Abstract
Evidence implicating differential gene expression as a significant driver of evolutionary novelty continues to accumulate, but our understanding of the underlying sources of variation in expression, both environmental and genetic, is wanting. Heritability in particular may be underestimated when inferred from genetic mapping studies, the predominant “genetical genomics” approach to the study of expression variation. Such uncertainty represents a fundamental limitation to testing for adaptive evolution at the transcriptomic level. By studying the inheritance of expression levels in 10,495 genes (10,527 splice variants) in a threespine stickleback pedigree consisting of 563 individuals, half of which were subjected to a thermal treatment, we show that 74–98% of transcripts exhibit significant additive genetic variance. Dominance variance is also prevalent (41–99% of transcripts), and genetic sources of variation seem to play a more significant role in expression variance in the liver than a key environmental variable, temperature. Among-population comparisons suggest that the majority of differential expression in the liver is likely due to neutral divergence; however, we also show that signatures of directional selection may be more prevalent than those of stabilizing selection. This predominantly aligns with the neutral model of evolution for gene expression but also suggests that natural selection may still act on transcriptional variation in the wild. As genetic variation both within- and among-populations ultimately defines adaptive potential, these results indicate that broad adaptive potential may be found within the transcriptome.
Keywords: Gasterosteus aculeatus, gene expression, quantitative genetics, QST, transcriptome, microarray
Introduction
Recent research indicates that sequence variants linked to phenotypic differences are predominantly found in noncoding, regulatory regions of the genome (Flint and Mackay 2009; Hindorff et al. 2009; Ku et al. 2010; Wray 2013). Similarly, there is mounting evidence that gene regulatory networks, and differential patterns of expression in genes may be a significant cause of phenotypic novelty (Erwin and Davidson 2009; Davidson and Erwin 2010), suggesting that variation in gene expression is likely an important source of evolutionary potential. Given the relative ease and large scale at which it can be quantified, transcription is commonly used as a proxy for gene expression (Schena et al. 1995; Lockhart and Winzeler 2000). Although transcription represents only the first of many steps leading to an ultimate/functional gene product, even small variation in mRNA abundance can have a significant impact on the amount of protein (Kaern et al. 2005; Bar-Even et al. 2006; Ghazalpour et al. 2011), with 12–78% of variation in protein abundance explained by transcriptional variation, depending upon taxa, cellular location, and protein type (Greenbaum et al. 2003). Moreover, transcriptional variation can underlie complex phenotypic variation (Ayroles et al. 2009; Liao et al. 2010; Hines et al. 2012; Richards et al. 2012). Thus, despite its “proxy” status with respect to gene expression, transcriptional variation itself is also a likely source of evolutionary potential.
Transcriptional variation can also be viewed as a phenotype (Houle et al. 2010), and as with any phenotype, it is subject to varying degrees of genetic and environmental control, and its inherent variation may be shaped by natural selection (Whitehead and Crawford 2006b). It is generally held that transcriptional variation is additive, with an underlying polygenic architecture (Gilad et al. 2008; Kim and Gibson 2010). Indeed, results emerging from association studies in humans are consistent with the view that a large fraction of transcriptomic variation is heritable (Dixon et al. 2007; Powell et al. 2013). Line cross analyses of both cereal crops (Swanson-Wagner et al. 2006; Chelaifa et al. 2013) and diverse animal taxa (Rottscheidt and Harr 2007; Wayne et al. 2007; Debes et al. 2012; Gao et al. 2013) are also suggestive of widespread genetic variance for transcription, though few report estimates of variance and/or heritability (but see Wayne et al. 2004). In comparison, transcriptome-wide variance components analyses and/or pedigree-based studies into the quantitative genetics of transcriptional variance in model organisms have been limited to mice (Cui et al. 2006), yeast (Brem et al. 2002), Caenorhabditis elegans (Li et al. 2006), and Drosophila (Ayroles et al. 2009). For wild, outbred populations, potential for heritable variation in transcription has been inferred on the basis of high levels of within-population variance, but precise estimates of genetic variance are rarely available, and when attempted, most suffer from low statistical power due to small sample sizes (Whitehead and Crawford 2006b). Moreover, there may be considerable plasticity in patterns of expression (Aubin-Horth and Renn 2009), which if left uncontrolled, can equally lead to biased estimates of heritability by confounding environmental and additive genetic sources of variation (Visscher et al. 2008). This represents a fundamentally important problem in elucidating the evolutionary potential of transcriptomic variation as our ability to predict a response to changing selective pressures—a particularly compelling challenge in an age of rapid habitat/environmental change—is dependent upon the degree to which we can disentangle heritable and environmental contributions to trait variation (Gienapp et al. 2008; Hoffmann and Willi 2008; Hoffmann and Sgrò 2011). Clearly a more controlled experimental evaluation is required.
Our understanding of how selective processes have shaped transcriptional variation also remains a point of contention. To date the literature is dominated by two competing viewpoints, both empirically derived from comparative analyses across broad taxonomic groups. One posits that gene expression differences are effectively neutral (Oleksiak et al. 2002; Khaitovich et al. 2004, 2006; Whitehead and Crawford 2006a). The other suggests that stabilizing selection is the dominant force shaping the evolution of gene expression (Rifkin et al. 2003, 2005; Lemos et al. 2005; Gilad, Oshlack, and Rifkin 2006; Bedford and Hartl 2009), based on assumptions that energetic costs of transcription should favor an optimal level of expression (Wagner 2005, 2007), and that purifying selection should act to conserve transcript function (Brawand et al. 2011). Mutation accumulation experiments offer support for both views, demonstrating that significant expression divergence can emerge by random processes, but that purifying and/or stabilizing selection also acts upon expression variation in animals (Denver et al. 2005; Rifkin et al. 2005). However, a detailed analysis of mutation accumulation lines has revealed that not all genes are equally mutable, depending to a large extent on their regulatory properties and the transmutational target size (Landry et al. 2007). Similarly, it has been shown that genes expressed in multiple tissues—likely to be facultative, housekeeping genes—tend to be less divergent among species than tissue-specific genes (Khaitovich et al. 2005; Gilad, Oshlack, and Rifkin 2006). Moreover, divergence in cis-regulated expression may frequently be under positive selection (Emerson et al. 2010). Thus, although the debate regarding the predominant mode of evolution continues, it is becoming increasingly clear that other nonneutral evolutionary dynamics are at work on the transcriptome.
Complicating the study of transcriptomic evolution is the lack of a consensus model of its neutral evolution, stemming from discrepancies in data and assumptions regarding the rate of change in expression divergence (Khaitovich et al. 2004; Ogasawara and Okubo 2009). Equally problematic is the contradictory nature of its mutational variance: Transcription has been associated with both increased and decreased mutation rate—transcription-associated mutagenesis and transcription-coupled repair, respectively—and there is conflicting evidence as to whether levels of gene expression are positively or negatively correlated with mutation rate (Hanawalt and Spivak 2008; Kim and Jinks-Robertson 2012; Martincorena et al. 2012; Park et al. 2012). In the absence of a reliable/validated neutral model, comparison against a distribution of putative neutral divergence provides an expedient and informative alternative approach. This is the rationale underlying a well-studied quantitative approach to the problem of trait divergence (Spitze 1993; Leinonen et al. 2013), yet surprisingly few studies to date have actually applied this to transcriptional data (Roberge et al. 2007; Kohn et al. 2008).
In this article, we undertake a quantitative genetics approach to study the inheritance and evolution of transcriptional variation, addressing a number of issues earlier highlighted as central for resolving this topic (Gibson and Weir 2005; Gibson 2008). We do so in a model teleost fish, the threespine stickleback (Gasterosteus aculeatus). Teleosts such as the stickleback represent ideal candidates to decompose phenotypic variation into its genetic and environmental components. Almost all are obligate ectotherms; thus, metabolic/transcriptional variation may be induced by thermal treatment (Nikinmaa et al. 2013). Moreover, relatively large clutch sizes and external fertilization facilitate the creation of complex breeding designs with multiple related individuals which permit an accurate estimation of genetic variance components for focal traits.
The threespine stickleback is also a particularly interesting model to address many outstanding questions related to transcriptional variation and its role in the adaptive evolutionary process given that adaptive morphological variation associated with the species’ expansion into novel habitats has been tied to differential expression of candidate genes and putative regulatory elements in its genome (Chan et al. 2010; Kitano et al. 2010; Jones et al. 2012). Adaptive divergence has also been linked to transcriptional differences in candidate genes for osmoregulation (McCairns and Bernatchez 2010), and in pigmentation differences between freshwater and marine populations (Greenwood et al. 2012). Moreover, examples are beginning to accumulate in which trends in transcription profiles are corroborated by functional assays. For example, differential expression of thyroid-stimulating hormone mRNA between stream-resident and marine sticklebacks reflects differences in lower plasma levels of thyroid hormone and reduced metabolic rate, hypothesized to underlie part of the physiological adaptation to permanent freshwater residency (Kitano et al. 2010). Additionally, transcript expression accurately reflects a functional reaction for some components of hypoxia response (Leveelahti et al. 2011), and is reasonably predictive of gene expression/activity with respect to an oxidative stress response in steady-state systems (Nikinmaa et al. 2013).
