Abstract
RNA sequencing measures the quantitative change in gene expression over the whole transcriptome, but it lacks spatial context. On the other hand, in situ hybridization provides the location of gene expression, but only for a small number of genes. Here we detail a protocol for genome-wide profiling of gene expression in situ in fixed cells and tissues, in which RNA is converted into cross-linked cDNA amplicons and sequenced manually on a confocal microscope. Unlike traditional RNA-seq our method enriches for context-specific transcripts over house-keeping and/or structural RNA, and it preserves the tissue architecture for RNA localization studies. Our protocol is written for researchers experienced in cell microscopy with minimal computing skills. Library construction and sequencing can be completed within 14 d, with image analysis requiring an additional 2 d.
Keywords: In situ, RNA-seq, gene expression, FISSEQ, RNA localization, sequencing-by-ligation
INTRODUCTION
Background
Cell type and function in tissues can be inferred from RNA or protein markers1, 2, but this approach to functional classification requires well-characterized biomarkers. Ideally, it would be preferable to define cell or tissue types using high-throughput molecular profiling in situ with high-resolution imaging. Indeed, several studies have surveyed global gene expression in situ, in which hundreds of organ tissue slices from multiple animals were individually interrogated using gene-specific probes3–6; however, such approaches represent a massive experimental undertaking and produce only an average view of tissue-specific gene expression.
In theory multiplexed in situ RNA detection demands fewer samples, but so far this approach is limited by the number of spectrally distinct fluorophores and the optical diffraction limit of microscopy7–11. Alternatively, padlock probes12–16 can capture specific RNA sequences from dozens of genes in parallel for targeted sequencing in situ12; however, padlock probes can have a significant amount of probe-specific bias17, and the approach cannot easily be scaled to the transcriptome. Given these challenges, in situ RNA profiling is typically restricted to a small number of well-annotated genes, and they can miss differences arising from unexpected signaling pathways or non-coding RNAs. In contrast we wanted to develop an unbiased and transcriptome-wide sampling method for quantitative visualization of RNA in situ, preferably using direct molecular sequencing18, 19 for detecting tissue-specific gene expression, RNA splicing and post-transcriptional modifications while preserving their spatial context.
Overview of the procedure
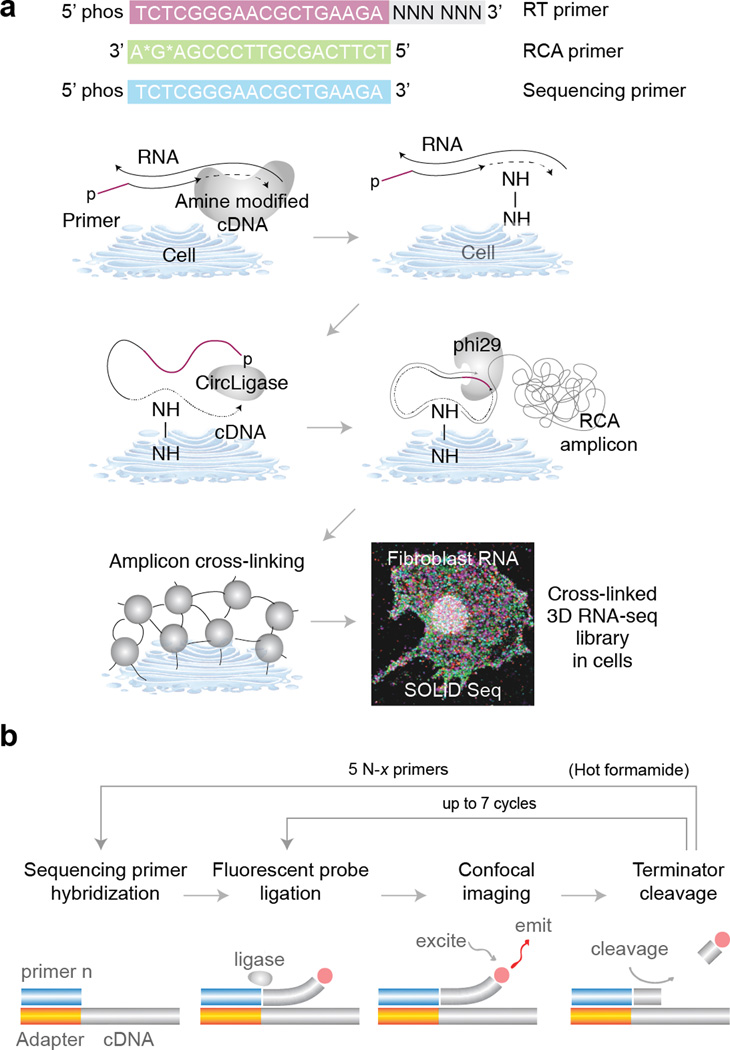
We begin by fixing cells on a glass slide and performing reverse transcription (RT) in situ. Following RT we degrade the residual RNA to prevent it from competitively inhibiting CircLigase, and cDNA fragments are circularized at 60°C. To prevent cDNA fragments from diffusing away, we incorporate primary amines in cDNA fragments during RT via aminoallyl dUTP, and we then cross-link the primary amines using BS(PEG)9. Each cDNA circle is linearly amplified using RCA into a single molecule containing multiple copies of the original cDNA sequence, and the amine-modified RCA amplicons are cross-linked to create a highly porous and three-dimensional nucleic acid matrix inside the cell (Fig. 1a).
Figure 1.
Schematic overview of FISSEQ library construction and sequencing. (a) Fixed cells or tissues are permeabilized and reverse transcribed in situ in the presence of aminoallyl dUTP and adapter sequence-tagged random hexamers. The cDNA fragments are fixed to the cellular protein matrix using a non-reversible amine cross-linker and circularized after degrading the RNA. The circular templates are amplified using RCA primers complementary to the adapter sequence in the presence of aminoallyl dUTP and stably cross-linked. The nucleic acid amplicons in cells are then ready for sequencing and imaging (fibroblast shown). (b) Each amplicon contains numerous tandem copies of the cDNA template and adapter sequence. A sequencing primer hybridizes to the adapter sequences in individual amplicons, and fluorescent eight base probes interrogate the adjacent dinucleotide pair. After imaging, the three bases attached to a fluorophore are cleaved, generating a phosphorylated 5’ end at the ligation complex suitable for additional ligation cycles interrogating every fifth dinucleotide pairs. The whole process is repeated using four other sequencing primers with an offset to interrogate intervening base positions.
In SOLiD sequencing-by-ligation, critical enzymatic steps can be performed directly on a standard microscope at room temperature. First, a sequencing primer is hybridized to multiple copies of the adapter sequence in RCA amplicons, followed by ligation of dinucleotide-specific fluorescent oligonucleotides. After imaging, the fluorophores are cleaved from the ligation complex, and ligation of fluorescent oligonucleotides is repeated six more times to interrogate dinucleotide pairs at every fifth position (Fig.1 b). To fill in the gaps between dinucleotide pairs, the whole ligation complex is stripped off, and four additional sequencing primers with a single base offset are used to repeat dinucleotide interrogation starting from positions n-1, n-2, n-3 and n-4, generating up to 35 raw 3D image stacks representing dinucleotide compositions at all base positions over time.
The raw images are enhanced using standard 3D deconvolution techniques to reduce the background noise, and our MATLAB script performs image alignment to produce TIFF images that are then used for base calling using a separate Python script. The base calls from individual pixels are then aligned to the reference transcriptome using Bowtie, and neighboring pixels with highly similar sequences are grouped into a single object generating a consensus sequence. The final dataset includes the number of individual pixels per object, gene ID, consensus sequence, x and y centroid positions, number of mismatches, base call quality and alignment quality.
One of the key considerations early in the development of FISSEQ was imaging. Biological patterns, including RNA localization, occur in a scale-dependent manner, in which some patterns are visible at one scale but disappear at another. Therefore, we developed our sequencing method specifically for confocal microscopy using a wide range of objectives, magnification, numerical apertures, scanning speed and depth. Also, autofluorescence, cell debris, and background noise are common in cell imaging, unlike in standard Next-generation sequencing. So we developed an approach to classify individual pixels based on their specific color transitions to detect true signals even in the noisy and/or low intensity environment. Finally, we also developed a way to control the imaging density of single molecules, enabling one to sequence the sequence a large number of molecules in single cells regardless of the microscopy resolution.
Comparisons to single cell RNA Seq
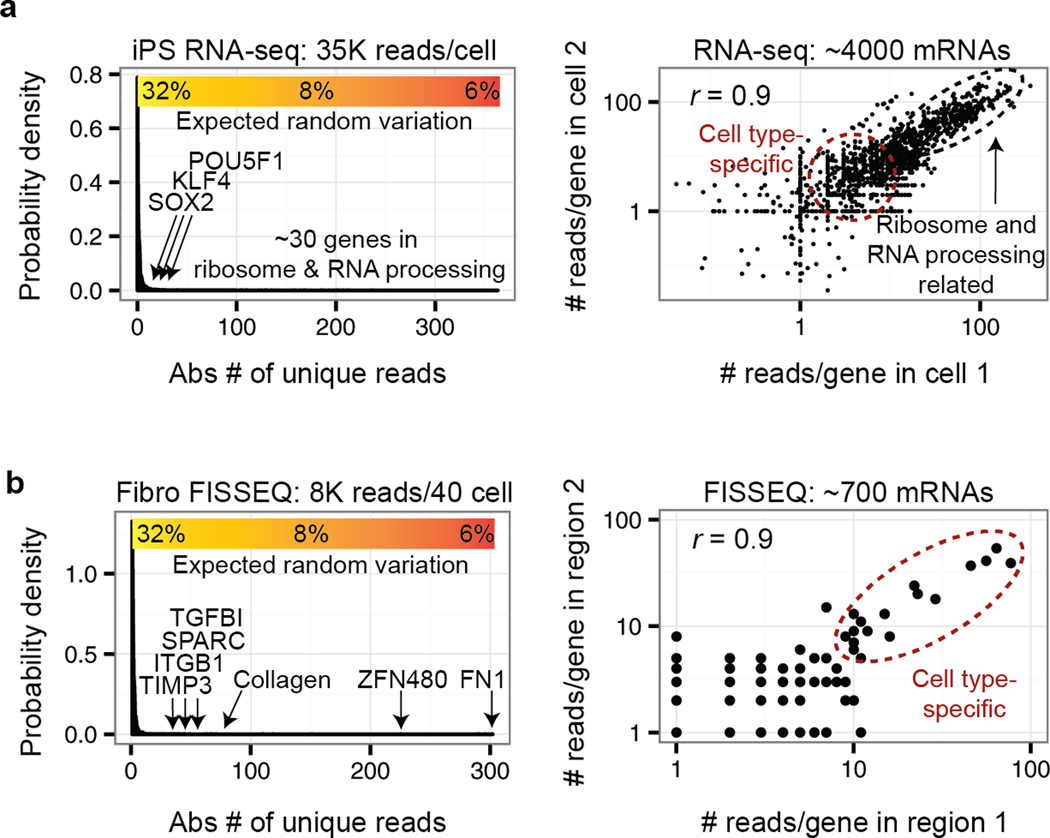
More than 1 million mRNA reads per cell can be obtained from a single cell RNA-seq experiment20, but typically <100,000 reads per cell are from unique cDNA fragments, and PCR amplification accounts for the remainder20–22. In one study the detection sensitivity of single-cell RNA-seq was estimated as ~10% or ~3% compared to single molecule FISH or spiked-in controls, respectively20. This means that only ~300 genes are expected to have a coefficient of variation (CV) of <23% based on Poisson distribution; however, such genes are generally uninformative and include many housekeeping genes such as ribosomal subunit proteins (Fig. 2a), requiring one to combine reads from multiple cells to detect biologically meaningful gene expression differences between groups of single cells.
