Abstract
Fragile X is the most common known inherited cause of intellectual disability and autism, and it typically results from transcriptional silencing of FMR1 and loss of the encoded protein, FMRP (fragile X mental retardation protein). FMRP is an mRNA-binding protein that functions at many synapses to inhibit local translation stimulated by metabotropic glutamate receptors (mGluRs) 1 and 5. Recent studies on the biology of FMRP and the signaling pathways downstream of mGluR1/5 have yielded deeper insight into how synaptic protein synthesis and plasticity are regulated by experience. This new knowledge has also suggested ways that altered signaling and synaptic function can be corrected in fragile X, and human clinical trials based on this information are under way.
Introduction
This year we expect to learn the outcome of clinical trials for potentially disease-modifying treatments of fragile X (FX). Three important developments outside the realm of basic neuroscience paved the way for this progress: First, careful clinical observation defined the syndrome and suggested a genetic etiology (Martin & Bell 1943); second, mutations that silenced a single gene (FMR1) on the X chromosome were discovered to be the major cause (Pieretti et al. 1991, Verkerk et al. 1991); and third, the generation and widespread dissemination of an Fmr1-knockout (KO) mouse enabled studies of pathophysiology (Dutch-Belgian Fragile X Consort. 1994) (Figure 1). FMR1 encodes fragile X mental retardation protein (FMRP), an mRNA-binding protein that is highly expressed in neurons. As with most neurobehavioral disorders of genetic origin, it was assumed that development of the brain in the absence of this key protein irrevocably alters neuronal connectivity to produce the devastating behavioral symptoms, including intellectual disability and autism, that are characteristic of this disease.
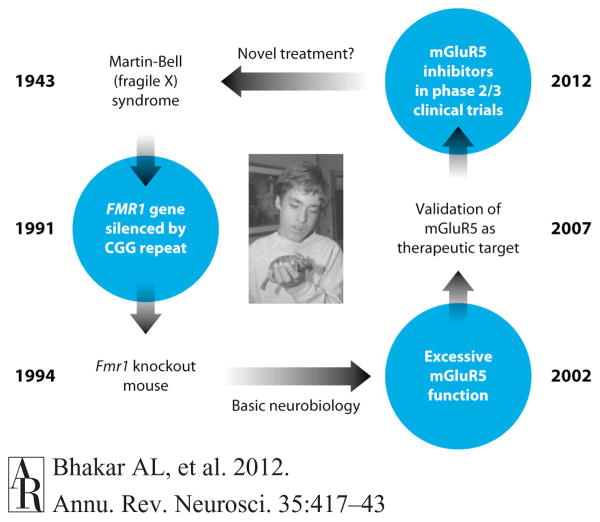
Figure 1.
Fulfilling the promise of molecular medicine in FX. Martin & Bell (1943) described a group of patients characterized by a common set of features that included intellectual disability and social withdrawal. The causative gene mutation was discovered in 1991 (Pieretti et al. 1991, Verkerk et al. 1991). The FMR1 gene on the X chromosome is silenced, and the protein FMRP is not produced. Shortly thereafter, the Fmr1-KO mouse model was generated (Dutch-Belgian Fragile X Consort. 1994) and has been intensively studied by neurobiologists interested both in the disease and FMRP. In 2002, it was discovered that a form of synaptic plasticity—mGluR LTD—was exaggerated in the Fmr1 KO mouse (Huber et al. 2002). This led to the mGluR theory of fragile X (Bear et al. 2004), which posits that many symptoms of the disease are due to exaggerated responses to activation of mGluR5. The theory was definitively validated in 2007 with the demonstration that multiple FX phenotypes are corrected in the Fmr1-KO mouse by genetic reduction of mGluR5 protein production (Dolen et al. 2007). In addition, numerous animal studies showed that pharmacological inhibition of mGluR5 ameliorates FX mutant phenotypes. In 2009, inhibitors of mGluR5 entered into human phase 2 trials (http://clinicaltrials.gov). If successful, these trials will represent the first pharmacological treatment for a neurobehavioral disorder that was developed from the bottom up: from gene discovery to pathophysiology in animals to novel therapeutics in humans. Abbreviations: CGG, cytosine-guanine-guanine; FMRP, fragile X mental retardation protein; FX, fragile X; mGluR5, metabotropic glutamate receptor 5; KO, knockout; LTD, long-term synaptic depression. Image courtesy of FRAXA Research Foundation, with permission.