Specifically, we address the three following questions: 1) How heritable is transcriptional variation; 2) to what extent does environmental variation affect transcript abundance, and are there genetically based differences in this plasticity of expression; and 3) to what extent has transcriptional variation been shaped by natural selection? To this end, we apply rigorous and statistically powerful quantitative genetic approaches to mRNA profiles of lab-reared threespine sticklebacks originally derived from wild source populations. To address the question of heritable variation in transcript expression—a critical element in evaluating the adaptive potential of any phenotype—we employ a half-sib breeding design of first generation fish (n = 563) representative of the putative ancestral, marine population, and quantify mRNA expression using an oligo microarray featuring approximately 15,000 transcripts. The experiment also includes an environmental manipulation (an acute rapid temperature increase) in order to facilitate an evaluation of plasticity in expression, as well as family-based variation in environmental sensitivity, a proxy for genotype–environment interaction (G×E). To test for signatures of selection on the transcriptome, we use the index of quantitative trait divergence (QST) to compare expression variation within and among three discrete populations representing a range of diverse environmental/habitat conditions: One marine and two lakes of differing size and community complexity with pronounced latitudinal difference (supplementary fig. S1A, Supplementary Material online).
Results
Partitioning of Transcriptional Variance
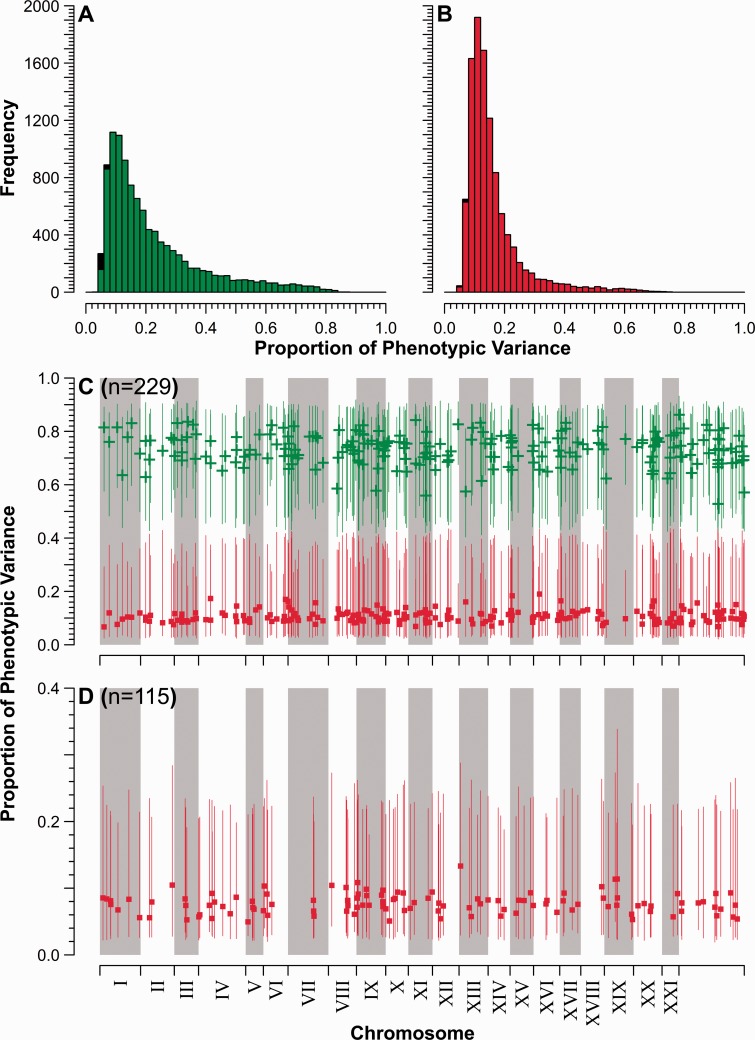
The vast majority (10,384) of the 10,527 transcripts detected above background levels exhibited significant additive genetic variance (VA; estimates presented in supplementary table S1, Supplementary Material online). Significant dominance variance (VD) was also found in 10,499 transcripts. After correcting for potential “false positives” (i.e., Type I errors), estimated through phenotypic simulation (supplementary figs. S2A and B and appendix S1, Supplementary Material online), this would suggest significant genetic variation underlying the expression of at least 7,813 transcripts (74.2%)—heritability estimates for 8,593 transcripts (81.6%) exceeded the mean of possible false positive estimates (h2 > 0.094). The proportion of total phenotypic variance explained by either additive genetic or dominance variance was quite similar: The mean estimate of narrow-sense heritability (h2) for transcript abundance was 0.226 (0.070–0.596, 95% confidence intervals [CIs]; fig. 1A), whereas the mean proportion of variance explained by dominance variance (d2) was 0.161 (0.078–0.361, 95% CIs; fig. 1B). Posterior density interval estimates overlapped for 10,155 transcripts, suggesting approximately equal contributions of dominance and additive genetic variance for most transcripts. In contrast, additive genetic effects were significantly greater than dominance variance for 229 transcripts (fig. 1C); and for 115 transcripts, significant dominance variance was detected in the absence of significant additive genetic variance (fig. 1D).
Fig. 1.
Proportion of phenotypic variance explained by additive genetic and dominance variance. (A) Frequency distribution of narrow-sense heritability (h2) estimates for all 10,527 transcripts. Nonsignificant estimates are represented in the black fraction, whereas statistically significant estimates are plotted in gray (green online). (B) Distribution of estimates for the dominance proportion of total phenotypic variance (d2). Significant estimates are plotted in gray (red online), nonsignificant in black. (C) Chromosomal location of the 229 transcripts for which h2 is significantly greater than d2. Point estimates of h2 are plotted as “+” (green online); d2 are squares (red online). Vertical lines denote 95% PDIs. Mitochondria and unlinked scaffold are combined in the region to the right of chromosome XXI. (D) Chromosomal location of the 115 transcripts with putatively significant d2 and nonsignificant h2 estimates—note that these may represent “false positive” estimates on the basis of phenotypic simulations (d2 ≤ 0.144). Symbols and chromosomal arrangement as in (C).
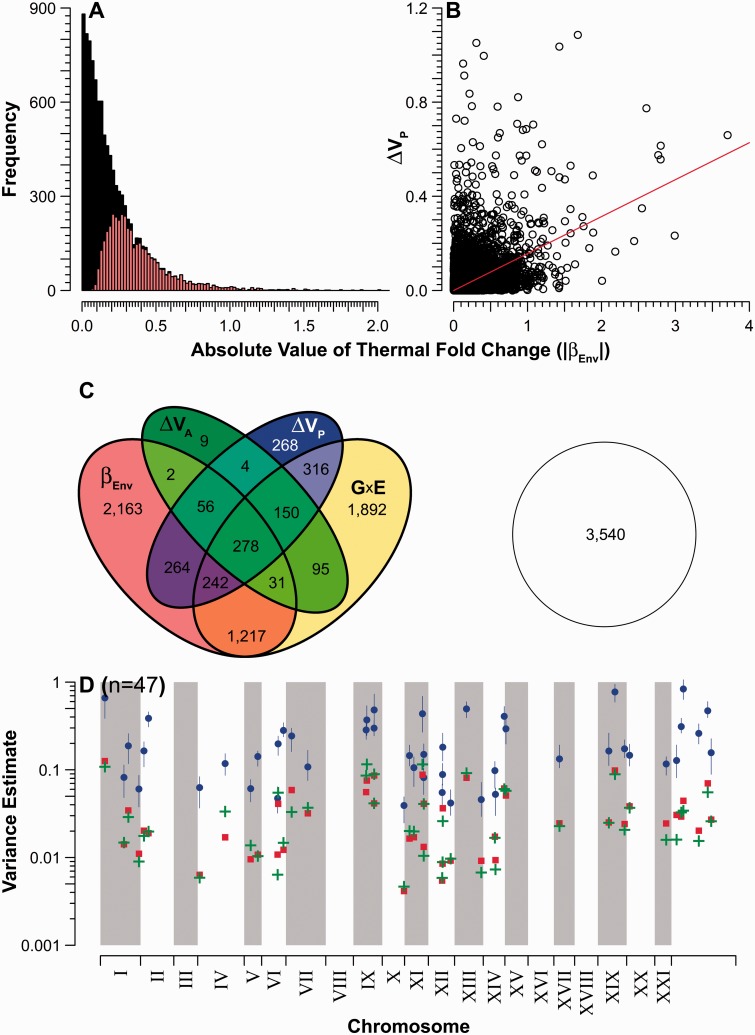
Environmental (i.e., temperature) effects were less prevalent than genetic variance, irrespective of estimation method. When assessed as simple fixed effects, model coefficients describing average differences between temperature treatments were deemed significant for 4,253 transcripts (fig. 2A), following false discovery rate (FDR) correction for multiple comparisons (5,550 uncorrected). Note that in the two environment case, the coefficient describing this effect (βEnv) represents the mean difference in log2 transcript abundance between 17 and 23 °C treatments; thus, the mean fold change. Incorporation of variation among families when modeling temperature effects (random slopes; family × temperature) suggested 4,221 transcripts in which thermal environmental effects are likely mediated through G×E (fig. 2C). Comparing temperature-specific estimates of total phenotypic variance revealed 1,578 transcripts with significantly different estimates of VP (i.e., nonoverlapping posterior density intervals [PDIs]) in each thermal environment (ΔVP; fig. 2C). In total, 66.4% of transcripts showed at least one signal of thermal sensitivity, significantly less than the 74.2% (FDR corrected) with additive genetic variation for expression (χ2[1] = 154.8; P < 0.001). Moreover, in only 47 cases (0.4%) did the absolute range of ΔVP exceed the PDI estimates of either VA or VD (fig. 2D). Finally, for all transcripts with a significant response to the temperature treatment, absolute values of βEnv were significantly correlated with ΔVP (r = 0.347; P < 0.001; fig. 2B).