Figure 2.
Comparing single cell RNA-seq to FISSEQ. (a) A typical single cell RNA-seq20 can generate >1 million reads per cell, but less than 10% represent unique reads from cDNAs, and they are comprised largely of structural and/or house-keeping genes (i.e. ribosome-related). Many genes of interest are found near the detection limit with a large coefficient of variation (CV), and the high correlation reported for single cell RNA-seq is typically due to house-keeping genes. (b) The current version of FISSEQ combines mRNA reads from ~40 cells to obtain a comparable result, but the high correlation between biological replicates in FISSEQ results from mostly cell type-specific expression markers.
In FISSEQ only ~200 mRNA reads per cell are obtained without ribosomal RNA depletion23 (vs. ~40,000 in single-cell RNA-seq); however, functionally important transcripts are enriched in FISSEQ by >10-fold compared to single-cell RNA-seq (Fig. 2b). When examining a single spatial region of ~40 cells (~8000 mRNA reads) the top ranked genes lie significantly above the detection threshold and form highly reproducible cell type-specific annotation clusters23. Because of the relative absence of housekeeping genes, the high correlation (Pearson’s r > 0.9) between biological replicates in FISSEQ are driven by cell type- and/or function-specific genes rather than housekeeping genes.
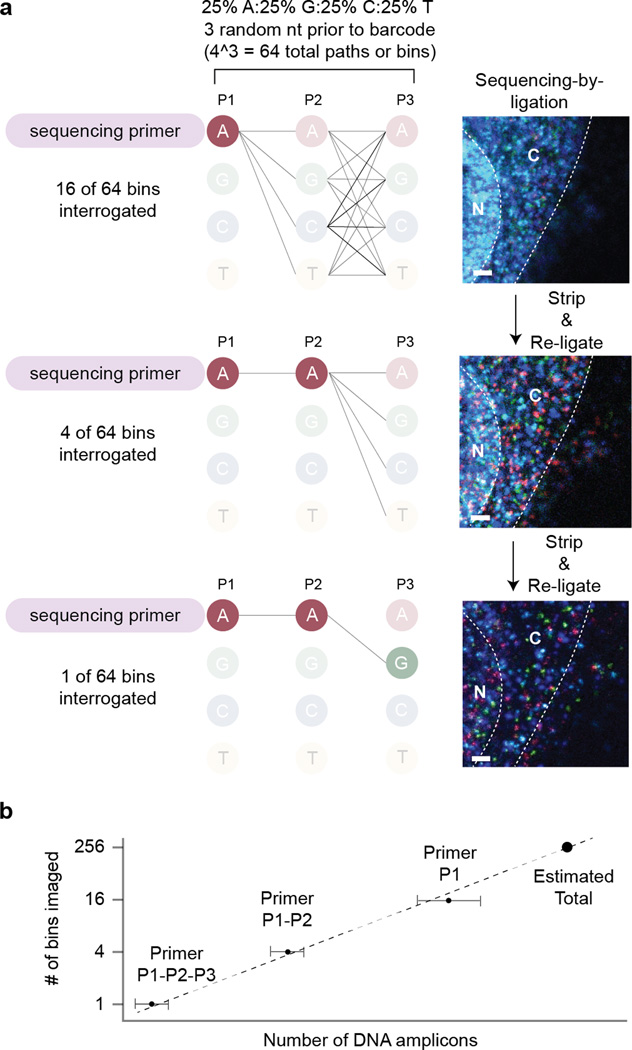
In order to attain truly single cell gene expression profiling that is biologically meaningful, FISSEQ may require a read depth per cell that is ~40-times deeper (~8,000 amplicons per cell). Since the ribosomal RNAs comprise >80% of the reads in FISSEQ23, it may be possible to increase the read depth by ~5-fold by simply depleting ribosomal RNA in situ24. We expect another ~5-fold increase in the amplicon density by optimizing our reaction condition, and a read depth of ~5,000 non-ribosomal RNA reads per cell may soon be possible. Since individual amplicons of any density can be discriminated using partition sequencing23 (Fig. 3), the actual size of each amplicon now becomes a limiting factor in the number of reads generated per cell.
Figure 3.
Counting resolution-limited amplicons using partition sequencing. (a) The cDNA or padlock probe template can include 3 random nucleotides in equal proportions. By controlling the length of the complementary portion of the sequencing primer to the random bases, one can ligate fluorescent probes to different amplicon pools of varying sizes (fibroblasts; bar: 1 µm). This scheme works for single-base sequencing-by-ligation, and the SOLiD sequencing chemistry requires additional modifications to the bridge oligonucleotide. (b) Serial ligation reactions using the sequencing primers with 0 to 3 complementary bases to the random partitioning bases are analogous to doing a serial dilution experiment. The average count from each primer category can be used to extrapolate and estimate the actual amplicon count, regardless of the limitations in optical microscopy.
Single-cell RNA-seq and FISSEQ are fundamentally limited by the efficiency of mRNA to cDNA conversion. In single-cell RNA-seq this is estimated to be ~10% compared to single molecule FISH20, with a detection threshold of ~5–10 mRNA molecules per cell21. This means that most low abundance genes are not detected in single-cell RNA-seq for a given cell. For FISSEQ this value is harder to determine because not all genes are enriched in the same manner, but we estimate the current detection threshold at ~200–400 mRNA molecules per cell. After ribosomal RNA depletion and other improvements, the detection threshold may improve to ~10–20 mRNA molecules per cell; however, a large fraction of low abundance genes will still remain undetected.
Comparisons with other approaches
Compared to microdissection25, 26 or photo-activated mRNA capture27-based single-cell RNA-seq21, 28–31, FISSEQ scales to large tissues more efficiently32, and it can compare multiple RNA localization patterns in a non-destructive manner23. Also, other methods require RNA isolation and PCR that can introduce a significant amount of technical variability20–22, assuming a Poisson distribution model of transcript abundance. In contrast, all samples can be processed together in a single well from cell culture to sequencing in FISSEQ.
Single molecule FISH remains a gold standard for high sensitivity detection of RNA in single cells7–9, 33–37; however, spectral discrimination of hybridized probes can be difficult to multiplex and require high resolution microscopy. Recently, highly scalable FISH was demonstrated in single cells, in which sequential hybridization is used to barcode a color sequence for each transcript10. In theory only seven hybridization cycles are required to interrogate 47 or >16,000 genes using four colors; however, this approach is limited by the sheer number of probes needed, and the optical diffraction limit prevents accurate quantification of highly abundant or aggregated transcripts.
The sensitivity of padlock probes is two orders of magnitude higher than FISSEQ for a given gene12, 13, but the use of locked nucleic acid (LNA) makes this approach prohibitively expensive for multiplexing, and individual probes must be calibrated for measuring the relative RNA abundance. For certain applications it may be possible to combine FISSEQ and padlock probes to interrogate a large number of loci in situ. In a recent study sequencing was limited to short barcodes from dozens of gene-specific padlock probes12, but now hundreds of thousands of padlock probes17, 38–41 can be discriminated using a 20-base barcode. In the same study the microscopy resolution limited the number of targeted genes12, but our partition sequencing23 bypasses such limitations for highly multiplexed amplicon discrimination in situ.
Limitations
On a practical level, equipping a microscope for four-color imaging can cost up to $20,000 for a new filter set and a laser. Most users will need to reserve the microscope for 2–3 weeks so that sequencing can proceed uninterrupted. We have used laser scanning confocal, wide-field epifluorescence and spinning disk confocal microscopes and obtained comparable sequencing data that differ mainly in the read density. With the laser scanning confocal microscope, imaging can take over 30 minutes per stack, but wide-field or spinning disk confocal microscopes can image the same volume in 1–2 minutes. Reagent exchanges are done manually in the current protocol, but FISSEQ samples can remain on the microscope and be sequenced over 2–3 weeks.
On a technical level, a major limitation of our current protocol is the lack of ribosomal RNA depletion. Initially we used ribosomal RNA as an internal control for library construction, sequencing and bioinformatics; however, this reduced the number of mRNA reads per cell. In primary fibroblasts the ribosomal RNA reads comprised 40–80% of the total23; therefore, if one were to deplete the ribosomal RNA24, it may be possible to increase the number of mRNA reads per cell by ~5 fold.
