Abstract
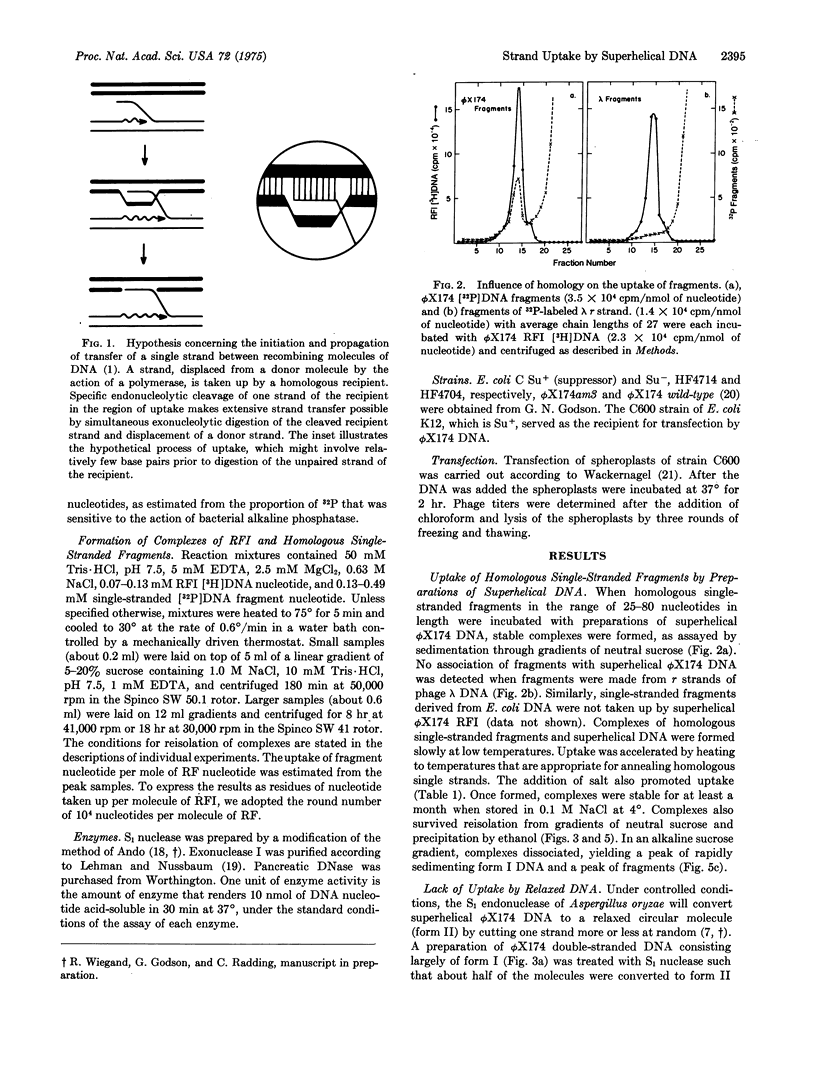
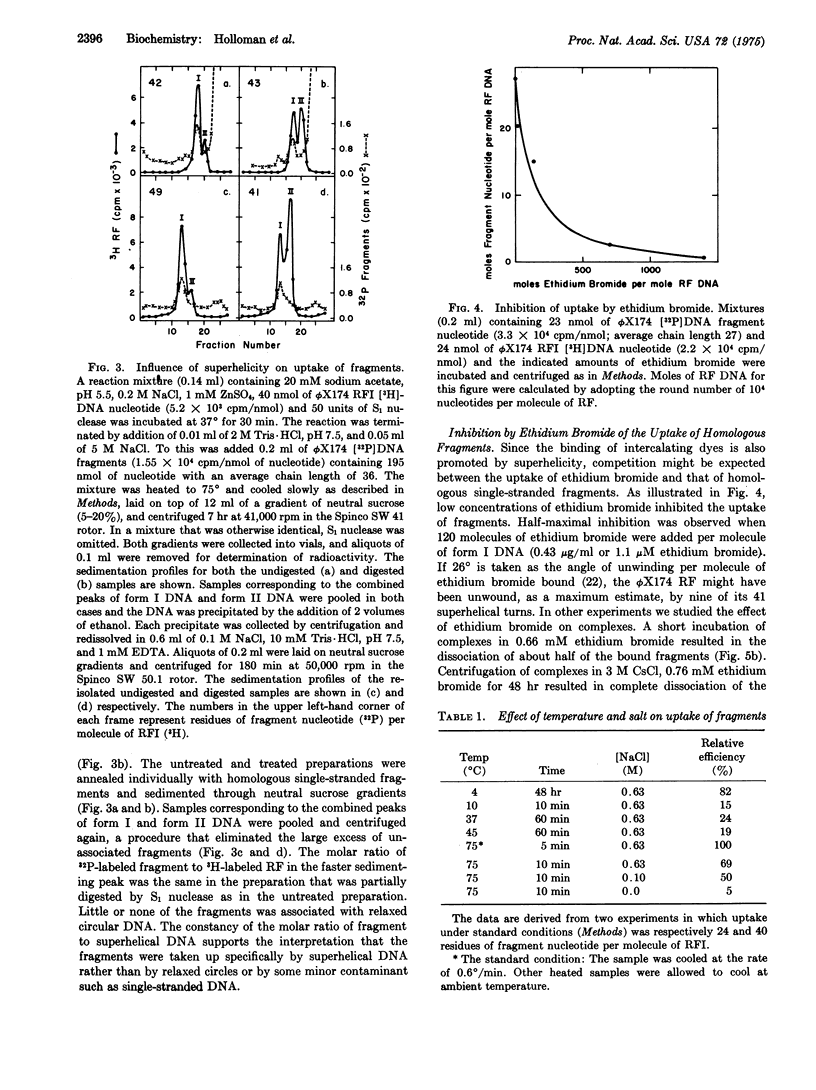
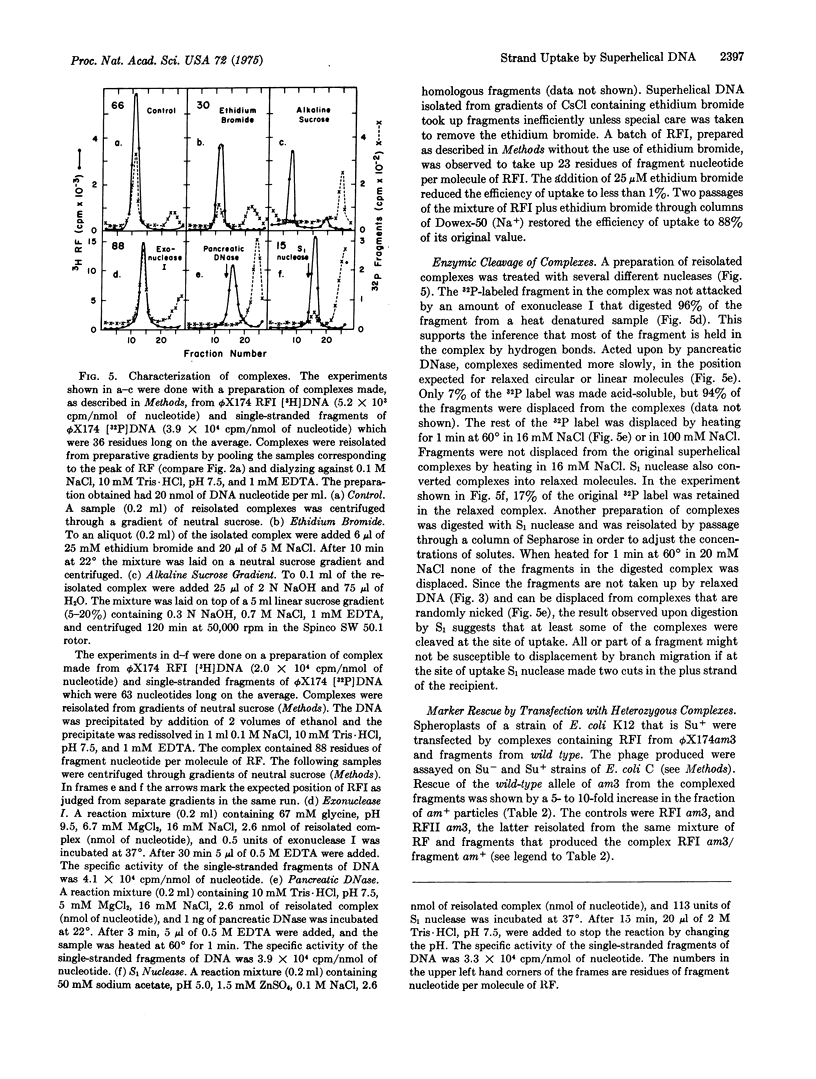
Superhelical [3-H]DNA (replicative form I, RFI) of bacteriophage phiX174 slowly but spontaneously took up 32-P-labeled homologous single-stranded fragments at 4 degrees. Uptake was accelerated by heating to 75 degrees. RFI did not take up single-stranded fragments derived from DNA of Escherichia coli or from separated strands of phage lambda. Uptake was inhibited by low concentrations of ethidium bromide. Relaxed circular phiX174 DNA did not take up homologous fragments. Per molecule of RFI, the complexes contained as much as 90 nucleotide residues of homologous fragment. The 32-P-lebeled fragments were largely resistant to digestion by exonuclease I, and were not displaced by heating complexes at 60 degrees for 1 min in 16 mM or 100 mM NaCl. Under comparable conditions of temperature and salt all of the fragments were displaced from complexes in which at least one phosphodiester bond was cleaved by pancreatic DNase, but a significant fraction of the fragments was retained in complexes that were relaxed by digestion with S1 nuclease. These observations are interpreted to mean that S1 nuclease digested the plus (viral) strand of the recipient RF at the site of uptake in some instances. Transfection of E. coli by heterozygous complexes produced recombinant progeny, thereby showing that genetic information can be transferred from the fragment of plus strand to progeny plus strands. We propose that both uptake of a third strand by superhelical DNA and the action of nucleases on the resulting complex may simulate early steps in genetic recombination.