Fig. 2.
Significant temperature effects on transcript expression. (A) Distribution of fixed-effects coefficients describing the effect of thermal treatment on mean transcript expression (βEnv). The gray fraction (colored online) denotes statistically significant values. (B) Relationship between βEnv and ΔVP, the difference in total phenotypic variance estimated in the two temperature treatments. In total, 6,987 transcripts with a temperature response are plotted. (C) Union of transcripts based on their significance with respect to various indicators of environmental effects. Indicators include βEnv, differences in temperature-specific estimates of additive genetic variance (ΔVA), G×E, and ΔVP. Overlapping regions denote multiple significant effects. The open circle represents the proportion of nonsignificant transcripts. (D) Chromosomal location of the 47 transcripts for which the magnitude of ΔVP exceeded interval estimates of both VA and VD. For clarity, only point estimates are provided for VA (+) and VD (squares); circles represent point estimates of ΔVP, bounded by 95% PDIs (vertical lines). Chromosomal arrangement as in figure 1.
Signatures of Selection on the Transcriptome
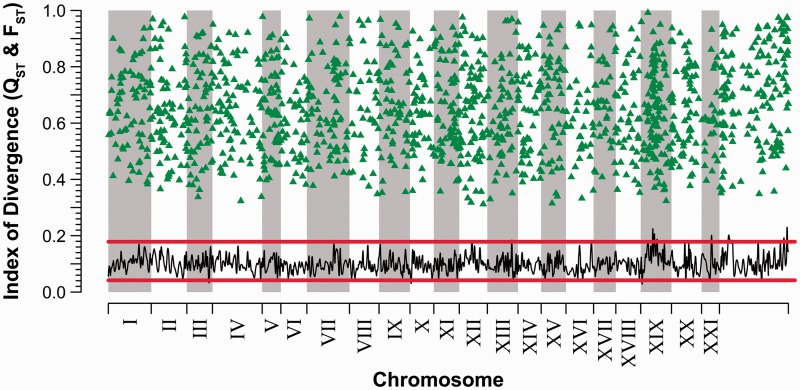
For 84.2% of transcripts, PDIs for the index of quantitative divergence (QST) overlapped with those of neutral expectation. Estimates of neutrality were significantly higher based on the rate of divergence (Δ) of gene expression: 99.2% of transcripts (χ2[1] = 1,319; P < 0.001). Signatures of directional selection—QST estimates significantly greater than neutral expectation (fig. 3)—were inferred for 1,411 transcripts (15.8%). In contrast, estimation of Δ suggested only 71 transcripts (0.8%) under directional selection: 58 of these were also inferred to be under selection in the QST-based analysis, but 13 were inferred neutral by QST. Neither analysis detected a signal of stabilizing selection: Zero transcripts displayed an upper PDI or CI less than the lower ranges defining neutral expectation.
Fig. 3.
Among-population divergence in transcript abundance, plotted as a function of genomic location. Loess smoothing (5-kb intervals) of lower 95% posterior density interval values indicate putative genomic regions rich in adaptive expression divergence. Triangles denote QST point estimates for transcripts which significantly exceed a baseline of neutral divergence, as defined by 17 microsatellite markers (range defined by horizontal lines; see Materials and Methods for details). For clarity, probes whose levels of expression divergence fell within the range of neutral differentiation are not shown. Chromosomal arrangement as in figures 1 and 2.
The per chromosome proportion of transcripts showing signals of selection differed significantly across the 21 linkage groups (χ2[20] = 68.8; P < 0.001), ranging from 10.1% on chromosome VIII to 25.5% on chromosome XIX. 17.1% (156/912) of transcripts on unlinked scaffolds exhibited signals of selective divergence, a proportion not significantly different from that on known linkage groups (χ2[1] = 1.07; P = 0.302). None of the ten mitochondrial transcripts present in the analysis showed signatures of selection.
We detected four putative peaks of high selective divergence, excluding those located on unlinked scaffolds (fig. 3). Three peaks were located on chromosome XIX: A 460-kb window 7.9 Mb from the beginning of the chromosome, a 500-kb window around position 9 Mb, and a 390-kb window at 10.7 Mb (supplementary fig. S3A, Supplementary Material online). The remaining peak mapped to a 485-kb window around position 6.5 Mb of chromosome XXI (supplementary fig. S3B, Supplementary Material online).
Discussion
Our results raise two interesting observations relevant to the evolution of transcriptional variation, namely: 1) The vast majority of transcripts exhibit significant genetic variance in expression, indicating that within-population transcriptional variation is heritable and thus potentially responsive to selection; and 2) although the majority of differences in transcript expression among populations are likely neutral, directional selection is also evident. Although observations here regarding the relative fraction of selected transcripts may be species and/or population specific, inferential discrepancies arising from different analytical frameworks may help bring new perspective on the broader debate regarding the roles of neutral evolution and stabilizing selection in transcriptomic evolution.
Pervasive Heritability—Evidence for Broad Adaptive Potential in the Transcriptome
Whether one considers all transcripts with evidence of significant VA, or a more conservative FDR corrected number, results indicate that a majority of transcripts active in stickleback liver tissue exhibit heritable variation in expression. If the behavior of these transcripts is representative of all 29,245 splice variants in the stickleback genome, this would suggest that at least 74% of the stickleback transcriptome—perhaps as high as 98%—may have heritable variation underlying patterns of expression. This is substantially greater than initial expectations: In the first known synthesis of available data, Gibson and Weir (2005) concluded that irrespective of tissue or organism, 10–50% of transcripts would likely vary due to heritable differences. The reason for this discrepancy may lie in the fact that much of the work describing heritability of transcriptional variance is dominated by quantitative trait locus (QTL)-based estimation. For example, early eQTL studies of human lymphoblastoid cell lines suggested that only 31% of transcripts display heritable variation in expression (Monks et al. 2004), with similar results (28% of transcripts) observed in a an independent genome-wide association study (GWAS; Dixon et al. 2007). Although QTL and related GWAS approaches may provide significant insights into the genetic architecture underlying expression variation, associated estimates of heritability may suffer from both overestimating the proportion of variance explained by individual loci, and a failure to capture all additive genetic variance due to the statistical exclusion of minor-effect loci (Petretto et al. 2006; Manolio et al. 2009). Yet in the context of transcriptional variation, an additional “missing heritability” problem may be an inability to correctly identify the number of transcripts with heritable variation. In a recent study of gene expression in whole blood, Powell et al. (2013) demonstrate a marked reduction in the number of heritable transcripts when estimated based on an eQTL approach (21%) compared with a pedigree-based analysis of same data (64%)—in fact QTL could be mapped to only 30% of the probes for which VA was judged significant under a robust pedigree and analytical framework.
Our findings with respect to the number of transcripts with heritable variation in expression are, however, comparable with those from similar pedigree-based analyses. Heritable variation has been reported in 60% of transcripts expressed in the livers of inbred mouse strains (Cui et al. 2006), and for 42% and 63–82% of transcripts for human adipose tissue and blood, respectively (Price et al. 2011; Powell et al. 2013). And though constraints imposed by their study system preclude the estimation of additive genetic variance, Ayroles et al. (2009) nevertheless reveal broad-sense heritability for 68% of transcripts expressed during development in inbred Drosophila lines. Together, these results suggest that it is likely the majority fraction of the transcriptome for which patterns of differential expression have some genetic basis.