Another limitation is the lack of information on biases in our method. FISSEQ enriches for biologically active genes, enabling discrimination of cell type-specific processes with a small number of reads23; however, it is not clear how such enrichment occurs. We hypothesize that active RNA molecules are more accessible to FISSEQ, whereas RNA molecules involved in ribosome biogenesis, RNA splicing, or heat-shock response are trapped in ribonucleoproteins, spliceosomes, or stress granules. It is now important to investigate and understand the molecular basis of such enrichment across multiple cell types and conditions and correlate the result with the observed cellular phenotype.
Applications
The current FISSEQ protocol is suitable for most cultured cells and tissue sections, including formalin-fixed and paraffin-embedded (FFPE) tissue sections. Whole mount Drosophila embryos, iPS-cell derived embryo bodies (EBs) and organoids are also compatible (Table 1). In FISSEQ each sequencing read has a spatial coordinate, and the reads are binned according to the cellular morphology, sub-cellular location, protein localization, or GFP fluorescence. A statistical test is then applied to identify enriched genes and pathways de novo and discover possible biomarkers of the cellular phenotype23. This approach may be combined with padlock probes to detect evolving mutations and RNA biomarkers in cancers12, 13 or to compare gene expression in asymmetric cells or tissues.
Table 1.
A list of the specimens tested for FISSEQ library construction.
| Types | Fixation | Mounting substrate |
Permeabilization | Notes |
|---|---|---|---|---|
| HeLa, 293A, COS1, U2OS, iPSC, primary fibroblasts, bipolar neurons | 10% formalin or 4% PFA | Poly lysine- coated cover slip (Matrigel for iPSCs) | 70% EtOH or 0.25% Triton- X100 (0.1N HCl optional) | Changes in temperature can cause altered mRNA localization |
| Mouse embryo FFPE section (20-um) | Already fixed | Superfrost Plus glass slide | 0.1% Pepsin in 0.1N HCl | Use silicone isolators (Grace Bio-Labs) |
| Mouse brain fresh frozen section (20-um) | 10% formalin | Poly lysine- coated cover slip | 0.1% Pepsin in 0.1N HCl | Use silicone isolators (Grace Bio-Labs) |
| iPS-derived 3D organoids | 10% formalin | Poly lysine- coated cover slip (embed in Matrigel & fix with 4% PFA) | 0.25% Triton- X100 & 0.1N HCl | 10% formalin less effective for fixing Matrigel |
| Dechorionated whole mount Drosophila embryos | 10% formalin | Poly lysine- coated cover slip (embed in Matrigel & fix with 4% PFA) | 100% MeOH, then PBS with 0.2% Triton- X100 & 0.2% Tween-20 | 10% formalin less effective for fixing Matrigel |
FISSEQ may also sequence molecular barcodes in individual cells and transcripts, where expression or reporter (i.e. cDNA, promoter-GFP) libraries are examined in a pool of single cells for massively parallel functional assays and cell lineage tracing. In essence a practically unlimited number of DNA-associated cellular features may now be imaged, enumerated and analyzed across multiple spatial scales using the DNA sequence as a temporal barcode.
Experimental design
General considerations
This protocol details the method described in our original report23, where endogenous RNAs in cultured fibroblasts were sequenced on a confocal microscope. The availability of a microscope and computational resources will guide the general experimental approach (Table 2). We provide basic computational tools along with a sample dataset, but a background in python, MATLAB, ImageJ and/or R is helpful for analyzing a large number of images. If such expertise is not available, we recommend focusing on a few regions-of-interest with well-demarcated features for comparing gene expression using our custom scripts23. After outlining the experiment, one should download our sample image, software and dataset and become familiar with image and data analysis (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/). One should then finalize the experimental design and define the imaging parameters (i.e. area, thickness, resolution, magnification).
Table 2.
A comparison of the microscopy platforms tested for FISSEQ.
| Model | Pros | Cons | Uses | |
|---|---|---|---|---|
| Wide-field epifluorescence | Nikon TE-2000 | Fast imaging Simple set-up | Poor axial resolution Low S/N ratio Lower read depth | Thin cells & tissue sections Whole cell barcode labeling |
| Scanning confocal | Zeiss LSM 710 confocal Leica TCS SP5 Confocal | Good axial resolution Scanning zoom Flexible pixel density | Slow imaging | High resolution FISSEQ of a single region |
| Spinning disk confocal | Yokogawa CSU-W1 | Fast imaging Good axial resolution | Fixed pixel density | All-purpose |
Cell and tissue fixation
We have been able to fix and generate in situ sequencing libraries in a wide number of biological specimens (Table 1). The only case in which we failed was a hard piece of bone marrow embedded in Matrigel, which detached from the glass surface after several wash steps. Fixation artifacts can include changes in sub-cellular RNA localization, cell swelling, incomplete permeabilization and RNA leakage. Certain primary cell types are also sensitive to cold42, while transformed cell lines or stem cells appear to be less sensitive (Fig. S1). If using FISSEQ to study sub-cellular localization, we recommend fixing cells by adding warm formalin directly into the growth media to a final concentration of 10%.
Cell and tissue sample mounting
For high resolution imaging we recommend poly lysine- or Matrigel-coated glass bottom dishes, but 96-well plastic bottom plates can be used for simple protocol optimization. Tissue sections can be mounted using a standard mounting procedure, and we advise inexperienced users to consult those practiced in the art of tissue mounting. For non-adherent cell types and whole mount specimens, we recommend fixing samples embedded in Matrigel using 4% paraformaldehyde on a glass bottom dish.
Reverse transcription (RT) in situ
The length of RT primers should be less than 25 bases to prevent self-circularization. We perform RT overnight for most samples, but one hour is often sufficient for cell monolayers. A negative control without RT should be included to rule out self-circularization of the primer. A positive control primer with the adapter sequence plus a synthetic sequence (~30 additional bases) can be used to check rolling circle amplification (RCA) and imaging parameters. Other than the 5’ region of highly abundant mCherry transcripts23, we have not had consistent results with targeted RT. We typically see very few amplicons regardless of the primer design, while random hexamers (24 bases) and poly dT primers (33 bases) work well across all conditions. Some of the possible reasons for failure may include poor target accessibility and competitive inhibition of CircLigase by non-specifically bound sequence-specific RT primers capable of self-circularization. Possible solutions include using LNA-based RT primers for high temperature hybridization13, ligation of the adapter sequence post-RT and tiling multiple RT probes across a gene target. We have yet to try these alternatives.
Generation of amplicon matrix
Aminoallyl dUTP is a dTTP analog commonly used in fluorescence labeling of cDNA43, which we utilize for cross-linking nucleic acids; however, the efficiency of RT and RCA is inversely correlated with the concentration of aminoallyl dUTP23. The cross-linker, bis(succinimidyl)-nona-(ethylene glycol) or BS(PEG)9, is functionalized with NHS ester groups at both ends44, and it forms a stable covalent bond with primary amine groups provided by aminoallyl dUTP at pH 7–9. The cross-linking density can be enhanced by increasing the concentration of aminoallyl dUTP or BS(PEG)9, or by increasing the pH. Cross-linking after RT is optional, but cross-linking of RCA amplicons is essential for high quality sequencing reads.
Sequencing
We use sequencing-by-ligation18, 19, 45 (SOLiD46, 47) because it works well at room temperature and so a heated stage is not required. SOLiD uses a dinucleotide detection scheme where a base position is interrogated twice per sequencing run46, 47, and this can reduce the base calling error rate; however, converting the color sequence to the base sequence is not straightforward due to its propensity to propagate errors, and sequence analysis must remain in the color space (Box 1). In comparison, sequencing-by-synthesis (Illumina) works at 65°C for primer extension and cleavage and utilizes proprietary fluorophores, requiring a heated flow-cell and a custom imaging set-up. Since sequencing-by-synthesis can generally yield a much longer read length, we are currently investigating its compatibility with FISSEQ.
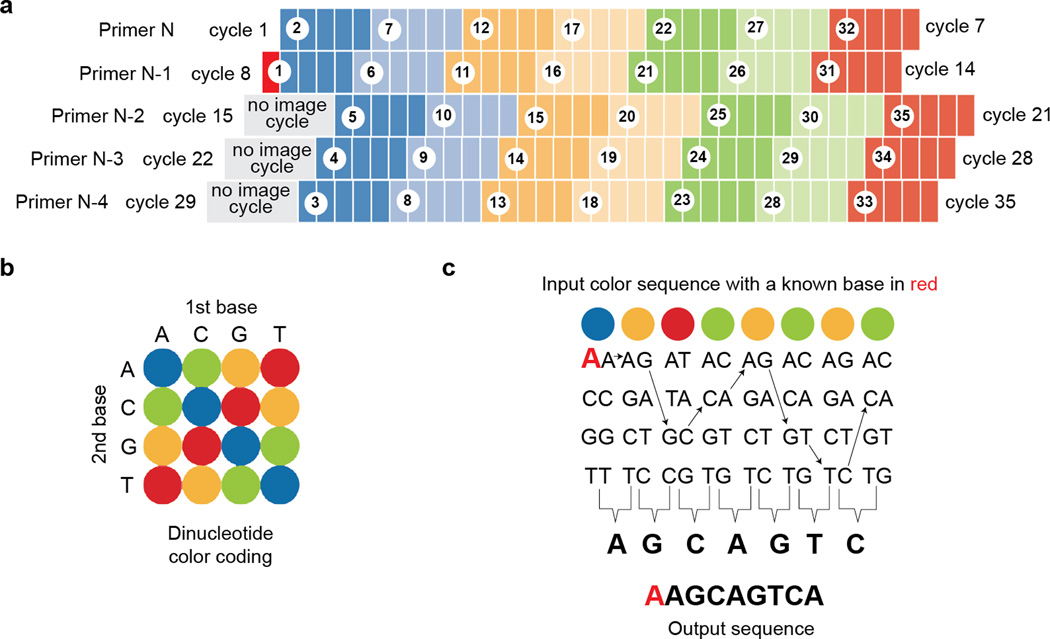
Box 1| SOLiD sequencing chemistry.