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim Biophys Acta. 1966 Jan 18;114(1):158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Asymmetric information transfer during phi X174 DNA replication. J Mol Biol. 1972 Feb 14;63(3):557–568. doi: 10.1016/0022-2836(72)90447-0. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman T. A., Lebowitz J. Further analysis of the altered secondary structure of superhelical DNA. Sensitivity to methylmercuric hydroxide a chemical probe for unpaired bases. J Mol Biol. 1973 Sep 25;79(3):451–470. doi: 10.1016/0022-2836(73)90398-7. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Hutchison C. A., Fabricant J. D., Sinsheimer R. L. Genetic Map of Bacteriophage phiX174. J Virol. 1971 May;7(5):549–558. doi: 10.1128/jvi.7.5.549-558.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow R. M., Zuccarelli A. J., Sinsheimer R. L. A role for single-strand breaks in bacteriophage phi-X174 genetic recombination. J Mol Biol. 1974 Sep 25;88(3):629–651. doi: 10.1016/0022-2836(74)90414-8. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Zuccarelli A. J., Sinsheimer R. L. Recombinant DNA molecules of bacteriophage phi chi174. Proc Natl Acad Sci U S A. 1975 Jan;72(1):235–239. doi: 10.1073/pnas.72.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAIRNS J. The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol. 1963 Mar;6:208–213. doi: 10.1016/s0022-2836(63)80070-4. [DOI] [PubMed] [Google Scholar]
- Carter D. M., Radding C. M. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971 Apr 25;246(8):2502–2512. [PubMed] [Google Scholar]