Although it is generally held that transcriptional variation is additive (Gilad et al. 2008; Kim and Gibson 2010), nonadditive sources of genetic variance have been somewhat neglected in the empirical literature. Plant-based research is one exception to this trend, perhaps because these systems are more amenable to the types of complex breeding designs which facilitate estimation of nonadditive effects and/or the viability of mixed-ploidy hybrids (Riddle et al. 2010). For example, line cross analyses between inbred strains of maize have revealed an abundance of transcripts exhibiting dominance variance for expression, though results are equivocal as to whether VA or VD is more prevalent: examples can be found supporting equal proportions of transcripts with both (Pea et al. 2008; Li et al. 2009), as well as only a minority fraction (10–30%) exhibiting VD (Swanson-Wagner et al. 2006; Stupar et al. 2007). Similar analyses of inter-specific wheat hybrids suggest that significantly fewer transcripts exhibit VD compared to those with significant VA (Qi et al. 2012; Chelaifa et al. 2013). Although low numbers of significant estimates imply a potential limitation of statistical power, one study of Arabidopsis nevertheless reveals 3% of transcripts with dominance variance in expression, compared with 8% with VA (Zhang et al. 2008). For vertebrates, we are aware of only four studies reporting the presence of VD for transcription. A line cross analysis of inter-specific Takifugu hybrids suggests the occurrence of VD, though details are insufficient to comment on its prevalence (Gao et al. 2013). Similarly, a cross between wild and domesticated Atlantic salmon revealed that a third of differentially expressed transcripts may have significant VD, compared to 66% with significant VA (Debes et al. 2012). Dominance variance is also far less prevalent than VA in mice, irrespective of tissue type (Rottscheidt and Harr 2007), and in one human study, only 5–32% of probes expressed in whole blood show significant dominance variance (Powell et al. 2013). That up to 99% of stickleback transcripts show evidence of significant VD is surprising, and likely indicative of abundant false positive estimates. Unlike for VA, we are unable to simulate phenotypes with “known” VD using the experimental pedigree; thus, a formal power analysis cannot be performed. However, we attempted to estimate the FDR by simulating over a range of known h2, with d2 (VD) implicitly zero. Deviance Information Criterion (DIC)-based selection favored a model including VD in nearly all (1,009) 1,100 simulations, and mean d2 of these false estimates was 0.144 (supplementary fig. S2D, Supplementary Material online). This would suggest that any “significant” VD estimate for which d2 ≤ 0.144 should be regarded as suspect. Only 4,323 transcripts with putatively significant VD exceed this threshold (41.1%), suggesting a far more conservative estimate for the proportion of the transcriptome with dominance variance for expression.
The Genetic Basis of Transcriptional Variance
The range of heritability estimates observed for stickleback liver transcripts is remarkably similar to that reported in diverse study systems: A mean of point estimates of 0.226 (median = 0.169; fig. 1A). Cui et al. (2006) report a median transcript heritability of 0.22 in mouse liver, with 75% of transcripts having a heritability greater than 0.01. In stickleback liver, the minimum point estimate for all transcripts with significant heritability was 0.041, with a minimum lower 95%PDI estimate of 0.018—although based on phenotypic simulations, estimates less than 0.094 should be viewed with some skepticism. A median heritability of 0.35 for transcript expression has been reported for human lymphoblastoid cell lines, though this estimate may be upwardly biased given that heritability was estimated only for differentially expressed transcripts (Monks et al. 2004). Other human studies show that average transcript heritability differs across tissues: 0.37 in blood and only 0.24 in adipose (Price et al. 2011)—95% quantiles of nonzero estimates from this study overlap (blood = 0.018–0.497; adipose = 0.042–0.539), both with each other and with those reported here for stickleback liver transcripts (0.070–0.596).
Surprisingly, the proportion of variance described by dominance variance (d2) was similar to h2. When the same probes in human blood exhibit both significant VA and VD, the dominance fraction of phenotypic variance is much less than h2 (Powell et al. 2013). In contrast, less than 3% of stickleback liver transcripts had heritability significantly larger than the dominance fraction (fig. 1C), although this is significantly greater than the fraction of probes with only dominance variance (fig. 1D; χ2[1] = 37.7; P < 0.001). This may be due to a possible conflation of maternal effects and dominance variance in half-sib breeding designs (Lynch and Walsh 1998), or perhaps more likely due to the wide/conservative parameter ranges described by the 95%PDIs of the estimates. If only point estimates are considered, 906 stickleback transcripts (8.6%) exhibit h2 > 0.5, a significantly larger fraction than the 213 transcripts (2.0%) for which d2 is greater than 0.5 (χ2[1] = 451; P < 0.001). This may be indicative of a greater role for cis-regulation of gene expression (Lemos et al. 2008), although this remains to be confirmed at the genomic level. It is interesting to note, however, that transcripts with only dominance variance, thus putatively under trans-regulatory control, are largely categorized by genes involved in cellular organization and function, whereas transcripts with h2 significantly greater than d2 seem to represent genes of relatively higher function (supplementary table S2, Supplementary Material online). This is a pattern consistent with the view of cis-regulatory sequences contributing important genetic variation for adaptation through their influence on morphology, physiology, and behavior (Wray 2007). We reiterate, however, that at present this conjecture is speculative, though we suggest that it certainly warrants future consideration.
Although the majority of transcripts appear to have genetic variation for expression, less than 50% of the total phenotypic variation is typically explained by additive effects, and h2 exceeds 0.5 for only 8.6% of transcripts. Our estimates are not substantially different than those derived from human studies which estimate that only 5% of the transcriptome exhibits h2 > 0.5 (Gilad et al. 2008). Dominance variance does not appear to capture the remaining unexplained phenotypic variance. Cumulatively, h2 and d2 explain on average only 38.3% of total phenotypic variance (0.154–0.778, 95% CIs; supplementary fig. S4A, Supplementary Material online). Although the PDIs of the combined estimates are also quite wide, even the upper limit only rarely captures 100% of the phenotypic variance (0.830 on average; supplementary fig. S4C, Supplementary Material online), suggesting that at least 17% of expression variance remains unexplained for most transcripts. Other nonadditive genetic effects (e.g., genetic maternal effects, epistasis) may contribute significantly to transcriptional variation. Maternal effects may be a less likely source of variance as they generally diminish over time, having their greatest influence on traits early in life (Roff 1997). Epistasis, however, is a frequently overlooked effect potentially leading to bias in estimates of transcriptional heritability (Haig 2010; MacLean 2010; Zuk et al. 2012). However, highly specialized breeding designs are required to accurately quantify these nonadditive genetic effects (Rosa et al. 2006). Constraints imposed by female fecundity necessitated use of a half-sib design for this study which precludes our ability to address these issues reliably, though we strongly encourage future research in this direction.
Environmental Variance
Environmental effects are also a likely candidate to explain residual variance, and the ability to explicitly model critical environmental factors, in this case temperature, is a major advantage of experimental systems. In stickleback livers, 66.4% of transcripts showed some evidence of significant temperature sensitivity over a wide range of indicators. Moreover, 60.4% of these 6,987 transcripts exhibited evidence of G × E (n = 4,221; fig. 2C), suggesting a large potential influence of genotype-specific sensitivity to environmental effects. It is noteworthy that the among-family variation in the response to the type of temperature effects studied here is also suggestive of broad-sense heritable variation in plasticity for transcript abundance, a phenomenon reported in another model ectotherm, C. elegans (Li et al. 2006).
With only two treatments, we cannot explicitly estimate the phenotypic variance component due to temperature. However, our measure of phenotypic differences between environments, ΔVP, may be a useful proxy in this regard: It correlates reasonably well with model coefficients describing temperature effects (fig. 2B), but has the advantage of being measured in units of variance. Moreover, when expressed as a ratio relative to total phenotypic variance, we observe the expected, albeit weak, negative correlation with the cumulative genetic proportion of phenotypic variance (r = −0.087; P < 0.001; supplementary fig. S4B, Supplementary Material online). Furthermore, when ΔVP relative to total VP is modeled as a function of genomic position, chromosomal “valleys” of low broad-sense genetic variance appear to coincide with many “peaks” of our proxy “VE(temp)” metric (supplementary fig. S4D, Supplementary Material online). However, the grand cumulative proportion of phenotypic variance, combining relative genetic and ΔVP values, explains only a 0.496 proportion of total phenotypic variance in expression for an average transcript (supplementary fig. S4D, Supplementary Material online)—based on the CIs of all estimates (0.206–0.905), nearly 10% of expression variance remains unexplained.
Although reasonable to expect temperature to be a major contributor to expression variance in an ectotherm, this does not rule out other environmental factors. For example, in yeast lines transcription appears to exhibit greater environmental variation than that of genetic effects in response to varying nutritional inputs (Landry et al. 2006), and in Daphnia pulex, 72–85% of the transcriptome is differentially expressed in response to abiotic environmental stressors (Colbourne et al. 2011). Manipulating other environmental variables would undoubtedly also induce some degree of expression variance, though we are hard-pressed to suggest one which would likely produce greater effects than temperature, given its direct effects on the metabolism and rate of transcription in ectotherms. Thus, in this context, it is striking that genetic effects appear to capture a significantly greater proportion of expression variance.
Neutrality and Directional Selection Reconsidered
Both QST results and divergence rate estimates (Δ) reveal that for the majority of transcripts, among-population differences in expression are reflective of neutral processes. Moreover, neither analysis could detect a significant signal of stabilizing selection. Together, these observations provide a case which challenges the view of stabilizing and/or purifying selection as the predominant mode of evolution of the transcriptome, and aligns with a neutral model of transcriptome evolution.
To some extent, the failure to detect stabilizing selection may be related to the tendency for dominance variance to upwardly bias estimates of QST, potentially leading to false signals of neutrality (reviewed in Leinonen et al. 2013). However, the same metric (Δ; Lemos et al. 2005) used to infer a majority fraction of transcripts under stabilizing selection across a diverse collection of taxa also points to neutral divergence between focal stickleback populations. This in turn points to a problem frequently encountered in studies employing metrics of the rate of evolution: A neglect to consider uncertainty in parameter estimates (Hendry and Kinnison 1999). In our application, we employ a simple bootstrapping procedure to express uncertainty in terms of CIs, and explicitly use this uncertainty when inferring significance of neutrality or selection. Others argue that selection of “conservative” estimates for parameters defining neutrality is sufficient (Lemos et al. 2005; Fay and Wittkopp 2008); however, if the magnitude of differences between upper and lower CIs exceeds that of the neutral region, point estimates may have a high probability of providing false signals. This certainly appears to be the case with these data, wherein the CIs for point estimates of Δ indicative of stabilizing selection range from approximately 30 to 100 times greater than the neutral range (supplementary fig. S5C, Supplementary Material online). Moreover, many transcripts identified to be under stabilizing selection by the average value of Δ have QST values well above neutrality (supplementary fig. S5B and D, Supplementary Material online), values extremely unlikely for a trait under stabilizing selection.