However, this dim view of FX has changed dramatically in the past ten years. It is now believed that many symptoms of FX could arise from modest changes in synaptic signaling—changes that can be corrected with targeted therapies such as those that are now in clinical trials. The origins of this new view can be traced to fundamental research on synaptic plasticity (Bear et al. 2004, Huber et al. 2002). Since this initial insight into how synaptic signaling is altered in FX, the progress toward developing therapeutics for FX has been explosive. It has been shown that seemingly unrelated symptoms of the disease can be corrected by manipulating a molecular target, mGluR5, that is amenable to drug therapy (Dolen et al. 2007). Furthermore, studies in multiple animal models of FX have shown that this core pathophysiology is evolutionarily conserved. This extraordinary progress has been the subject of a number of recent reviews (see e.g., Dolen et al. 2010, Krueger & Bear 2011, Levenga et al. 2010, Santoro et al. 2011).
Certainly research on synaptic plasticity has informed the understanding of FX pathophysiology; but it is also true that the biology of FX has informed the understanding of synaptic function and plasticity. This is the point of view we take in the present review.
Overview of Fragile X
In the majority of FX patients, a trinucleotide (CGG) repeat expansion leads to hypermethylation and transcriptional silencing of the FMR1 gene and subsequent loss of FMRP (Fu et al. 1991, Pieretti et al. 1991). In one identified patient, disease is caused by a point mutation in FMR1 that alters protein function (De Boulle et al. 1993). Disease severity varies with the expression level of FMRP, which can fluctuate as a result of germline mosaicism and, in females, X inactivation (De Boulle et al. 1993, Hatton et al. 2006, Kaufmann et al. 1999, Loesch et al. 1995, Lugenbeel et al. 1995, Reiss & Dant 2003). Accordingly, understanding the cellular function of FMRP has become an obvious priority.
Epidemiological studies conservatively estimate that FX occurs in 1:5000 males (and approximately half as many females), making it the leading cause of inherited intellectual disability (Coffee et al. 2009). FX was also the first recognized genetic disorder associated with autism, and despite expanding diagnostic criteria and newly discovered candidate genes, FX remains the most common known inherited cause of autism (Wang et al. 2010b). In addition to moderate to severe intellectual disability and autistic features (social/language deficits and stereotyped/restricted behaviors), the disease is characterized by seizures and/or epileptiform activity, hypersensitivity to sensory stimuli, attention deficit and hyperactivity, motor incoordination, growth abnormalities, sleep disturbances, craniofacial abnormalities, and macroorchidism. Because FX is a monogenic and relatively common cause of autism, it has been a useful model for dissecting pathophysiology that may apply to genetically heterogeneous autisms.
New Insights into the Biology of FMRP
Biochemical characterization of FMRP has provided key insights into the pathophysiology of FX, and after 20 years of research, we now know that FMRP is an RNA-binding protein that largely functions to negatively regulate protein synthesis in the brain. Recent work has led to the view that many symptoms of FX arise from a modest increase in synaptic protein synthesis, an aspect of cerebral metabolism that can continue to be corrected after birth to produce substantial benefit. Therefore, there is great interest in the question of how FMRP interacts with mRNA to regulate synaptic protein synthesis.
FMRP Binds RNA
Sequence analysis first identified three common RNA-binding domains in the protein structure of FMRP, providing the first suggestion of a direct interaction between FMRP and RNA (Ashley et al. 1993, Siomi et al. 1993). Two of the domains are hnRNP K-homology (KH) domains, and the third, located close to the C-terminal end, is an RGG box (Figure 2). KH domains are thought to recognize and bind “kissing-complex” tertiary motifs in RNA (Darnell et al. 2005), whereas the RGG box recognizes stem-G-quartet loops, possibly in a methylation-dependent manner (Blackwell et al. 2010). A stem loop SoSLIP motif, found in one target (Sod1 mRNA), has also been identified and can bind to the C-terminal RGG region (Bechara et al. 2009). In addition, U-rich sequences have been isolated as potential RNA-binding motifs, although no precise binding domains within FMRP have yet been described (Chen et al. 2003, Fahling et al. 2009).