The SOLiD sequencing chemistry consists of multiple reaction cycles in which a sequencing primer is extended using fluorescent eight-base probes via sequential DNA ligation. The fluorescent amplicons are then imaged, and the last three bases and the fluorophore are cleaved, followed by the ligation of another eight-base probes. These steps are repeated using 4 additional sliding primers to record the dinucleotide color values from starting positions 1-6-11-16-20-26-31 (Primer N), 0-5-10-15-20-25-30 (Primer N-1), 4-9-14-19-24-29-34 (Primer N-2), 3-8-13-18-23-28-33 (Primer N-3) and 2-7-12-16-22-27-32 (Primer N-4) (Fig. 4a). Most bases are represented by two sequential colors, and although each color represents up to four possible dinucleotide combinations, the exact nucleotide sequence can be determined if the identity of any one base is known (i.e. the base identity in the sequencing primer). For example AAGCAGTCA is equivalent to BORGOGOG (B: blue, O: orange, R: red, G: green) (Fig. 4b); however, the conversion table alone cannot assign the base identity from color codes. But if one base is known (i.e. 1st base is A in BORGOGOG), assigning the base identity is relatively straightforward (Fig. 4c). One disadvantage is that any missing or wrong base calls can affect the whole read, and it makes sequence-to-sequence comparisons impossible. Therefore, the SOLiD sequencing reads and the reference database must remain in the color space for sequence alignment, and the user should keep this in mind when designing a custom sequence analysis pipeline.
Partition sequencing
T4 DNA ligase has a single-base specificity at the ligation junction18, and sequencing primers differing by one base can recognize different sets of amplicons23. By dividing imaging over multiple separate runs, spatially overlapping amplicons can be enumerated using multiple sequencing primers even on a low resolution microscope; however, this requires full automation for the increased number of sequencing runs per sample. Without automation partition sequencing is better suited for quantifying short barcode sequences rather than full RNA sequences in situ12 (Fig. 3).
Imaging
Epifluorescence microscopy can generate a reasonable number of alignable reads from relatively thin specimens (<5-um), such as HeLa cells23, but thicker samples require confocal microscopy to obtain high density reads. Spinning disk confocal microscopy is significantly faster than laser scanning confocal microscopy, and it has a good balance of imaging speed and axial resolution. An automated stage capable of finding a z-stack across multiple x-y tiles is highly desirable (Table 2).
In FISSEQ individual amplicons can be detected using objectives with N.A. 0.4 or greater. The magnification required is determined by the biological question and the amplicon density48. Typically we use a 20× N.A. 0.75 objective to examine tissue sections and cultured cell mono-layers, while 40× N.A. 0.8 and 63× N.A. 1.2 water immersion objectives are used for high-resolution imaging of single cells. We have observed noticeable chromatic aberration in our experiments, depending on the objectives used. The degree of chromatic aberration should be measured using image calibration beads (i.e. FocalCheck Fluorescence Microscope Test Slide) prior to sequencing and calibrated by the microscope vendor if necessary.
For each imaging set-up the user should determine the ideal Nyquist rate. This value can be calculated using http://www.svi.nl/NyquistCalculator. The x-y pixel and z-step sizes should not be greater than 1.7 times the Nyquist value for image deconvolution. Four color imaging should proceed from the longest to shortest wavelength (i.e. Cy5, Texas Red, Cy3, FAM), and an intensity histogram should be used to adjust laser power to prevent saturated pixels. The intensity histogram should be consistent across fluorescence channels and sequencing cycles. To use our software the image file name must be standardized: <Position>_<Primer #>_ <Ligation #>_<Date_Time>.extension (e.g. 06_N1_2_2013_10_25_11_57_18.czi).
Image analysis tools
In practice the extent of image processing and analysis is dictated by the available imaging tools and computing resources49. We use Bitplane Imaris for data visualization and movie creation and SVI Huygens for 3D deconvolution. While they are easy to use, scalable and relatively fast, their cost may be out of reach for small labs; however, free and/or open source alternatives are also available49–51.
Image deconvolution
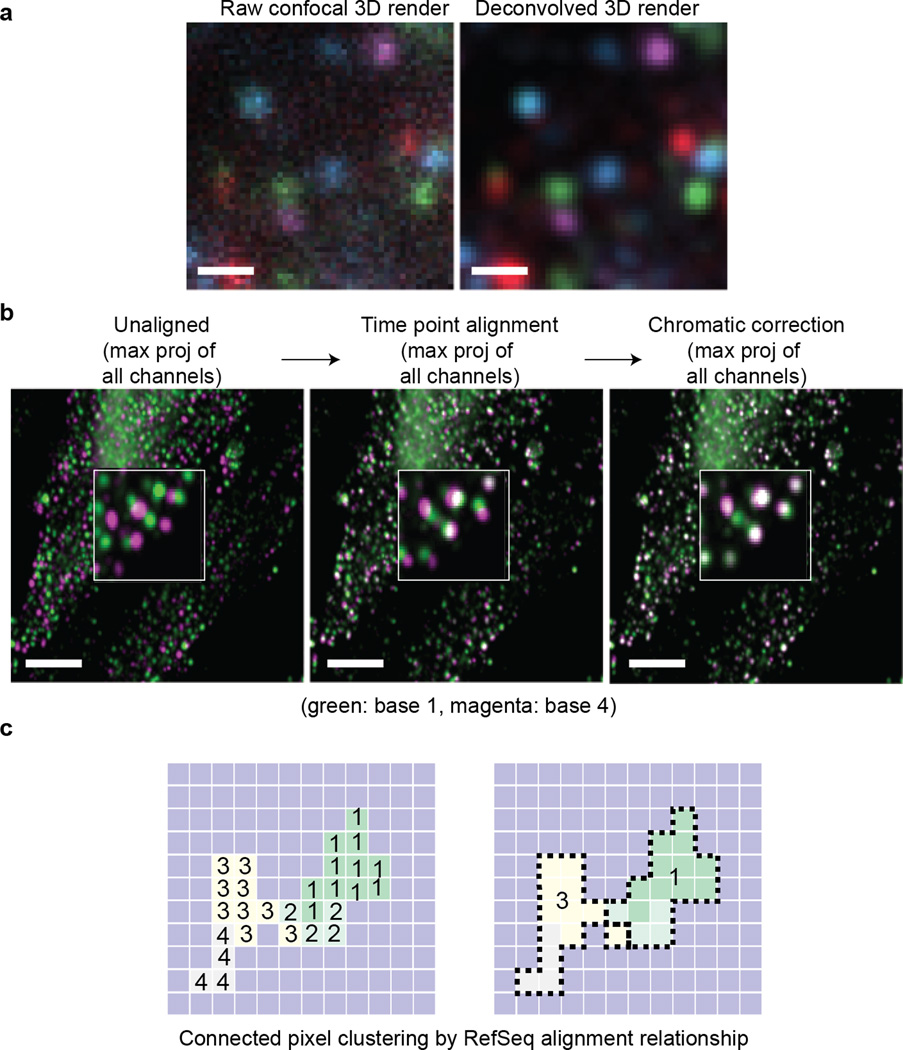
We use 3D deconvolution52 to reduce the out-of-focus background and improve the quality of base calls (Fig. 5a). High quality 3D deconvolution requires sampling near the Nyquist rate, but this increases the image acquisition and deconvolution time as well as the file size. We generally recommend using high-quality confocal imaging and minimal 3D deconvolution for FISSEQ. Using 3D deconvolution to compensate for low quality imaging will not necessarily improve the quality or number of sequencing reads. We provide a sample dataset containing raw and deconvolved image stacks from a successful 30-base sequencing experiment for practice (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/).
Figure 5.
Example of image analysis, registration and sequence clustering. (a) A four color confocal image stack is deconvolved using a CLME algorithm with 10 iterations and a signal-to-noise ratio of two (bar: 1 µm). (b) Sequencing images for base 1 and base 4 (left) are aligned using a composite channel across various time points (middle) and then using a composite time projection across various channels (right) (bar: 5 µm). (c) Individual non-zero pixels are aligned to the reference sequence database (i.e. human RefSeq). Highly related sequences connected to the neighboring pixels are then grouped into a single cluster.
Image registration, base calling and sequence alignment
As long as the input image files are correctly named, our software will generate the maximum intensity projection, register images and correct for chromatic shifts23 (Fig. 5b). The resulting images are used for base calling and sequence alignment to human RefSeq (Fig. 5c), but our software does not generate z-coordinates for sequencing reads, as it uses maximum intensity projection for base calling. We provide a sample data output and screen logs for troubleshooting our bioinformatics software (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/).
Data analysis
Our software generates a tab delimited text file containing 10,000 to 50,000 aligned reads per field of view. We recommend RStudio with the latest version of R installed for plotting reads by RNA classes, position, cluster size, quality, gene name, strand, etc. We provide a sample R session file used for FISSEQ data analysis as an introduction to statistical computing and assessing the quality of FISSEQ dataset (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/).