It is perhaps noteworthy that most empirical studies which conclude that stabilizing and/or purifying selection is the predominant mode of transcriptional evolution are based on interspecific comparisons (Lemos et al. 2005; Gilad, Oshlack, Smyth, et al. 2006; Bedford and Hartl 2009; Brawand et al. 2011), though see Nuzhdin et al. (2004) who demonstrate substantial adaptive/selected variation between Drosophila simulans and D. melanogaster. Yet simulations—the results of which closely match those of observed data—suggest that gene expression divergence is not a simple linear function of generation time: Initially it occurs rapidly, but tends toward an asymptote (Ogasawara and Okubo 2009). Paradoxically then, the rate of change of gene expression divergence when compared among extremely phylogenetically distant taxa would be less than that among more recently diverged groups. To some extent this is reflected by comparing trends between intra- and interspecific contrasts, the former typified by greater rates of drift and directional selection, though the largest fraction of transcripts show signatures of stabilizing selection (Lemos et al. 2005). The implications of this are 2-fold: 1) Comparisons across broader taxonomic groups may be most likely to reveal signatures of stabilizing selection; and 2) the generations immediately following—perhaps even during—divergence, may represent a dynamic phase when mutation, drift, and selection may have the most profound effects on expression divergence.
Adaptive differentiation between populations of the same species would certainly fit the description of a dynamic phase in a taxon’s history. The populations used in this study show adaptive phenotypic and genetic divergence typical for this species (Cano et al. 2006). Moreover, divergence has occurred relatively recently, likely within the past approximately 8,000 years following retreat of Pleistocene ice sheets (Mäkinen and Merilä 2008). And although the majority of transcriptional divergence is neutral, a relatively high proportion of transcripts (15.8%) show signatures consistent with divergent selection. Irrespective of the actual mechanisms and selection pressures which underlie this divergence, it is clear that some degree of adaptive differentiation has occurred at the level of the transcriptome.
It should be noted, however, that transcript abundance differs among tissues, and that tissue-specific (i.e., specialized) expression patterns are more likely to be subject to positive selection (Brawand et al. 2011). Whether liver tissue represents a potentially biased sample—either over- or underrepresentative of potential adaptive differences among habitats sampled—remains open for debate. However, killifish populations adapted to a natural temperature cline also show expression divergence in the liver, but in these populations, the majority of transcripts showing signatures of natural selection are under stabilizing selection (Whitehead and Crawford 2006a). This, and the multifaceted functional role of the liver, leads us to believe tissue-related bias should be minimal.
Another potential source of concern is the discrepancy between QST and Δ in the number of transcripts showing signals of directional selection. This is unlikely due to the use of only two populations in the rate divergence analysis: Our previous work clearly shows that among-population differences in both expression and enzymatic activity are largely driven by the Lake Vättern population (Nikinmaa et al. 2013), a population which was included in the analysis due to its unambiguous evolutionary relationship with the Baltic Sea population. Rather, we would suggest that differences between methods may be reflective of difficulties in analysis of variance (ANOVA)-based comparisons which do not correct for nonindependent phylogenetic associations: Simulations show that failure to reject neutrality, even under strong selective pressures may be common for such tests (Eng et al. 2009). We would argue that QST-based analyses implicitly include a phylogenetic correction in that neutrality is inferred from the same types of genetic data which would be used to generate the phylogenetic covariance matrices in admixture modeling (Eng et al. 2009). Moreover, the wide PDIs bounding QST estimates represent a conservative comparison against neutral parameter space (supplementary fig. S5D, Supplementary Material online). Consequently, we are confident that results of QST are truly reflective of recent evolutionary history in this system.
Further support for our inference of selection can be gleaned from its genomic locations, and the nature of its putative functional significance. For example, we show a peak of high selective divergence mapping to a 485-kb window around position 6.5 Mb of chromosome XXI (fig. 3 and supplementary fig. S3B, Supplementary Material online). This also corresponds to a chromosomal inversion associated with global patterns of marine–freshwater divergence in the species (Jones et al. 2012). Additionally, we show several peaks on chromosome XIX (fig. 3 and supplementary fig. S3B, Supplementary Material online), the stickleback sex chromosome. Interestingly, sex differentiation is one of the higher-ranked functional clusters identified in an enrichment analysis of putatively selected transcripts. On their own, such analyses are merely suggestive of further avenues of research, and are not confirmatory of functional significance. Nevertheless, they form one line of evidence which is consistent with selection on transcriptional variation playing a role in the process of adaptive differentiation in this system.
Finally, we do not wish to espouse a pan-selectionist view of transcriptional divergence—far from it given that neutral divergence was predominant. Nevertheless, our results show that a relatively larger-than-expected proportion of the transcriptome has diverged in response to directional selection. Taken together, evidence of within population heritable variation and selective variation among populations provide convincing supporting for the contention of broad adaptive potential within the stickleback transcriptome.
Materials and Methods
Broodstock and Husbandry
Crosses for among-Population Comparisons
Broodstock were obtained by sampling mature sticklebacks from three wild, Fennoscandian populations: Lake Pulmanki (Pulmankijärvi; PUL) in Finnish Lapland; the Baltic Sea in the vicinity of Helsinki, Finland (HEL); and Lake Vättern (VAT) from south-central Sweden (supplementary fig. S1A, Supplementary Material online). Full-sib F1 families were created by crossing parental fish at the sampling sites. Fertilized eggs were transported to the laboratory at the University of Helsinki. Offspring were initially maintained at 17 ± 1 °C, with a photoperiod of 18 h light and 6 h darkness. Six months after hatching, environmental conditions were gradually changed to simulate wintering conditions (24 h darkness; water temperature 9 ± 1 °C). After 5 months, fish were stimulated into breeding condition by gradually reverting environmental conditions to an 18:6 h light/dark photoperiod, and water temperature to 17 ± 1 °C.
Mating pairs were chosen randomly, and new crosses were made to obtain F2 offspring from within each population. At the time of the experiments, F2 offspring were approximately 20 months old. Although sexually mature, all experimental fish were reproductively inactive. Twelve F2 fish from each population (N = 36) were randomly selected and assigned to separate, screened containers in one of two identical tanks (six fish per population per tank). Fish were acclimated overnight. One tank was maintained as a control at 17 °C. At the beginning of the next photoperiod, water in the second tank was heated approximately 1 °C per hour for 6 h to a final temperature of 23 °C. After 1 h at final temperature, each fish was euthanized in a lethal concentration of MS-222 in water, and its liver removed and immediately frozen in liquid nitrogen for storage at −80 °C.
Crosses for Quantitative Genetics
Thirty mature males and 60 gravid females were collected from the Baltic Sea for use as broodstock in a paternal, half-sib design (HEL; supplementary fig. S1B, Supplementary Material online). A more detailed description of crossing protocols has been described elsewhere (Leinonen et al. 2011), but in brief, each male was artificially crossed (in vitro, Barber and Arnott 2000) with two separate females, producing 30 unique half-sib blocks (supplementary fig. S1B, Supplementary Material online), 60 families in total. Groups of 15 individuals from each of the 60 families were randomly assigned to separate 10-l tanks. Fish were reared for 6 months at 17 ± 1 °C and 12:12 h light/dark photoperiod.
After 6 months, two individuals from each family were randomly selected and arbitrarily assigned to control or treatment. Fish were placed in the tanks in the evening and allowed to acclimate overnight. The treatment fish were subjected to the same temperature treatment as described above and sampled. This was conducted for five consecutive days, so that five treated and five control fish were sampled for each of the 60 families (N = 563, see Sample Preparation). Analysis of power and bias was performed by simulating phenotypic data, based on the final pedigree and over a range of known heritabilities (Morrissey et al. 2007), and iterative estimation with the same analytical method used on experimental data (supplementary fig. S2 and appendix S1, Supplementary Material online).
Sample Preparation
Total RNA was isolated from liver tissue by means of Tri Reagent (Sigma, St Louis, MO and Ambion, Austin, TX), following manufacturer’s protocols. RNA was treated with DNase (Promega, Madison, WI) and reisolated using Tri Reagent. RNA concentration was quantified using a Nanodrop ND-1000 (Thermo Scientific, Waltham, MA), and RNA quality was assessed using an Experion automated electrophoresis system (Bio-Rad, Hercules, CA). RNA was extracted from 35 of 36 individuals used for population comparison (one PUL control missing), and all 600 individuals of known pedigree comprising the quantitative genetics data set. RNA labeling, hybridizations, and scanning were performed by Agilent certified commercial service providers: The Finnish Microarray and Sequencing Centre (Turku, Finland; array 1), and the University Health Network Microarray Centre (Toronto, Canada; array 2). Thirty-seven individuals were removed from the quantitative genetic analyses due to either missing tissue, bad quality RNA, or bad array hybridization. The final data set contained 563 individuals in total, 283 controls (158 females and 125 males) and 280 thermally treated (152 females and 128 males).