- Francke B., Ray D. S. Fate of parental phi X174-DNA upon infection of starved thymine-requiring host cells. Virology. 1971 Apr;44(1):168–187. doi: 10.1016/0042-6822(71)90163-2. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Boyer H. Susceptibility of the phiX-like phages G4 and G14 to R-EcoRi endonuclease. Virology. 1974 Nov;62(1):270–275. doi: 10.1016/0042-6822(74)90321-3. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Vapnek D. A simple method of preparing large amounts of phiX174 RF 1 supercoiled DNA. Biochim Biophys Acta. 1973 Apr 11;299(4):516–520. doi: 10.1016/0005-2787(73)90223-2. [DOI] [PubMed] [Google Scholar]
- Gschwender H. H., Haller W., Hofschneider P. H. Large-scale preparations of viruses by steric chromatography on columns of controlled pore glass. Phi-X174, M13, M12, Q-beta and T4 bacteriophages. Biochim Biophys Acta. 1969 Oct 22;190(2):460–469. doi: 10.1016/0005-2787(69)90095-1. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Vinograd J. Replication of circular DNA in eukaryotic cells. Annu Rev Biochem. 1974;43(0):695–719. doi: 10.1146/annurev.bi.43.070174.003403. [DOI] [PubMed] [Google Scholar]
- Kato A. C., Bartok K., Fraser M. J., Denhardt D. T. Sensitivity of superhelical DNA to a single-strand specific endonuclease. Biochim Biophys Acta. 1973 Apr 21;308(7):68–78. doi: 10.1016/0005-2787(73)90123-8. [DOI] [PubMed] [Google Scholar]
- LEHMAN I. R., NUSSBAUM A. L. THE DEOXYRIBONUCLEASES OF ESCHERICHIA COLI. V. ON THE SPECIFICITY OF EXONUCLEASE I (PHOSPHODIESTERASE). J Biol Chem. 1964 Aug;239:2628–2636. [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackernagel W. An improved spheroplast assay for lambda-DNA and the influence of the bacterial genotype on the transfection rate. Virology. 1972 Apr;48(1):94–103. doi: 10.1016/0042-6822(72)90117-1. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J Mol Biol. 1974 Aug 25;87(4):797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]