Level of expertise required for the protocol
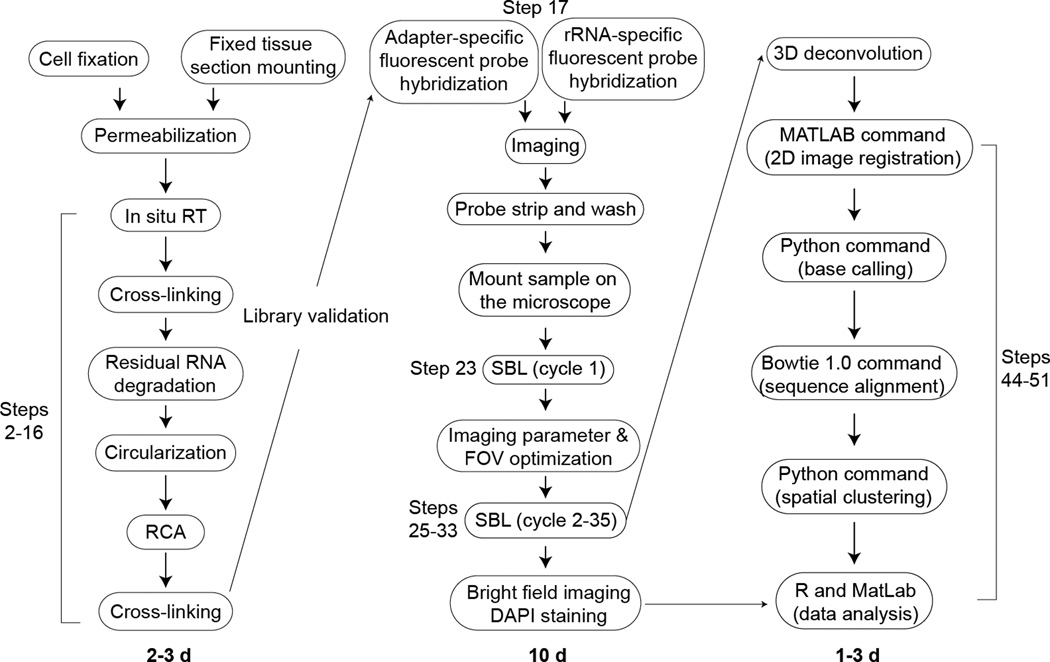
FISSEQ is at the interaction of cell imaging and functional genomics, and it has generated much interest from cell biologists not familiar with RNA-seq. Our protocol is aimed at such researchers, who are familiar with cell image analysis but have few computing skills (Fig. 6). FISSEQ library construction can be performed by anyone with basic molecular biology skills, but image acquisition is best done with help from an imaging core specialist for the initial set-up. Once the equipment, software, imaging and deconvolution parameters are finalized, a capable technician, graduate student, or post-doc can perform manual sequencing on a microscope with some training and practice. Image and sequence analysis using our software can be performed by anyone familiar with the Unix environment, but statistical data analysis requires either a graduate student or post-doc familiar with statistical tools and concepts.
Figure 6.
Schematic overview of FISSEQ experimental and analysis steps.
Considerations about the laboratory facilities
All steps in FISSEQ library construction can be carried out in a standard laboratory setting. A vacuum line facilitates solution aspiration and reagent exchanges, and we do not find RNA degradation or PCR contamination to be a significant problem in our method. We advise having a dedicated microscope with proper excitation and emission filters on a vibration isolation table in a low traffic area.
MATERIALS
REAGENTS
Starting material of interest. The Procedure is written for cultured cells in glass bottom dishes or for tissue sections on glass bottom dishes or cover slips. However, it can be adapted for use on a range of starting materials (see Table 1).
-
Acetone (for detaching a Petri dish glued to the microscope stage)
CAUTION Highly flammable. Work in a well-ventilated area.
Aminoallyl dUTP, 4 mM (Anaspec, part no. 83203)
Betaine, 5 M (included in CircLigase II kit, Epicentre, part no. CL9025K)
-
BS(PEG)9, 100 mg (Thermo Scientific, part no. 21582)
CRITICAL STEP BS(PEG)9 loses its effectiveness 1 month after reconstitution in DMSO. Prepare a fresh batch every month, especially if it has been frozen and thawed repeatedly.
CircLigase II kit (Epicentre, part no. CL9025K)
Cleave Solution 1 (Applied Biosystems, part no. 4406489)
-
Cleave Solution 2.1 Kit (Applied Biosystems, part no. 4445677)
CAUTION Contains toxic organoamine. Wear gloves and work in a well-ventilated area.
Cyanoacrylate adhesive, optical grade (VWR, part no. 19806-00-1)
DEPC-treated water (Santa Cruz Technologies, part no. sc-204391)
DMSO (Sigma, part no. D8418)
dNTP, 25 mM (Enzymatics, part no. N2050L)
Ethanol, 70% (in DEPC water)
-
Formalin, 10% (Electron Microscopy Science, part no. 15740)
CAUTION Wear gloves and work in a well-ventilated area. Dispose of waste per institutional guideline.
Formamide (Sigma, part no. 221198)
HCl, 0.1 N (in DEPC water)
Immersol W 2010 (ne=1.33) for water immersion lens (Zeiss, part no. 444969-0000-000)
Instrument Buffer, 10X (Applied Biosystems, part no. 4389784)
M-MuLV reverse transcriptase (Enzymatics, part no. P7040L)
Mineral oil (Sigma, M5904)
MnCl2 (included in CircLigase II kit, Epicentre, part no. CL9025K)
Nuclease-free water, not DEPC-treated (Life Technologies, part no. AM9932)
Pepsin, 1 g (dissolve in 10 ml H2O and store at −20°C; Affymetrix, part no. 20010)
Phi29 DNA polymerase (Enzymatics, part no. P7020-HC-L)
Phosphate-buffered solution (Life Technologies, part no. 10010023)
RNase, DNase-free (Roche Applied Science, part no. 11579681001)
RNase H (Enzymatics, part no. Y9220F)
RNase inhibitor (Enzymatics, part no. Y9240L)
RNaseZap (Life Technologies, part no. AM9780)
Silicone isolator (Grace Bio-Labs, part no. 664304)
Sodium acetate, 3 M (pH 7.5)
SOLiD™ ToP Sequencing Kit Fragment Library F3 Tag MM50 (Applied Biosystems, part no. 4449388)
SSC, 20X (Roche Applied Science, part no. 11666681001)
Streptavidin Alexa Fluor 647 (Life Technologies, part no. S32357)
T4 DNA ligase (Enzymatics, part no. L6030-LC-L)
Tris solution, 1 M (G Biosciences, part no. R002)
Trisodium citrate dihydrate (Sigma, part no. C8532)
Triton X-100, 10% solution (Sigma, part no. 93443)
RT, RCA and SEQUENCING PRIMERS
Random hexamer RT primer, 100 uM in nuclease-free H2O (/5phos/TCTCGGGAACGCTGAAGANNNNNN; hand-mixed, IDT)
RCA primer, 100 uM (TCTTCAGCGTTCCCGA*G*A; * is phosphothioate)
Sequencing primer N : /5phos/TCTCGGGAACGCTGAAGA (HPLC-purified)
Sequencing primer N-1 : /5phos/CTCGGGAACGCTGAAGA (HPLC-purified)
Sequencing primer N-2 : /5phos/TCGGGAACGCTGAAGA (HPLC-purified)
Sequencing primer N-3 : /5phos/CGGGAACGCTGAAGA (HPLC-purified)
Sequencing primer N-4 : /5phos/GGGAACGCTGAAGA (HPLC-purified)
CONTROL PRIMERS
Adapter-specific probes, 100 uM (/56-FAM/TCTCGGGAACGCTGAAGA)
Adapter-specific probes, 100 uM (/5TYE563/TCTCGGGAACGCTGAAGA)
Adapter-specific probes, 100 uM (/5TEX615/TCTCGGGAACGCTGAAGA)
Adapter-specific probes, 100 uM (/5TYE665TCTCGGGAACGCTGAAGA)
18S rRNA detection primer 1: /5biotin/GCTACTGGCAGGATCAACCAGGTA
18S rRNA detection primer2: /5biotin/TACGCTATTGGAGCTGGAATTACC
18S rRNA detection primer3: /5biotin/GTTGAGTCAAATTAAGCCGCAGGC
18S rRNA detection primer4: /5biotin/TTGCAATCCCCGATCCCCATCACG
28S rRNA detection primer1: /5biotin/CCACGTCTGATCTGAGGTCGCG
28S rRNA detection primer2: /5biotin/CACGCCCTCTTGAACTCTCTCTTC
28S rRNA detection primer3: /5biotin/CTCCACCAGAGTTTCCTCTGGCT
28S rRNA detection primer4: /5biotin/TGAGTTGTTACACACTCCTTAGCG
28S rRNA detection primer5: /5biotin/CGACCCAGCCCTTAGAGCCAATC
28S rRNA detection primer6: /5biotin/GACAGTGGGAATCTCGTTCATCCA
28S rRNA detection primer7: /5biotin/GCACATACACCAAATGTCTGAACC
EQUIPMENT
4°C and −20°C storage units
Centrifuge for 1.5- and 2-ml tubes
Dry block heater for microtubes at 80°C
Falcon conical centrifuge tubes (15 ml and 50 ml) (Fisher Scientific, cat. nos. 14-959-49B and 14-432-22)
-
Flexible plastic I.V. catheter for reagent aspiration (Terumo, part no. SR*FF2419)
CAUTION The catheter comes with a plastic outer sheath and a sharp needle in the middle. The needle must be carefully removed and discarded into a sharps container.
FocalCheck Fluorescence Microscope Test Slide (Life Technologies, part no. F36909)
Glass bottom Mattek dish (Poly lysine-treated: part no. P35GC-1.5-14-C, Poly lysine-treated 96-well plate: part no. P96GC-1.5-5-F)
Glass Pasteur pipettes (autoclaved)
Incubators at 30°C, 37°C (humidified) and 60°C
Inverted confocal microscope, PC and image acquisition software
Microscope stage insert, metal (for securely gluing the specimen holder)
Non-sterile syringes, 10 ml (BD Biosciences, part no. 301029)
RNase-free microtubes (Eppendorf, part no. 0030 121.589)
Sealable plastic Tupperware container or Ziploc bags (for CircLigase reaction at 60°C)
Vacuum flask, trap and tubing
REAGENT SETUP
0.25% Triton X-100 Dilute 0.25 ml 10% Triton X-100 in DEPC-treated H2O to a total volume of 10 ml. Store at RT for 6 months.
2X SSC Dilute 20X SSC in H2O and make up to a total volume of 50 mL. Store at RT for 6 months.
1X SSC Dilute 20X SSC in H2O and make up to a total volume of 50 mL. Store at RT for 6 months.
5X SASC Make 0.75 M sodium acetate, 75 mM tri-sodium citrate, pH to 7.5 using acetic acid in H2O to a final volume of 50 ml. Store at RT for 6 months.
RCA primer hybridization buffer Dilute 20X SSC2x SASC, 30% formamide in H2O. Store at RT for 6 months.
Strip Buffer (80% formamide in H20, 0.01% Triton-X100 in a final volume of 50 ml. Store at RT for 6 months.
Cleave Solution 2.1, reconstituted Mix 1 ml Cleave Solution 2.1 Part 1 with 2.75 ml Cleave Solution 2.1 Part 2. Store at 4°C in dark for up to 24 hours.