Microarray Design and Data Normalization
Two microarrays, based on user-defined 60-bp oligonucleotide probes, were designed using the custom gene expression platform from Agilent (Santa Clara, CA). The first—used for among-population comparisons (i.e., QST estimation)—was a 4 × 44K array consisting of 43,605 probes representing 19,274 unique genes, including most known splice-variants (27,723 transcripts; Leder et al. 2009), with most transcripts replicated at least twice. Full technical details are provided in previously published work (Leder et al. 2009, 2010), but in brief, total RNA (400 ng) was amplified and Cy3 labeled. cRNA was examined with the Nanodrop ND-1000 and the Experion Automated Electrophoresis System to assess the concentration and quality of labeling. Each sample (1.65 µg) was hybridized to its array at 65 °C overnight (17 h). Arrays were scanned with Agilent Technologies Scanner (model G2505B). Spot intensities and other quality control features were extracted with Agilent’s Feature Extraction Software (v9.5.3.1), and array quality was assessed through the use of Agilent control features, as well as spike-in controls.
For the second array, the probe set was reduced by removing several replicate probes, as well as many of the probes that did not appear to hybridize to liver RNA in the first experiment, in order to fit on the newer 8 × 15K array format. RNA quality was determined using the Agilent Bioanalyzer 2100, and only RNA with integrity numbers greater than 7.0 were used for labeling. RNA (500 µg) was labeled (Cy3 or Cy5) using the Agilent QuickAmp Kit for two-color microarray-based gene expression analysis. Equal numbers of individuals within family groups (control and temperature-treated; males and females) were assigned to each dye, and alternately labeled. Control and treated individuals were selected at random for hybridization to a given array (Agilent Hi-RPM kit). Images of the arrays were acquired using a G2565BA DNA Scanner, and image analysis was performed using Agilent’s Feature Extraction Software (v9.5; protocol GE2-v5_95_Feb07 with default settings).
Postprocessed signals were standardized across arrays using a supervised normalization approach, implemented in the package “snm” for R/Bioconductor (Mecham et al. 2010), which has been shown to remove technical artifacts without altering the underlying variance structure of expression data (Qin et al. 2013)—the latter being essential for variance components analysis, as undertaken in this study (for an expanded explanation of the normalization procedure, see supplementary appendix S1, Supplementary Material online). For estimating quantitative genetic parameters, the entire second data set (i.e., all probes present on the second microarray) was normalized across arrays/slides: “Biological variables” included sex, family, and temperature treatment; “adjustment variables” included dye, array, and batch (i.e., slide). Following normalization, data were screened for potential outliers. Individual intensity values more than 2 SDs from their family-by-treatment mean were flagged and removed. Any probes with missing values for an entire family, or for more than 56 individuals (i.e., 10% of data), were removed from the final data sets. For transcripts with two or more replicate probes, we calculated pairwise correlations between probe sets. Replicate probes with a correlation coefficient less than 0.9 were discarded, and replicates which remained were averaged by transcript for each individual. Raw and normalized data, in addition to R scripts describing the normalization procedure, are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3098.
For among-population comparisons, nonnormalized transcripts common to both arrays were merged into a single data set for a broader normalization across studies/array designs, thereby providing standardized data sufficient for estimating both within- and among-population components of genetic variance for inferring selection (see Final Data and Analyses and/or supplementary appendix S1, Supplementary Material online). Biological variables included population, family, and temperature treatment; adjustment variables included dye, array, batch (i.e., slide), and array design; previously identified outliers were removed from the final data set. Data and R scripts are also archived on ArrayExpress (accession E-MTAB-3099).
Microsatellite Data
Details of genotyping are outlined elsewhere (Cano et al. 2006, 2008). Briefly, DNA was extracted from pectoral fin tissue using fine silica 96-well filter plates. Seventeen putatively neutral microsatellite loci, that is, markers approximately 3 cM or more from a mapped QTL region, were genotyped for 28 individuals in each of the three populations (HEL, VAT, and PUL); see Cano et al. (2006) for a list of loci. Forward primers were labeled with fluorescent dyes (FAM, TET, and HEX), and a GTTT-tail was added to the 5′-end of the reverse primers to enhance 3′-adenylation (Brownstein et al. 1996). Polymerase chain reaction products were diluted (1:50), mixed with ROX size standard, and run on a MegaBACE 1000 capillary sequencer. Fragment Profiler (v1.2) was used to score genotypes, with manual corrections as needed.
We evaluated the data set through outlier analysis to ensure that all markers fell within a simulated range of neutral expectation (Antao et al. 2008). As the index of quantitative divergence (QST) has been shown to behave similarly to a single-locus estimate of FST, it should be compared against a distribution of single-locus estimates, rather than the CI of mean FST (Whitlock 2008). We defined a conservative parameter space indicating neutral differentiation based on the full range (minimum and maximum) of FST values observed in all 17 neutral markers.
Final Data and Analyses
Quantitative Genetics
Expression levels for 10,527 transcripts (approximately 36% of the stickleback transcriptome), comprising 10,495 predicted/projected genes (Ensembl Stickleback Genome version 65.1, updated May 2010), were retained for analysis after normalization and quality control checks. We decomposed expression variation into additive genetic (VA) and dominance (VD) variance using a linear mixed-effects model (i.e., the “animal model”). Fixed effects included dye (Cy3 or Cy5), sex, temperature treatment (17 or 23 °C), and their interaction terms; random effects were assigned by the relationship matrix (i.e., pedigree) for the estimation of VA and VD. We employed a Bayesian implementation of the animal model in the R packages “MCMCglmm” and “nadiv” (Hadfield 2010; Wolak 2012). Variance components were estimated by MCMC (Markov chain Monte Carlo) sampling of their posterior distributions, after first removing effects of fixed model terms (temperature, sex, and dye), with parameter estimates based on the posterior mode, and bounded by 95% PDIs. We ran a series of three nested models to test the posterior significance of both VA and VD: A simple “null” model included only fixed effects; a second model included random variation attributable to the individual (i.e., “animal”) level weighted by the half-sib pedigree structure (VA); and a final model also including dominance, obtained by inversion of the pedigree matrix to obtain a “fraternity matrix” from which VD can be estimated (Ovaskainen et al. 2008)—matrix inversion was implemented using the “nadiv” package (Wolak 2012). Models were run with an initial burn-in of 50,000 iterations, followed by an additional 200,000 iterations from which each 200th point on the Markov Chain were sampled. We used the DIC to infer significance of a given variance component by comparing the more “complex” model containing the term with a simpler model without: Significance of VA was determined by comparison with the “null” model, and a “maximal” model (VA and VD) was evaluated against both the VA only model and the null model.
Within our analytical framework, likelihood ratio testing and associated p-values for the significance of random effects (i.e. variance estimates) are not possible. Nor can we conduct probability-based significance testing by profiling their posterior distributions, since variance estimates are constrained to be positive. This precludes the use of standard adjustments for multiple comparisons (e.g., FDR correction). Consequently, we used phenotypic simulations to determine the reliability of estimates through an evaluation of “local” false discoveries. We simulated 1,000 trait values with no additive genetic variance using the package “pedantics” (Morrissey et al. 2007), and compared the animal model DIC to a null model—selection of the animal model indicated a false positive results. We then calculated the mean heritability estimate for all false positives (0.094; n = 247) to define a threshold value indicative of a reliable estimate. Likewise, we simulated data under the assumption of no dominance variance over a range of known heritabilities (h2 = 0–1 in 0.1 intervals; 100 simulations each). As with the transcriptional data, models including VD estimates were compared with both a simpler animal model and a null model. For all false positives, we calculated the mean estimate of d2 to define the significant threshold value (0.144; n = 1,009).
As only two temperature treatments were employed, we could not directly estimate environmental variance associated with thermal effects (herein simplified as VE(temp)) as a random effect. However, we attempted to discern the relative contributions of environmental effects on phenotypic variance in a number of ways. First, we estimated both total phenotypic variance (VP) and VA separately in each “environment” (i.e., thermal treatment): We reasoned that if VP was significantly different between environments, this could be attributable to a G×E and/or to nongenetic effects—given the thermal treatment imposed on an ectothermic organism, it stands to reason that an environmental effect associated with temperature (VE(temp)) would be a likely source of this nongenetic variation. We estimated the relative magnitude of this effect as the difference between parameter estimates (i.e., 1,000 sampling points from the Markov Chains) in each thermal treatment. Significant differences in phenotypic (ΔVP) and/or additive variance (ΔVA) were assigned if the 95% PDIs of the difference profiles excluded zero. To simplify comparisons with estimates of VA and VP, we converted ΔVP estimates to their absolute values, reversing lower and upper PDIs for negative estimates, and assuming a lower estimate of zero for nonsignificant differences. Finally, we evaluated the potential for G×E by testing for broad-sense genetic variation in expression reaction norms (family × temperature). We ran an additional series of mixed-effects models (dye, sex, and temperature as fixed effects): A minimal model included random variation among families only in the intercept, and a second model which also included random variation among families in the coefficient describing temperature (i.e., environmental) effects (random slopes; family × temperature). Significance of G×E was evaluated by comparison of model DICs. Relative effect sizes of temperature, sex, and their interaction were estimated from the posterior distributions of their coefficients; P values were also profiled from the posterior distributions. Correction for multiple comparisons was performed using a local FDR procedure implemented in the R package “fdrtool” (Strimmer 2008).
