Abstract
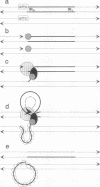
Insertion sequence 91 (IS91) inserts specifically at GTTC or CTTG target sequences without duplication of the target. After insertion, the right inverted repeat (IRR) lies adjacent to the 3' end of the target sequences (or 5' to the complementary sequence CAAG or GAAC). We have analyzed the effects of alteration of each terminus of IS91 on transposition activity in Escherichia coli. IRR is absolutely required for transposition. Deletion analysis indicates that a 14-bp segment is not sufficient, but an 81-bp sequence within the IRR region is sufficient. Furthermore, the GTTC/CTTG target site is also required. The left inverted repeat (IRL) of IS91 is dispensable. Plasmid fusions originated by one-ended transposition of IS91 derivatives lacking IRL occur at about the same frequency as cointegrate formation observed for the wild-type element. In the one-ended-type fusions, the inserted fragment of donor DNA is flanked at one end (constant end) by IRR and at the other end by a GTTC or CTTG sequence present in the donor (variable end) in a way that usually results in multiple tandem insertions of the donor plasmid in the target site. These results are easily accommodated by a rolling-circle replicative transposition mechanism. This model also draws support from the finding that the IS91 transposase is related in sequence to the superfamily of rolling-circle replication proteins and the observation that IRR shows some conservation in sequence and secondary structure with the origins of replication of some rolling-circle replication plasmids.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballester S., Lopez P., Espinosa M., Alonso J. C., Lacks S. A. Plasmid structural instability associated with pC194 replication functions. J Bacteriol. 1989 May;171(5):2271–2277. doi: 10.1128/jb.171.5.2271-2277.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé B., Jubete Y., Martínez E., de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991 Jun 15;102(1):75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- Bolland S., Llosa M., Avila P., de la Cruz F. General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J Bacteriol. 1990 Oct;172(10):5795–5802. doi: 10.1128/jb.172.10.5795-5802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Diaz-Aroca E., de la Cruz F., Zabala J. C., Ortiz J. M. Characterization of the new insertion sequence IS91 from an alpha-hemolysin plasmid of Escherichia coli. Mol Gen Genet. 1984;193(3):493–499. doi: 10.1007/BF00382089. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. On the molecular mechanisms of transposition. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4858–4862. doi: 10.1073/pnas.78.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A., Kokke M., van de Putte P. Transposition of mini-Mu containing only one of the ends of bacteriophage Mu. EMBO J. 1986 Dec 20;5(13):3687–3690. doi: 10.1002/j.1460-2075.1986.tb04700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina T. V., Koonin E. V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992 Jul 11;20(13):3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneluk R. G., Quan F., Gravel R. A. Rapid and reliable dideoxy sequencing of double-stranded DNA. Gene. 1985;40(2-3):317–323. doi: 10.1016/0378-1119(85)90055-1. [DOI] [PubMed] [Google Scholar]
- Martin C., Grinsted J., de la Cruz F. Effects of variation of inverted-repeat sequences on reactions mediated by the transposase of Tn21. J Bacteriol. 1989 Jul;171(7):3996–4001. doi: 10.1128/jb.171.7.3996-4001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola M. V., Jubete Y., de la Cruz F. DNA sequence of IS91 and identification of the transposase gene. J Bacteriol. 1992 Feb;174(4):1345–1351. doi: 10.1128/jb.174.4.1345-1351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola M. V., de la Cruz F. IS91 transposase is related to the rolling-circle-type replication proteins of the pUB110 family of plasmids. Nucleic Acids Res. 1992 Jul 11;20(13):3521–3521. doi: 10.1093/nar/20.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola M. V., de la Cruz F. Specificity of insertion of IS91, an insertion sequence present in alpha-haemolysin plasmids of Escherichia coli. Mol Microbiol. 1989 Jul;3(7):979–984. doi: 10.1111/j.1365-2958.1989.tb00247.x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Mötsch S., Schmitt R., Avila P., de la Cruz F., Ward E., Grinsted J. Junction sequences generated by 'one-ended transposition'. Nucleic Acids Res. 1985 May 10;13(9):3335–3342. doi: 10.1093/nar/13.9.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Romantschuk M., Richter G. Y., Mukhopadhyay P., Mills D. IS801, an insertion sequence element isolated from Pseudomonas syringae pathovar phaseolicola. Mol Microbiol. 1991 Mar;5(3):617–622. doi: 10.1111/j.1365-2958.1991.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Rose R. E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988 Jan 11;16(1):355–355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Moscoso M., Espinosa M. Rolling circle-replicating plasmids from gram-positive and gram-negative bacteria: a wall falls. Mol Microbiol. 1993 May;8(5):789–796. doi: 10.1111/j.1365-2958.1993.tb01625.x. [DOI] [PubMed] [Google Scholar]