EQUIPMENT SETUP
Microscope setup
Configure a 4 channel microscope with appropriate excitation light sources and emission filters: FITC-488 ex, 490–560 nm em; Cy3–561 nm ex, 563–593 nm em; Texas Red-594 nm ex, 597–647 nm em; Cy5–633 nm ex, 637–758 nm em. Suggested microscope objectives are Plan-Apochromat dry 20× NA 0.75, dry 40× NA 0.8 and water immersion 63× NA 1.3.
Software installation
Verify that Bio-Formats (http://loci.wisc.edu/software/bio-formats) plug-ins are available for Fiji/ImageJ. Download a free academic version of Canopy Python 2.7 (https://www.enthought.com/downloads) in the home directory on the remote host, and follow the installation instruction (http://docs.enthought.com/canopy/quick-start/install_linux.html). Canopy Python 2.7 is easy to install and has all the required packages for our FISSEQ software. Install the latest version of ggplot2 and data.table packages in RStudio.
PC setup
Access to a high performance computing cluster (remote host)
-
Bowtie 1.0 or earlier (http://bowtie-bio.sourceforge.net) on the remote host
CRITCAL Bowtie 2.0 or higher does not work with SOLiD sequencing.
Fiji/ImageJ (http://fiji.sc/Fiji) on a PC
MATLAB (http://www.mathworks.com) on the remote host
-
Python 2.7 (https://www.enthought.com/products/canopy/) on the remote host
CRITCAL Other versions of Python lack the required modules for running our script.
R (http://www.r-project.org) and RStudio (http://www.rstudio.com) on a PC
Windows PC or Mac with 16GB RAM minimum
Optional: SVI Huygens 3D deconvolution software (commercial), Bitplane Imaris 3D rendering software (commercial)
PROCEDURE
FISSEQ library construction in cultured cells or tissue sections •TIMING 2–3 d
-
1|This step can be performed using option A or B depending on the type of a specimen examined. All reagents and washes are at RT unless indicated otherwise.
- Cultured adherent cells on a glass bottom dish.
- Fix cells using 2 ml of 10% formalin in PBS for 15 min at 25°C.
- Wash with 2 ml PBS three times.
- Add 2 ml of 0.25% Triton-X100 in DEPC-PBS for 10 min, or 70% ethanol for 2 min. Triton-X100 tends to maintain the subcellular structures better than 70% ethanol.
- Wash with 2 ml of PBS three times.
- Some cell types may require acid-treatment for improved permeabilization: add 0.1 N HCl in DEPC-treated H2O for 10 min, followed by three PBS washes (Fig. S2).
TROUBLESHOOTING
-
Tissue sections on a glass bottom dish.
- Mount 10–20 µm thick formalin-fixed tissue sections onto an RNase-free cover glass slip using a standard mounting procedure.
- Remove the glass cover slip attached to a Mattek glass bottom dish by gently pressing around the cover slip with a razor blade.
- Attach the glass cover slip with a mounted tissue section to the Mattek dish using double-sided adhesive tape.
- Wash twice using DEPC-treated H2O for 5 min each.
- Add 0.25% Triton X-100 in DEPC-treated H2O for 15 min and aspirate.
- Wash with DEPC-treated H2O twice.
- Add 200 µl 0.1% pepsin in 0.1 N HCl for up to 10–30 min. Most tissue sections are permeabilized after 10–15 minutes. We recommend optimizing the permeabilization conditions for each tissue type.
- Wash with 2 ml PBS three times to inactivate pepsin.
TROUBLESHOOTING
-
2|
Prepare a reverse transcription mixture on ice, as indicated below.
CRITICAL STEP Chilling the assembled mix to 4°C prior to RT improves the efficiency of primer annealing.Component Amount (µl) Final DEPC-H2O 159 M-MuLV RT buffer, 10× 20 1× dNTP, 25 mM 2 250 µM Aminoallyl dUTP, 4 mM 2 40 µM RT primer, 100 µM (/5Phos/TCTCGGGAACGCTGAAGANNNNNN) 5 2.5 µM RNase Inhibitor (40 U µl−1) 2 0.4 U µl−1 M-MuLV reverse transcriptase (100 U µl−1) 10 5 U µl−1 Total 200 -
3|
Incubate the specimen with the reaction mixture for 10 min at 4°C, then transfer to 37°C overnight. Typically 1–2 hours is sufficient, but more time may be required for thicker samples. Aspirate and wash with PBS once.
TROUBLESHOOTING
-
4|
To cross-link cDNA molecules containing aminoallyl dUTP, add 20 µl reconstituted BS(PEG)9 in 980 µl PBS to sample for 1 hr at RT.
-
5|
Aspirate and wash with PBS and quench with 1 M Tris pH 8.0 for 30 min.
PAUSE POINT The sample can be stored in PBS for up to a week at 4°C.
-
6|
Aspirate and add 10 µl DNase-free RNase and 5 µl RNase H in 1× RNase H buffer for 1 hr at 37°C (Fig. S3).
CRITICAL STEP Skipping this step results in few amplicons.
-
7|
Rinse with 2 ml nuclease-free H2O twice to remove traces of phosphate.
TROUBLESHOOTING
-
8|Prepare a CircLigase reaction mixture on ice as tabulated below, and add to the glass bottom dish containing fixed cells.
Component Amount (µl) Final Nuclease-free H2O 128 CircLigase buffer, 10× 20 1× MnCl2, 50 mM 10 2.5 mM Betaine, 5 M 40 0.5 M CircLigase II (100 U µl−1) 2 1 U µl−1 Total 200 -
9|
Place the glass bottom dish in a tightly sealed plastic container or a Ziploc bag with moist wipes and incubate at 60°C for 1 hr. If a longer reaction time is desired, 1 ml of mineral oil can be layered on top of the cells.
-
10|
Aspirate the reaction mixture, and wash with PBS. Mineral oil can be removed using PBS with 0.1% Triton X-100.
PAUSE POINT The sample can be stored in PBS at 4°C indefinitely.
-
11|
Add 200 µl RCA primer hybridization buffer containing 500 nM RCA primer to the glass bottom dish and incubate at 60°C for 1 hr.
-
12|
Aspirate and wash with RCA hybridization buffer at 60°C for 10 min.
-
13|
Aspirate and wash with 2X SSC, 1X SSC and PBS once each.
-
14|
Prepare an RCA reaction mixture on ice as tabulated below. Add to the sample and incubate overnight at 30°C. Additional dNTP (up to 10 µl) and Phi29 DNA polymerase (up to 10 µl) can enhance the fluorescence signal from DNA amplicons.
CRITICAL STEP Aminoallyl dUTP is required for cross-linking and should not be omitted.Component Amount (µl) Final Nuclease-free H2O 174 Phi29 buffer, 10× 20 1× dNTP, 25 mM 2 250 µM Aminoallyl dUTP, 4 mM 2 40 µM Phi29 DNA polymerase (100 U µl−1) 2 1 U µl−1 Total 200 -
15|
To cross-link cDNA molecules containing aminoallyl dUTP, wash gently with PBS, add 20 µl reconstituted BS(PEG)9 in 980 µl PBS to sample and incubate for 1 hr at RT.
CRITICAL STEP BS(PEG)9 expires after 2–3 wks with multiple freeze-thaw cycles, and using expired BS(PEG)9 can lead to unstable amplicons and poor sequencing results.
-
16|
Wash with PBS, aspirate and add 1 M Tris pH 8.0 for 30 min.
PAUSE POINT Store in PBS at 4°C for up to 4 weeks.
-
17|
Aspirate and add 2.5 µM control probe in 200 µL 5X SASC, pre-heated to 80°C, to the sample and incubate for 10 minutes at RT. Use the adapter- or ribosomal RNA-specific probes to image all or rRNA amplicons respectively. RT-negative controls should not produce any amplicons.
-
18|
Wash two times for 1 minute each with 1 mL 1X Instrument Buffer. If using adapter sequence-specific probe, proceed directly to Step 19 for imaging. If using the biotinylated ribosomal RNA probes, incubate in 2 µg ml−1 Streptavidin Alexa Fluor in PBS for 5 min, followed by three 2 ml PBS washes before continuing with Step 19.
-
19|
Image on a microscope and inspect the amplicon density and distribution. Amplicons should be distributed uniformly throughout the sample across the glass bottom dish. Obtain an axial view and check to see if the amplicon density is similar between regions near the glass and cell surface.
CRITICAL STEP The sample can be imaged while immersed in 1X Instrument Buffer. If alternative immersion liquid is used, do not add Tris-EDTA or other chelating agents.
-
20|
Aspirate and incubate two times for 5 minutes each in 1 mL Strip Buffer at RT, pre-heated to 80°C.
-
21|
Wash two times for 5 minute each with 1 mL 1X Instrument Buffer at RT.
PAUSE POINT We have kept samples in 1X Instrument Buffer at 4°C for up to several months without suffering a significant loss in the fluorescence signal.
SOLiD sequencing-by-ligation •TIMING 10 days for 30 cycles
-
22|
Clamp the sample firmly to the microscope stage, and use cyanoacrylate adhesive to secure any potential sources of movement, such as adjustable stage inserts. Cyanoacrylate adhesive can be applied directly to metal components and removed with acetone after sequencing.
CRITCAL STEP Use only optical-grade cyanoacrylate adhesive, as standard cyanoacrylate adhesives de-gas and ruin nearby objectives.
-
23|
Add 2.5 µM sequencing primer N in 200 µL 5X SASC, pre-heated to 80°C, to the sample and incubate for 10 minutes at RT. Aspiration can be performed using a vacuum aspirator or a flexible plastic catheter attached to a syringe.
-
24|
Wash two times for 1 minute each with 1 mL 1X Instrument Buffer at RT.
-
25|Sequence the sample by adding a freshly prepared T4 DNA ligation mixture and incubating for 45 minutes at RT.
Component Amount (µl) Final Nuclease-free H2O 165 T4 DNA ligase buffer, 10× 20 1× T4 DNA ligase, 120 U µl−1 10 6 U µl−1 SOLiD sequencing oligos (dark purple tube from the SOLiD ToP sequencing kit) 5 Total 200 -
26|
Wash four times for 5 minutes each with 1 mL 1X Instrument Buffer at RT.