Comparison among Populations
Following normalization and quality control 8,904 transcripts, representing 8,882 predicted/projected genes (Ensembl Stickleback Genome version 65.1, updated May 2010) common to both microarrays were retained for analysis. To infer whether among-population differences were best explained by neutral or putative selective processes, we contrasted an index of quantitative genetic differentiation (QST) for each probe with that of neutral genetic divergence (Spitze 1993; Leinonen et al. 2013). In this approach, an estimate of within population variation equivalent to VA is required for robust inference. By combining array data (i.e., pooling and normalizing probes / transcripts common to both arrays), we were able to include a broader range of pedigrees, thereby permitting a more accurate and analytically comparable estimate of VA for calculating QST. Variance components (i.e., QST parameters) were estimated through mixed-model analysis using MCMCglmm: Temperature and dye effects were partitioned out as fixed model terms, with variance among (σ2GB) and within populations (σ2GW; VA) estimated as random effects. Confidence intervals (95% PDIs) were obtained by sampling the posterior distribution of 1,000 QST estimates sampled from the Markov chain. Significance of QST was inferred for those estimates with PDIs that did not overlap with the region of neutral differentiation, as defined by FST of neutral microsatellites. Additionally, we attempted to identify potential genomic regions of high selective divergence by modeling lower interval estimates (PDI0.05) as a function of transcript position within the stickleback genome with loess regression over 5-kb intervals.
Finally, we contrasted inference from QST-based analyses with that obtained by estimating rate of expression divergence. We used the divergence parameter (Δ) proposed by Lynch (1990) and adapted by Lemos et al. (2005) for application to mRNA expression data: Under this framework a parameter space between 10−4 and 10−2 represents neutral evolution, based on assumed rates of mutational variance. Estimates of Δ above and below neutrality are indicative of directional and stabilizing selection, respectively. As this metric requires an estimation of generation time between groups, we focused analyses on the two populations with the least ambiguous colonization histories: Baltic Sea and Lake Vättern (supplementary fig. S1, Supplementary Material online)—their stickleback populations share a recent genetic origin (Mäkinen et al. 2006), and the waterbodies themselves share a geological history. Following glacial retreat of the Fennoscandian peninsula, the area currently occupied by Lake Vättern was a bay of the Baltic Sea during its Yoldia Sea stage (approximately 10 ka BP), but by the end of the Ancylus Lake stage (approximately 8 ka BP), Lake Vättern was no longer contiguous with the Baltic (Stålberg 1939; Björck 1995). As sticklebacks breed only once per year, with new recruits added annually, these dates were used to bound estimates of the numbers of generations since divergence. We generated five balanced replicate blocks within each population–temperature treatment by stratified bootstrapping to estimate standard mean-square error within and between populations in a nested ANOVA. Generation time was sampled from a uniform distribution of integers between 8,000 and 10,000. Confidence intervals of Δ were profiled by bootstrapping the entire estimation procedure 1,000 times. Selection was inferred on the basis of CI exclusion from the parameter space defining neutral evolution (10−4 to 10−2).
Functional Annotation Analysis
A complete functional enrichment analysis is beyond the main scope of this article. Nevertheless, such analyses may provide preliminary interpretive value of current results, and more importantly, may be of utility for other researchers. As such, they are included as supplementary material, Supplementary Material online. We used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to determine whether candidate transcript sets were significantly overrepresented by genes of particular functional categories (Huang et al. 2007). Although DAVID will accept several probe identifiers, Entrez Gene identifiers generally produce the most comprehensive annotation data (Maglott et al. 2005; Sherman et al. 2007). Thus, to facilitate analyses, stickleback genes were assigned an Entrez GeneID based on their human orthologs, determined using BioMart (Durinck et al. 2005; Smedley et al. 2009) and BLAST search. Of the 10,527 transcripts detected above background, 92% (9,661) were assigned an Entrez GeneID. Input for analysis consisted of a list of candidate transcripts contrasted with a customized “background” set including only those transcripts represented on the custom array and detected above background. Though DAVID can integrate nonredundant functional annotations across multiple databases, we focused on Gene Ontology (GO) annotations at the levels of “biological process” and “molecular function” (Harris et al. 2004). We further restricted output by grouping enriched terms into functional annotation clusters, using default settings (Huang et al. 2009). Finally, we excluded those clusters with an overall enrichment score less than 0.5, and for which the mean fold enrichment of constituent GO terms was significantly less than 1.
Supplementary Material
Supplementary appendix S1, figures S1–S5, and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Hannu Mäkinen, Linda Uoti, and numerous others who helped in obtaining broodstock and maintaining the fish used in this study. This work was principally funded by the Academy of Finland (grant no. 129662), through the Centre of Excellence in Evolutionary Genetics and Physiology (2006–2011). Additional support was provided by the Academy of Finland to J.M. (grant nos. 134728, 250435, and 265211), C.P. (grant nos. 133875 and 141231), and E.L. (grant no. 136464). Computing resources were provided by the Finnish Centre for Scientific Computing. Research was conducted under an ethical license from the University of Helsinki (HY 121-06). The authors declare no conflict of interest.
References
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol. 2009;18:3763–3780. doi: 10.1111/j.1365-294X.2009.04313.x. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Barber I, Arnott SA. Split-clutch IVF: a technique to examine indirect fitness consequences of mate preferences in sticklebacks. Behaviour. 2000;137:1129–1140. [Google Scholar]
- Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci U S A. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björck S. A review of the history of the Baltic Sea, 13.0-8.0 ka BP. Quat Int. 1995;27:19–40. [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by tag DNA polymerase: primer modifications that facilitate genotyping. Biotechniques. 1996;20:1004–1006. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- Cano JM, Mäkinen HS, Leinonen T, Freyhof J, Merilä J. Extreme neutral genetic and morphological divergence supports classification of Adriatic three-spined stickleback (Gasterosteus aculeatus) populations as distinct conservation units. Biol Conserv. 2008;141:1055–1066. [Google Scholar]
- Cano JM, Matsuba C, Mäkinen H, Merilä J. The utility of QTL-Linked markers to detect selective sweeps in natural populations: a case study of the EDA gene and a linked marker in threespine stickleback. Mol Ecol. 2006;15:4613–4621. doi: 10.1111/j.1365-294X.2006.03099.x. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H, Chagué V, Chalabi S, Mestiri I, Arnaud D, Deffains D, Lu YH, Belcram H, Huteau V, Chiquet J, et al. Prevalence of gene expression additivity in genetically stable wheat allohexaploids. New Phytol. 2013;197:730–736. doi: 10.1111/nph.12108. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XQ, Affourtit J, Shockley KR, Woo Y, Churchill GA. Inheritance patterns of transcript levels in F1 hybrid mice. Genetics. 2006;174:627–637. doi: 10.1534/genetics.106.060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Evolutionary innovation and stability in animal gene networks. J Exp Zool B Mol Dev Evol. 2010;314B:182–186. doi: 10.1002/jez.b.21329. [DOI] [PubMed] [Google Scholar]
- Debes PV, Normandeau E, Fraser DJ, Bernatchez L, Hutchings JA. Differences in transcription levels among wild, domesticated, and hybrid Atlantic salmon (Salmo salar) from two environments. Mol Ecol. 2012;21:2574–2587. doi: 10.1111/j.1365-294X.2012.05567.x. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KCC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, Wang TY, Huang CJ, Lu HHS, Lu MYJ, Wu SH, Li WH. Natural selection on cis and trans regulation in yeasts. Genome Res. 2010;20:826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng KH, Bravo HC, Keles S. A phylogenetic mixture model for the evolution of gene expression. Mol Biol Evol. 2009;26:2363–2372. doi: 10.1093/molbev/msp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang H, Gao Q, Wang LL, Zhang FC, Siva VS, Zhou Z, Song LS, Zhang SC. Transcriptome analysis of artificial hybrid pufferfish Jiyan-1 and its parental species: implications for pufferfish heterosis. PLoS One. 2013;8:e58453. doi: 10.1371/journal.pone.0058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Gienapp P, Teplitsky C, Alho JS, Mills JA, Merilä J. Climate change and evolution: disentangling environmental and genetic responses. Mol Ecol. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AK, Cech JN, Peichel CL. Molecular and developmental contributions to divergent pigment patterns in marine and freshwater sticklebacks. Evol Dev. 2012;14:351–362. doi: 10.1111/j.1525-142X.2012.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw. 2010;33:1–22. [Google Scholar]
