extent to which adult, somatic cells are rejuvenated may also be explained by the distinct protocols and materials used. Regardless, it is apparent that some iPSCs and their differentiated progeny may exhibit age-related defects. Whether these defects are cell line specific or a broader problem with iPSCs requires additional data to determine. Moreover, aging is a complex disease marked by innumerable changes, and thus far, only a small set of age-related alterations has been investigated in iPSCs. To properly assess whether iPSCs exhibit an aging signature and are less youthful than ESCs, additional research into other aspects of aging is required.
Acknowledgments
Thanks go to Mr. Pooyan Naghsh for graphical design and making tables. The work presented in this paper was made possible by funding from the German Federal Ministry of Education and Research (BMBF 1315883).
Author contributions
L. R., A. A., and A. A. J. involved in collection and/or assembly of data. L. R involved in graphical design and making tables. A. A. J. involved in editing. A. S. provided the concept and conclusions as well as major editing of the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
Table S1 Aging signatures of different iPS cell lines and derivatives.
Table S2 iPS cell lines generated from different cell types or species.
References
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, Ng HH, Keefe DL, Goldman FD, Klingelhutz AJ, Liu L, Daley GQ. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, Giri N, Wernig M, Alter BP, Cech TR, Savage SA, Reijo Pera RA, Artandi SE. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, Rodolfa CT, Dimos JT, Mikkilineni S, MacDermott AB, Woolf CJ, Henderson CE, Wichterle H, Eggan K. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, Nerlov C, Leutz A, Andrade-Navarro MA, Jacobsen SE, Rosenbauer F. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res. 2007;35:7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ito S, Nishio N, Xiao H, Zhang R, Suzuki H, Okawa Y, Murohara T, Isobe K. Establishment of induced pluripotent stem cells from aged mice using bone marrow-derived myeloid cells. J. Mol. Cell. Biol. 2011;3:91–98. doi: 10.1093/jmcb/mjq044. [DOI] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park dJ, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 2006;348:1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann. N. Y. Acad. Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- Gokoh M, Nishio M, Nakamura N, Matsuyama S, Nakahara M, Suzuki S, Mitsumoto M, Akutsu H, Umezawa A, Yasuda K, Yuo A, Saeki K. Early senescence is not an inevitable fate of human-induced pluripotent stem-derived cells. Cell Reprogram. 2011;13:361–370. doi: 10.1089/cell.2011.0004. [DOI] [PubMed] [Google Scholar]
- González F, Boué S, Izpisúa Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat. Rev. Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 1962;4:256–273. doi: 10.1016/0012-1606(62)90043-x. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA. The aging stress response. Mol. Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Ho JC, Zhou T, Lai WH, Huang Y, Chan YC, Li X, Wong NL, Li Y, Au KW, Guo D, Xu J, Siu CW, Pei D, Tse HF, Esteban MA. Generation of induced pluripotent stem cell lines from 3 distinct laminopathies bearing heterogeneous mutations in lamin A/C. Aging (Albany NY) 2011;3:380–390. doi: 10.18632/aging.100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear transplantation: lessons from frogs and mice. Curr. Opin. Cell Biol. 2002;14:741–748. doi: 10.1016/s0955-0674(02)00380-0. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, Zuo B, Li M, Liang P, Ge WW, Tsibris JC, Keefe DL, Liu L. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat. Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AA, Akman K, Calimport SR, Wuttke D, Stolzing A, de Magalhães JP. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 2012;15:483–494. doi: 10.1089/rej.2012.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, Simon M, Lander C, Boardman LA, Cunningham JM, Smith GE, Litchy WJ, Boes B, Atkinson EJ, Middha S, B Dyck PJ, Parisi JE, Mer G, Smith DI, Dyck PJ. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, Lehmann S, Lemaitre JM. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MJ, Tang LY, Reddy MN, Shen CK. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J. Biol. Chem. 2005;280:861–864. doi: 10.1074/jbc.C400477200. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Mathew R, Jia W, Sharma A, Zhao Y, Clarke LE, Cheng X, Wang H, Salli U, Vrana KE, Robertson GP, Zhu J, Wang S. Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. FASEB J. 2010;24:2702–2715. doi: 10.1096/fj.09-148973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva O, Bourdeau V, Roux A, Deschênes-Simard X, Ferbeyre G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell. Biol. 2009;29:4495–4507. doi: 10.1128/MCB.01868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu A, Ikeya M, Yamamoto T, Watanabe A, Jin Y, Matsumoto Y, Hayakawa K, Amano N, Sato S, Osafune K, Aoyama T, Nakamura T, Kato T, Toguchida J. Genetically matched human iPS cells reveal that propensity for cartilage and bone differentiation differs with clones, not cell type of origin. PLoS ONE. 2013;8:e53771. doi: 10.1371/journal.pone.0053771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF. Stem cells: the dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Prigione A, Hossini AM, Lichtner B, Serin A, Fauler B, Megges M, Lurz R, Lehrach H, Makrantonaki E, Zouboulis CC, Adjaye J. Mitochondrial-associated cell death mechanisms are reset to an embryonic-like state in aged donor-derived iPS cells harboring chromosomal aberrations. PLoS ONE. 2011;6:e27352. doi: 10.1371/journal.pone.0027352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, Stewart R, Hoare S, Stojkovic M, Armstrong L, von Zglinicki T, Lako M. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26:455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, Murthy S, Cibelli JB. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE. 2009;4:e8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Tjong J, Alcasid N, Perkins GA, Goissis MD, Ellisman MH, Perez GI, Cibelli JB. Mitochondrial rejuvenation after induced pluripotency. PLoS ONE. 2010;5:e14095. doi: 10.1371/journal.pone.0014095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, TaFda T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS ONE. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Chapman KB, Guigova A, Teichroeb J, Lacher MD, Sternberg H, Singec I, Briggs L, Wheeler J, Sampathkumar J, Gonzalez R, Larocca D, Murai J, Snyder E, Andrews WH, Funk WD, West MD. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regen. Med. 2010;5:345–363. doi: 10.2217/rme.10.21. [DOI] [PubMed] [Google Scholar]
- Wang B, Miyagoe-Suzuki Y, Yada E, Ito N, Nishiyama T, Nakamura M, Ono Y, Motohashi N, Segawa M, Masuda S, Takeda S. Reprogramming efficiency and quality of induced Pluripotent Stem Cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr. 2011;3:RRN1274. doi: 10.1371/currents.RRN1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Cloning Stem Cells. 2007;9:3–7. doi: 10.1089/clo.2006.0002. [DOI] [PubMed] [Google Scholar]
- Yagi T, Kosakai A, Ito D, Okada Y, Akamatsu W, Nihei Y, Nabetani A, Ishikawa F, Arai Y, Hirose N, Okano H, Suzuki N. Establishment of induced pluripotent stem cells from centenarians for neurodegenerative disease research. PLoS ONE. 2012;7:e41572. doi: 10.1371/journal.pone.0041572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehezkel S, Rebibo-Sabbah A, Segev Y, Tzukerman M, Shaked R, Huber I, Gepstein L, Skorecki K, Selig S. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics. 2011;6:63–75. doi: 10.4161/epi.6.1.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Schöler HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo B, Yang J, Wang F, Wang L, Yin Y, Dan J, Liu N, Liu L. Influences of lamin A levels on induction of pluripotent stem cells. Biol. Open. 2012;1:1118–1127. doi: 10.1242/bio.20121586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Aging signatures of different iPS cell lines and derivatives.
Table S2 iPS cell lines generated from different cell types or species.