-
27|
Acquire images.
CRITICAL STEP The first ligation cycle for recessed primers N-2, N-3 and N-4 produces a fluorescence signal in just one channel. These images should NOT be included in the final dataset.
-
28|
Aspirate and cleave the fluorophore by incubating the sample two times for 5 minutes each in Cleave Solution 1 and then two times for 5 minutes each in reconstituted Cleave Solution 2.1 mix at RT.
-
29|
Aspirate and wash three times for 5 minutes each with 1 mL 1X Instrument Buffer.
PAUSE POINT The sample is stable for 2–3 days in 1X Instrument Buffer at RT.
-
30|
Go to Step 25 and repeat 6 times for a total of 7 cycles.
-
31|
Incubate four times for 5 minutes each in 1 mL Strip Buffer, pre-heated to 80°C.
-
32|
Wash two times for 1 minute each with 1 mL 1X Instrument Buffer.
PAUSE POINT The sample is stable for 2–3 days in 1X Instrument Buffer at RT.
-
33|
Repeat steps 23–32 using different sequencing primers (N-1, N-2, N-3 and N-4).
Image pre-processing •TIMING 6–12 h
-
34|
If necessary use ImageJ to crop image stacks for faster 3D deconvolution.
-
35|
Determine optimal 3D deconvolution parameters using a smaller cropped test image from the experiment. In Huygens Professional we typically use a Nyquist sampling rate of 1.7, CMLE mode, 5–10 iterations and a signal-to-noise ratio of 2–5.
-
36|Deconvolve all sequencing images, and save images as .ics/.ids files with the following names in a folder named ‘decon_images’ (Fig. S4).
- Filename: <Position>_<Primer #>_<Ligation #>_<Date__Time>.<ext>
- Position: Dinucleotide position as 2-digit integers 01 to 30
- Primer number: N followed by 1-digit integers N0 to N4
- Cycle number: Ligation cycle per primer from 1 to 7
- Date/time: An alphanumeric string using underscores
- File extension: .ics and .ids
TROUBLESHOOTING
Image analysis •TIMING 6–12 h
CRITICAL Some users of our method may have little or no background in bioinformatics. Here we introduce common computational environment and tools, but novice users should obtain additional help from experienced users, network administrators and online resources (i.e. http://www.ee.surrey.ac.uk/Teaching/Unix/).
-
37|Download fisseq.zip (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/) and copy to a remote host using a command line terminal on a PC.
local:~$ scp fisseq.zip <user@remote_host_name:~/>
CRITICAL STEP One must have an account to a designated remote host. Ask the network administrator at your institution.
-
38|Download and unzip decon_images.zip (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/). Copy decon_images folder (Step 36) to a scratch space on the remote host.
local:~$ scp –r ~/decon_images/ <user@remote_host_name:scratch_space/>
CRITICAL STEP Analyzing multiple high-resolution image stacks requires a large amount of disk space. Contact your network administrator for the location of a temporary scratch space.
-
39|Logon to the remote host, and submit a job request to work on a high memory queue interactively. We recommend at least 100GB (mem below is in MB).
local:~$ ssh <user@remote_host_name> remote:~$ bsub -R "rusage[mem=100000]" -q <queue_name> -Is bash
CRITICAL STEP Running CPU or memory intensive tasks incorrectly can bring down the remote host. Make sure that you are working on a designated node. Contact your network administrator for more information before proceeding.
-
40|Unzip fisseq.zip, and change the working directory to fisseq.
remote:~$ unzip fisseq.zip remote:~$ cd fisseq
CRITICAL STEP Working from folders other than ~/fisseq results in missing file errors when entering our commands as written below.
-
41|Download and decompress the RefSeq-to-Gene ID conversion table.
remote:~/fisseq$ wget ftp://ftp.ncbi.nih.gov/gene/DATA/gene2refseq.gz remote:~/fisseq$ gzip -d gene2refseq.gz
-
42|Download the organism-specific RefSeq RNA FASTA file, and unzip the file.
remote:~/fisseq$ wget ftp://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/mRNA_Prot/human.rna.fna.gz remote:~/fisseq$ gzip -d human.rna.fna.gz ## Use the following address for mouse or rat: ## ftp.ncbi.nlm.nih.gov/refseq/M_musculus/mRNA_Prot/mouse.rna.fna.gz ## ftp.ncbi.nlm.nih.gov/refseq/R_norvegicus/mRNA_Prot/rat.rna.fna.gz
-
43|Build the reference index of [ref_name] in color space. Here [ref-name] is refseq_human. This process can take several hours.
remote:~/fisseq$ bowtie-build -C -f human.rna.fna refseq_human
TROUBLESHOOTING
-
44|Start MATLAB, and add a search path:
remote:~/fisseq$ matlab >> addpath('~/fisseq', '~/fisseq/bfmatlab') -
45|Define the input and output directories and run image registration (Fig. S5).
>> input_dir='<scratch_space>/decon_images/' >> output_dir='registered_images/' >> register_FISSEQ_images(input_dir,output_dir,10,0.1,1) >> quit()
- Number of blocks per axis for local registration (default = 10)
- Fraction overlap between neighboring blocks (default = 0.1)
- Alignment precision where 10 will register images to 1/10 of a pixel. (default = 1)
TROUBLESHOOTING
-
46|Copy files in ~/fisseq/registered_images/ to a PC. Use ImageJ to open TIFF files (File > Import > Bio-Formats) as a time series, and check alignment in channel 4 by scrolling through the timeline (Supplementary Movies 1–4).
- Maximum-projected TIFF files (channel 4 is a composite of channel 0 to 3).
- Routput.mat: Block-wise registration offsets between bases.
- Rchadj.mat: Block-wise chromatics shifts as a matrix.
- Rtadj.mat: Registration offsets over time for the whole image (not block-wise).
TROUBLESHOOTING
-
47|Start Python, and write base calls to read_data_*.csfasta. The maximum number of missing base calls allowed per read is 6 by default. * denotes an automatically generated time stamp.
remote:~/fisseq$ python >>> import FISSEQ >>> FISSEQ.ImageData('registered_images','.',6) >>> quit()TROUBLESHOOTING
-
48|Align reads to refseq_human (Step 43) using Bowtie 1.0 or earlier, and write mapped reads to bowtie_output.txt. The exact name of read_data_*.csfasta can be determined by listing files in the directory (ls -l).
remote:~/fisseq$ bowtie -C -n 3 -l 15 -e 240 -a -p 12 -m 20 --chunkmbs 200 -f -- best --strata --refidx refseq_human read_data_*.csfasta bowtie_output.txt
-
49|Spatially cluster the Bowtie reads (Step 48), annotate clusters using gene2refseq (Step 41) and write to results.tsv. The default kernel size of 3 performs a 3×3 dilation prior to clustering.
remote:~/fisseq$ python >>> import FISSEQ >>> G = FISSEQ.ImageData('registered_images',None,6) >>> FISSEQ.AlignmentData('bowtie_output.txt',3,G,'results.tsv', 'human.rna.fna','gene2refseq','9606') >>> quit() ## Use the following command for mouse or rat: >>> FISSEQ.AlignmentData('bowtie_output.txt',3,G,'results.tsv', 'mouse.rna.fna','gene2refseq','10090') >>> FISSEQ.AlignmentData('bowtie_output.txt',3,G,'results.tsv', 'rat.rna.fna','gene2refseq','10116')
Data analysis •TIMING 1 d
CRITICAL Data analysis can be done on any software package, but R is convenient for interactive analysis and high-quality graphs23. Novice users may find RStudio more intuitive than the command line interface. We provide a sample R session containing a sample dataset and a list of commands (http://arep.med.harvard.edu/FISSEQ_Nature_Protocols_2014/).
-
50|
Open the FISSEQ RStudio project file (Menu > File > Open project…).
-
51|Find the HISTORY tab on the upper right console window and double-click on individual commands in order to re-execute the previous R session (Fig. S6) and learn how to:
- Import and filter data using a specific criterion (i.e. cluster size)
- Plot a distribution of reads by a specific criterion (i.e. RNA classes and strands)
- Convert a table of reads into a table of gene expression level
- Correlate gene expression from different images
- Find statistically enriched genes in different regions
Troubleshooting
TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3.