- Haig D. Does heritability hide in epistasis between linked SNPs? Eur J Hum Genet. 2010;19:123. doi: 10.1038/ejhg.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Kinnison MT. The pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines HM, Papa R, Ruiz M, Papanicolaou A, Wang C, Nijhout HF, McMillan WO, Reed RD. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics. 2012;13:288. doi: 10.1186/1471-2164-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nat Rev Genet. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S. A neutral model of transcriptome evolution. PLoS Biol. 2004;2:682–689. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Gibson G. Insights from GWAS into the quantitative genetics of transcription in humans. Genet Res. 2010;92:361–369. doi: 10.1017/S001667231000056X. [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Lema SC, Luckenbach JA, Mori S, Kawagishi Y, Kusakabe M, Swanson P, Peichel CL. Adaptive divergence in the thyroid hormone signaling pathway in the stickleback radiation. Curr Biol. 2010;20:2124–2130. doi: 10.1016/j.cub.2010.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn MH, Shapiro J, Wu CI. Decoupled differentiation of gene expression and coding sequence among Drosophila populations. Genes Genet Syst. 2008;83:265–273. doi: 10.1266/ggs.83.265. [DOI] [PubMed] [Google Scholar]
- Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? J Hum Genet. 2010;55:195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- Landry CR, Oh J, Hartl DL, Cavalieri D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–351. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Leder EH, Cano JM, Leinonen T, O’Hara RB, Nikinmaa M, Primmer CR, Merilä J. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol Biol Evol. 2010;27:1495–1503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Leder EH, Merilä J, Primmer CR. A flexible whole-genome microarray for transcriptomics in three-spine stickleback (Gasterosteus aculeatus) BMC Genomics. 2009;10:426. doi: 10.1186/1471-2164-10-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen T, Cano JM, Merilä J. Genetics of body shape and armour variation in threespine sticklebacks. J Evol Biol. 2011;24:206–218. doi: 10.1111/j.1420-9101.2010.02161.x. [DOI] [PubMed] [Google Scholar]
- Leinonen T, McCairns RJS, O’Hara RB, Merilä J. QST-FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat Rev Genet. 2013;14:179–190. doi: 10.1038/nrg3395. [DOI] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci U S A. 2008;105:14471–14476. doi: 10.1073/pnas.0805160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Cáceres M, Hartl DL. Rates of divergence in gene expression profiles of primates, mice, and flies: stabilizing selection and variability among functional categories. Evolution. 2005;59:126–137. [PubMed] [Google Scholar]
- Leveelahti L, Leskinen P, Leder EH, Waser W, Nikinmaa M. Responses of threespine stickleback (Gasterosteus aculeatus, L) transcriptome to hypoxia. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6:370–381. doi: 10.1016/j.cbd.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Lvarez OAA, Gutteling EW, Tijsterman M, Fu JJ, Riksen JAG, Hazendonk E, Prins P, Plasterk RHA, Jansen RC, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:2155–2161. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhang DF, Jia GQ, Dai JR, Wang SC. Genome-wide comparisons of gene expression for yield heterosis in maize. Plant Mol Biol Rep. 2009;27:162–176. [Google Scholar]
- Liao BY, Weng MP, Zhang JZ. Contrasting genetic paths to morphological and physiological evolution. Proc Natl Acad Sci U S A. 2010;107:7353–7358. doi: 10.1073/pnas.0910339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Lynch M. The rate of morphological evolution in mammals from the standpoint of the neutral expectation. Am Nat. 1990;136:727–741. [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sunderland (MA): Sinauer; 1998. [Google Scholar]
- MacLean RC. Predicting epistasis: an experimental test of metabolic control theory with bacterial transcription and translation. J Evol Biol. 2010;23:488–493. doi: 10.1111/j.1420-9101.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Genetic relationships among marine and freshwater populations of the European threespined stickleback (Gasterosteus aculeatus) revealed by microsatellites. Mol Ecol. 2006;15:1519–1534. doi: 10.1111/j.1365-294X.2006.02871.x. [DOI] [PubMed] [Google Scholar]
- Mäkinen HS, Merilä J. Mitochondrial DNA phylogeography of the three-spined stickleback (Gasterosteus aculeatus) in Europe: evidence for multiple glacial refugia. Mol Phylogenet Evol. 2008;46:167–182. doi: 10.1016/j.ympev.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Seshasayee ASN, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- McCairns RJS, Bernatchez L. Adaptive divergence between freshwater and marine sticklebacks: insights into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution. 2010;64:1029–1047. doi: 10.1111/j.1558-5646.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- Mecham BH, Nelson PS, Storey JD. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey MB, Wilson AJ, Pemberton JM, Ferguson MM. A framework for power and sensitivity analyses for quantitative genetic studies of natural populations, and case studies in Soay sheep (Ovis aries) J Evol Biol. 2007;20:2309–2321. doi: 10.1111/j.1420-9101.2007.01412.x. [DOI] [PubMed] [Google Scholar]
- Nikinmaa M, McCairns RJS, Nikinmaa MW, Vuori KA, Kanerva M, Leinonen T, Primmer CR, Merilä J, Leder EH. Transcription and redox enzyme activities: comparison of equilibrium and disequilibrium levels in the three-spined stickleback. Proc R Soc B Lond Biol Sci. 2013;280:20122974. doi: 10.1098/rspb.2012.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Ogasawara O, Okubo K. On theoretical models of gene expression evolution with random genetic drift and natural selection. PLoS One. 2009;4:e7943. doi: 10.1371/journal.pone.0007943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Ovaskainen O, Cano JM, Merilä J. A Bayesian framework for comparative quantitative genetics. Proc R Soc Lond B Biol Sci. 2008;275:669–678. doi: 10.1098/rspb.2007.0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Qian WF, Zhang JZ. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 2012;13:1123–1129. doi: 10.1038/embor.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pea G, Ferron S, Gianfranceschi L, Krajewski P, Pè ME. Gene expression non-additivity in immature ears of a heterotic F1 maize hybrid. Plant Sci. 2008;174:17–24. [Google Scholar]
- Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JE, Henders AK, McRae AF, Kim J, Hemani G, Martin NG, Dermitzakis ET, Gibson G, Montgomery GW, Visscher PM. Congruence of additive and non-additive effects on gene expression estimated from pedigree and SNP data. PLoS Genet. 2013;9:e1003502. doi: 10.1371/journal.pgen.1003502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Helgason A, Thorleifsson G, McCarroll SA, Kong A, Stefansson K. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B, Huang W, Zhu B, Zhong XF, Guo JH, Zhao N, Xu CM, Zhang HK, Pang JS, Han FP, et al. Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol. 2012;10:3. doi: 10.1186/1741-7007-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Kim J, Arafat D, Gibson G. Effect of normalization on statistical and biological interpretation of gene expression profiles. Front Genet. 2013;3:160. doi: 10.3389/fgene.2012.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Rosas U, Banta J, Bhambhra N, Purugganan MD. Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genet. 2012;8:482–495. doi: 10.1371/journal.pgen.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Jiang HM, An LL, Doerge RW, Birchler JA. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theor Appl Genet. 2010;120:341–353. doi: 10.1007/s00122-009-1113-3. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Roberge C, Guderley H, Bernatchez L. Genomewide identification of genes under directional selection: gene transcription QST scan in diverging Atlantic salmon subpopulations. Genetics. 2007;177:1011–1022. doi: 10.1534/genetics.107.073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff DA. Evolutionary quantitative genetics. New York: Springer; 1997. [Google Scholar]
- Rosa GJM, de Leon N, Rosa AJM. Review of microarray experimental design strategies for genetical genomics studies. Physiol Genomics. 2006;28:15–23. doi: 10.1152/physiolgenomics.00106.2006. [DOI] [PubMed] [Google Scholar]
- Rottscheidt R, Harr B. Extensive additivity of gene expression differentiates subspecies of the house mouse. Genetics. 2007;177:1553–1567. doi: 10.1534/genetics.107.076190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sherman BT, Huang DW, Tan QN, Guo YJ, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitze K. Population-structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics. 1993;135:367–374. doi: 10.1093/genetics/135.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stålberg N. Lake Vättern: outlines of its natural history, especially its vegetation. Uppsala (Sweden): Acta Phytogeographica Suecica; 1939. [Google Scholar]
- Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM. Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. 2007;145:411–425. doi: 10.1104/pp.107.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci U S A. 2006;103:6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy costs constrain the evolution of gene expression. J Exp Zool B Mol Dev Evol. 2007;308B:322–324. doi: 10.1002/jez.b.21152. [DOI] [PubMed] [Google Scholar]
- Wayne ML, Pan YJ, Nuzhdin SV, McIntyre LM. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics. 2004;168:1413–1420. doi: 10.1534/genetics.104.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Telonis-Scott M, Bono LM, Harshman L, Kopp A, Nuzhdin SV, McIntyre LM. Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:18577–18582. doi: 10.1073/pnas.0705441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci U S A. 2006a;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Mol Ecol. 2006b;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- Whitlock MC. Evolutionary inference from QST. Mol Ecol. 2008;17:1885–1896. doi: 10.1111/j.1365-294X.2008.03712.x. [DOI] [PubMed] [Google Scholar]
- Wolak ME. nadiv: an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol Evol. 2012;3:792–796. [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wray GA. Genomics and the evolution of phenotypic traits. Annu Rev Ecol Evol Syst. 2013;44:51–72. [Google Scholar]
- Zhang X, Byrnes JK, Gal TS, Li WH, Borevitz JO. Whole genome transcriptome polymorphisms in Arabidopsis thaliana. Genome Biol. 2008;9:R165. doi: 10.1186/gb-2008-9-11-r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.