Abstract
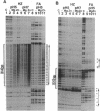
Chemical probing of two predominantly alternating purine-pyrimidine d(CA/TG)n repeats led us to propose previously that in supercoiled plasmids these elements adopt a non-B-DNA structure distinct from that of Z-DNA formed by d(CG)n sequences. Here, we present further evidence supporting this contention. Reactivity with the conformation-sensitive reagent chloroacetaldehyde, which reacts with unpaired adenines and cytosines, was confined strictly to adenines in the d(CA/TG)n repeat. In contrast, only bases outside the d(CG)n repeat exhibited chloroacetaldehyde reactivity. Two-dimensional gel analysis of topoisomers containing d(CA/TG)n tracts with bases out of strict purine-pyrimidine alteration revealed multiple superhelical-dependent transitions to an alternative left-handed structure. Within individual plasmid molecules, these multiple transitions resulted from the stepwise conversion of contiguous segments of alternating purine-pyrimidine sequence, which are delimited by bases out of alternation, to the full-length alternative conformation. When the left-handed helices increased in length to include more bases out of alternation, the average helical pitch changed substantially to produce a less tightly wound left-handed helix. Overall, these data indicate that d(CA/TG)n tracts adopt a left-handed conformation significantly different from that of the canonical Z-DNA structure of d(CG)n sequences.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaltonen L. A., Peltomäki P., Leach F. S., Sistonen P., Pylkkänen L., Mecklin J. P., Järvinen H., Powell S. M., Jen J., Hamilton S. R. Clues to the pathogenesis of familial colorectal cancer. Science. 1993 May 7;260(5109):812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. T., Walls J. D., Reifel-Miller A. E., Grinnell B. W. E1A-induced enhancer activity of the poly(dG-dT).poly(dA-dC) element (GT element) and interactions with a GT-specific nuclear factor. Mol Cell Biol. 1989 Nov;9(11):5248–5253. doi: 10.1128/mcb.9.11.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Wells R. D., Heintz N. H., Caddle M. S. Sequences near the origin of replication of the DHFR locus of Chinese hamster ovary cells adopt left-handed Z-DNA and triplex structures. J Biol Chem. 1990 Dec 15;265(35):21789–21796. [PubMed] [Google Scholar]
- Ellison M. J., Fenton M. J., Ho P. S., Rich A. Long-range interactions of multiple DNA structural transitions within a common topological domain. EMBO J. 1987 May;6(5):1513–1522. doi: 10.1002/j.1460-2075.1987.tb02394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M. J., Kelleher R. J., 3rd, Wang A. H., Habener J. F., Rich A. Sequence-dependent energetics of the B-Z transition in supercoiled DNA containing nonalternating purine-pyrimidine sequences. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8320–8324. doi: 10.1073/pnas.82.24.8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazka G., Palecek E., Wells R. D., Klysik J. Site-specific OsO4 modification of the B-Z junctions formed at the (dA-dC)32 region in supercoiled DNA. J Biol Chem. 1986 May 25;261(15):7093–7098. [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. A novel Z-structure for poly d(GC).poly d(GC). Biochem Biophys Res Commun. 1980 Jul 31;95(2):728–733. doi: 10.1016/0006-291x(80)90846-3. [DOI] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Conformational flexibility of DNA: polymorphism and handedness. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6486–6490. doi: 10.1073/pnas.77.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Petes T. D. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Jun;12(6):2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. C. Deoxyribonucleic acid structure: a new model. Science. 1981 Jan 16;211(4479):289–291. doi: 10.1126/science.7444467. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Johnston B. H., Quigley G. J., Ellison M. J., Rich A. The Z-Z junction: the boundary between two out-of-phase Z-DNA regions. Biochemistry. 1991 May 28;30(21):5257–5263. doi: 10.1021/bi00235a020. [DOI] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., McIntosh L. P., Arndt-Jovin D. J., Zarling D. A., Robert-Nicoud M., van de Sande J. H., Jorgenson K. F., Eckstein F. Left-handed DNA: from synthetic polymers to chromosomes. J Biomol Struct Dyn. 1983 Oct;1(1):21–57. doi: 10.1080/07391102.1983.10507425. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick M. W., Klysik J., Singleton C. K., Zarling D. A., Jovin T. M., Hanau L. H., Erlanger B. F., Wells R. D. Intervening sequences in human fetal globin genes adopt left-handed Z helices. J Biol Chem. 1984 Jun 10;259(11):7268–7274. [PubMed] [Google Scholar]
- Kladde M. P., D'Cunha J., Gorski J. Multiple transitions to non-B-DNA structures occur in the distal regulatory region of the rat prolactin gene. J Mol Biol. 1993 Jan 20;229(2):344–367. doi: 10.1006/jmbi.1993.1039. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Detection of non-B-DNA structures at specific sites in supercoiled plasmid DNA and chromatin with haloacetaldehyde and diethyl pyrocarbonate. Methods Enzymol. 1992;212:155–180. doi: 10.1016/0076-6879(92)12011-e. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Manes T., Kohwi Y. Unusual conformational effect exerted by Z-DNA upon its neighboring sequences. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2223–2227. doi: 10.1073/pnas.84.8.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Blaho J. A., Kilpatrick M. W., Wells R. D. Consecutive A X T pairs can adopt a left-handed DNA structure. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5884–5888. doi: 10.1073/pnas.83.16.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Lee J. W., Wells R. D. Characteristics of Z-DNA helices formed by imperfect (purine-pyrimidine) sequences in plasmids. J Biol Chem. 1988 May 25;263(15):7378–7385. [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of DNA sequence in the formation of Z-DNA versus cruciforms in plasmids. J Biol Chem. 1988 May 25;263(15):7370–7377. [PubMed] [Google Scholar]
- Naylor L. H., Clark E. M. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 1990 Mar 25;18(6):1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Santoro C., Costanzo F., Ciliberto G. Inhibition of eukaryotic tRNA transcription by potential Z-DNA sequences. EMBO J. 1984 Jul;3(7):1553–1559. doi: 10.1002/j.1460-2075.1984.tb02010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Thibodeau S. N., Bren G., Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993 May 7;260(5109):816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- Treco D., Arnheim N. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol. 1986 Nov;6(11):3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N., Marrot L., Rousseau N., Malfoy B., Leng M. Chloroacetaldehyde reacts with Z-DNA. J Mol Biol. 1988 Jun 20;201(4):773–776. doi: 10.1016/0022-2836(88)90474-3. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Frank-Kamenetskii M. D. Left-handed Z form in superhelical DNA: a theoretical study. J Biomol Struct Dyn. 1984 Jun;1(6):1325–1333. doi: 10.1080/07391102.1984.10507523. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]