Troubleshooting
| Step | Problem | Possible reasons | Possible solutions |
|---|---|---|---|
| 1 | Cells wash away during PBS washes or fixation | Cell attachment dependent on Ca++ | Use PBS with Ca++, or add formalin directly to growth media. |
| 1 | Membrane blebs | Diluting formalin using 1X PBS makes it hypotonic | Use 10X PBS and water to dilute formalin |
| 1 | Tissue sections falling apart or off slide | Pepsin over-digestion and/or small contact area between sample and glass | Shorter pepsin digestion; embed in Matrigel and re-fix |
| 19 | Few amplicons limited to the cell surface | Poor cell permeabilization | Use 0.1N HCl following Triton or ethanol-based cell permeabilization |
| 19 | Amplicons in no RT control | RT primer is too long | Use shorter RT primers |
| 19 | High background | Excess RCA primer | More stringent washes at Step 13 |
| 19 | Dim amplicons | Low template copy number per amplicon | Low template copy number per amplicon |
| 19, 27 | Dim, fuzzy, or stretched amplicons | Poor cross-linking | Fresh BS(PEG)9 at Step 15 |
| 28 | White precipitate build-up | Silver reacting with chloride | Eliminate chloride-containing buffers |
| 33 | Progressive loss of signa | Photo-damage to amplicons | Low laser exposure |
| 35 | Deconvolution takes too much time | Large images | Crop unused areas Smaller images Fewer iterations |
| 43 | Bowtie command not found | Bowtie v1.0 environment not set-up | Check Bowtie version (which bowtie); ask administrator |
| 45 | MATLAB out of memory error | Low RAM Low heap space for Java VM | Allocate >100GB RAM Increase Java VM in java.opts |
| 45 | Input or output folders not found | Incorrect slash use with folder name | No slash before and one slash after |
| 46 | ImageJ does not open TIFF files correctly | Incorrect slash use with folder name Image dimensions not correctly read | No slash before and one slash after Check ‘Group files…’, ‘Swap dim…’ and ‘Concatenate…’ when importing |
| 47 | Cannot find input images | Undefined path | Registered image directory must be in ~/fisseq |
| 47 | Extension error messages | Missing package | Use Canopy Python 2.7 |
| 48 | Bowtie not found | Bowtie 1.0 not loaded | Check available versions and load Bowtie 1.0 or earlier versions |
| 48 | Extra parameter(s) error | Typo, or option flags not in correct order | Copy and paste command from Step 48 |
| 51 | Unexpectedly high number of antisense mRNA reads | Noisy image, many missing or incorrect base calls at the 3’ end | Obtain better images, use deconvolution to reduce noise, sequence longer reads, trim reads, or increase the cluster size threshold. |
Timing
Steps 1–21: FISSEQ library construction: 2 d
Steps 22–33: Sequencing and imaging: 10 d
Steps 34–36: Image pre-processing: 6 h
Steps 37–49: Image analysis: 12 h
Steps 50–51: Data analysis: 2 d
ANTICIPATED RESULTS
The size of subcellular cDNA amplicons is slightly larger than the diffraction limit after 3D deconvolution. At 20× NA 0.75 the diameter of cDNA amplicons is approximately 400 to 800 nm after image deconvolution. A typical amplicon contains hundreds of fluorescent probe binding sites, and this results in images that are 20 to 50 times brighter and have a dramatically improved signal-to-noise ratio than single molecule FISH. A good FISSEQ library should yield many intensely bright amplicons that are distinct from cell debris and spurious amplification products. If long exposure time and high gain have to be used to visualize objects, it is likely that they represent contamination, reaction precipitates, or cell debris.
When fluorescent probes are stripped, nearly all of the fluorescence is completely removed except possibly in the nucleus. Stripping is a good way to distinguish a DNA amplicon from fluorescent debris, and we recommend alternately hybridizing the sample with FAM, Cy3, or Cy5 probes while the sample is still on the microscope. If the fluorescent object is a DNA amplicon, it should fluoresce in distinct colors sequentially with little or no cross-talk. The amplicon density varies depending on the cell size, but we typically see several hundreds of amplicons per cell in cultured cell lines (i.e. iPSC, fibroblasts, HeLa, bipolar neurons). We have detected up to 4,000 amplicons using synthetic DNA per cell in fibroblasts, suggesting that the RT efficiency may be a limiting factor.
The signal-to-noise ratio from SOLiD sequencing-by-ligation is high, especially for early ligation cycles. The quality drops after the fourth re-ligation cycle for each primer, and the image quality degrades significantly after 25 total cycles. Much of the image degradation results from the laser-induced damage during imaging. Typically, un-imaged regions remain pristine even after 30 cycles of sequencing, and it may be possible to obtain a longer read length with appropriate free-radical scavengers in the imaging buffer, but we have not attempted this yet.
Depending on the camera sensor size, density and bit depth, one image stack containing multiple optical planes across four channels can be 800MB to 2GB per field of view. Our image registration software then creates a separate folder containing TIFF images (5 channels per base) of 20M to 50M in size. Once our software processes and analyzes the images, it generates a tab delimited file containing the gene ID, name, cluster size, strand, class, base quality, alignment quality, color space sequence and x-y position. We recommend performing a quick data check by selecting a gene cluster size of >5 to compare the number of sense and anti-sense reads and also comparing the number of reads from different RNA classes. Typically, >90% of all reads should map to the positive sense strand. The ribosomal RNA read should comprise 50–80% of the total number of reads. We typically get 15,000 to 40,000 reads per image containing 30–50 cells. Regional or sub-cellular localization is measured in statistically significant enrichment scores, rather than absolute counts due to a small number of reads distributed over a large area. We recommend making B&W image masks based on the cell morphology, DAPI stains, immunohistochemistry and other types of spatial masks and measure the relative enrichment of individual genes using Fisher’s exact test or other similar tests23. With a high read density, it may be possible to use unsupervised local clustering of reads for regional identification of biological processes2.
Supplementary Material
Supplementary Movie 1. Image registration using 10 × 10 tiles, no overlap and upsampling factor of 1.
Supplementary Movie 2. Image registration using 10 × 10 tiles, 10% overlap and upsampling factor of 0.1.
Supplementary Movie 3. Image registration using 10 × 10 tiles, 10% overlap and upsampling factor of 1.
Supplementary Movie 4. Image registration using 10 × 10 tiles, 10% overlap and upsampling factor of 10.
Figure 4.
Schematic overview of the SOLiD color coding and decoding scheme. (a) The base position within the template sequence is enclosed by white circles and should be used for naming the image files, and the actual sequencing cycle numbers are noted on both sides. Each ligation extension is shown in different colors, and cycle 15, 22 and 29 are shown in gray as no images are acquired for these cycles. The red box at cycle 8 denotes a known base identity. (b) SOLiD dinucleotide coding scheme. (c) SOLiD color space decoding scheme (figures modified from the Life Technologies application note). As long as any one of the base identities are known (here in red), the color space sequence can be converted to the nucleotide sequence.
Acknowledgments
Funded by NIH CEGS grant P50 HG005550. J.H.L. and coworkers funded by NHBLI grant RC2HL102815, Allen Institute for Brain Science, and NIMH grant MH098977. E.R.D. funded by NIH grant GM080177 and NSF GRF grant DGE1144152.
Footnotes
Author contributions statement
J.H.L. and E.R.D. conceived FISSEQ library construction, sequencing, image analysis and bioinformatics. J.S., R.K., J.L.Y., B.M.T., H.S.L. and J.A. provided key feedbacks during the FISSEQ method development. R.T. and T.C.F. assisted with automated microscopy and image analysis. K.Z. and G.M.C. oversaw the project. J.H.L. wrote the paper, and E.R.D. wrote the FISSEQ software.
Competing financial interests
Potential conflicts of interests for G.M.C. are listed on http://arep.med.harvard.edu/gmc/tech.html. Other authors have no conflicts of interests.
REFERENCES
- 1.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods. 2013 doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- 4.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 5.Zeng H, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez-Roux G, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 8.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 10.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi HM, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2011;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke R, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013 doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 13.Larsson C, Grundberg I, Söderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nature methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 14.Larsson C, et al. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nature methods. 2004;1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 15.Lagunavicius A, et al. Novel application of Phi29 DNA polymerase: RNA detection and analysis in vitro and in situ by target RNA-primed RCA. RNA. 2009;15:765–771. doi: 10.1261/rna.1279909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkiene E, Gaidamaviciute E, Riauba L, Janulaitis A, Lagunavicius A. Direct detection of RNA in vitro and in situ by target-primed RCA: The impact of E. coli RNase III on the detection efficiency of RNA sequences distanced far from the 3'-end. RNA. 2010;16:1508–1515. doi: 10.1261/rna.2068510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, et al. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet. 2009;5:e1000718. doi: 10.1371/journal.pgen.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 19.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 20.Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014 doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 22.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adiconis X, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frumkin D, et al. Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues. BMC Biotechnol. 2008;8:17. doi: 10.1186/1472-6750-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovatt D, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid MW, et al. A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS One. 2012;7:e29685. doi: 10.1371/journal.pone.0029685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Avital G, Hashimshony T, Yanai I. Seeing is believing: new methods for in situ single-cell transcriptomics. Genome Biology. 2014;15:110. doi: 10.1186/gb4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buxbaum AR, Wu B, Singer RH. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itzkovitz S, Blat IC, Jacks T, Clevers H, van Oudenaarden A. Optimality in the development of intestinal crypts. Cell. 2012;148:608–619. doi: 10.1016/j.cell.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen CH, van Oudenaarden A. Allele-specific detection of single mRNA molecules in situ. Nat Methods. 2013 doi: 10.1038/nmeth.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 39.Kosuri S, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nature methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 43.Kaposi-Novak P, Lee JS, Mikaelyan A, Patel V, Thorgeirsson SS. Oligonucleotide microarray analysis of aminoallyl-labeled cDNA targets from linear RNA amplification. Biotechniques. 2004;37:580, 582–586, 588. doi: 10.2144/04374ST02. [DOI] [PubMed] [Google Scholar]
- 44.Nanda JS, Lorsch JR. Labeling a protein with fluorophores using NHS ester derivitization. Methods Enzymol. 2014;536:87–94. doi: 10.1016/B978-0-12-420070-8.00008-8. [DOI] [PubMed] [Google Scholar]
- 45.Mardis ER. Next-generation DNA sequencing methods. Annual review of genomics and human genetics. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 46.Massingham T, Goldman N. Error-correcting properties of the SOLiD Exact Call Chemistry. BMC Bioinformatics. 2012;13:145. doi: 10.1186/1471-2105-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Applied_Biosystems. ( https://www3.appliedbiosystems.com/cms/groups/global_marketing_group/documents/generaldocuments/cms_091372.pdf. [Google Scholar]
- 48.Itzkovitz S, van Oudenaarden A. Validating transcripts with probes and imaging technology. Nat Methods. 2011;8:S12–S19. doi: 10.1038/nmeth.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eliceiri KW, et al. Biological imaging software tools. Nat Methods. 2012;9:697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kankaanpaa P, et al. BioImageXD: an open, general-purpose and high-throughput image-processing platform. Nat Methods. 2012;9:683–689. doi: 10.1038/nmeth.2047. [DOI] [PubMed] [Google Scholar]
- 52.Pawley JB. Handbook of biological confocal microscopy. Edn. 3rd. New York, NY: Springer; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1. Image registration using 10 × 10 tiles, no overlap and upsampling factor of 1.
Supplementary Movie 2. Image registration using 10 × 10 tiles, 10% overlap and upsampling factor of 0.1.
Supplementary Movie 3. Image registration using 10 × 10 tiles, 10% overlap and upsampling factor of 1.
Supplementary Movie 4. Image registration using 10 × 10 tiles, 10% overlap and upsampling factor of 10.