Figure 2.
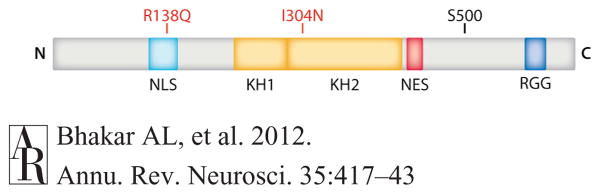
Functional domains of FMRP. Human FMRP, a 632 amino acid polypeptide (gray bar), has a nuclear localization signal (NLS; light blue), two K-homology domains (KH1 and KH2; orange), an RGG (arginine-glycine-glycine) box (dark blue), and a nuclear export sequence (NES; red). R138Q and I304N are naturally occurring mutations in patients with developmental delay and a severe form of FX, respectively. I304N abolishes polyribosome association. S500 is a major site of phosphorylation. Abbreviations: N, amino terminus; C, carboxy terminus; FMRP, fragile x mental retardation protein.
How FMRP associates with specific mRNAs is still under active investigation. A point mutation (I304N) within the second KH domain leads to a severe clinical presentation of the disease and has provided the first evidence that binding to mRNA and this domain in particular are critical to the function of FMRP (De Boulle et al. 1993, Feng et al. 1997a). Recent work using ultraviolet light to crosslink FMRP with endogenous mRNA in situ revealed, surprisingly, that FMRP binds largely within the coding regions of many mRNAs instead of the 5′ or 3′ untranslated regions (Darnell et al. 2011). Although this study did not reveal a specific consensus motif, synthetic kissing-complex RNA was still effective in competing with these target mRNAs for binding to FMRP, confirming that KH domains and kissing-complex motifs are critically involved. It has been estimated that ∼4% of total brain mRNA binds FMRP (Ashley et al. 1993, Brown et al. 2001, Darnell et al. 2011).
FMRP May Regulate RNA Transport
FMRP also contains a nuclear localization sequence and a nuclear export sequence (Ashley et al. 1993), and although its expression is largely cytoplasmic (found in the cell body, dendrites, and synapses), some FMRP can be found shuttling in and out of the nucleus (Feng et al. 1997b). To date, few data exist to support a role for FMRP in regulating transcription or RNA processing, but FMRP can be found bound to nuclear mRNA, a nuclear exporter protein (Tap/NXF1), and to pre-mRNA while it is being transcribed (Kim et al. 2009). A novel missense mutation (R138Q) was detected in the nuclear localization sequence of FMR1 in a patient with developmental delay (Collins et al. 2010), suggesting that nuclear FMRP is important for neuronal function.
Many in vitro studies have suggested a role for FMRP in transporting mRNA. The protein has been imaged in dynamic RNA granules that traffic from the soma to dendrites and axons (Antar et al. 2004, 2005, 2006; De Diego Otero et al. 2002). RNA granules are believed to be translationally repressed mRNP (messenger ribonucleoprotein) complexes. Granules are heterogeneous in their composition: P bodies and stress granules contain translational initiation machinery (e.g., monomeric ribosomal constituents, mRNA, and proteins) trapped before translational initiation, whereas high-density granules also contain elongation machinery (e.g., polyribosomes and ribosomal aggregates) whose translation has been stalled (Anderson & Kedersha 2006, Kiebler & Bassell 2006). Once localized to the synapse, mRNAs are released from the granules and subsequently translated in response to stimuli (Krichevsky & Kosik 2001). FMRP mRNPs have been found in all three types of RNA granules (Aschrafi et al. 2005, Barbee et al. 2006, Maghsoodi et al. 2008, Mazroui et al. 2002).
In some instances, FMRP trafficking into dendrites can be stimulated by neuronal activity (Antar et al. 2004, Gabel et al. 2004). However, it does not appear to be necessary for the steady-state maintenance or the constitutive localization of the majority of its target mRNAs in dendrites (Dictenberg et al. 2008, Steward et al. 1998). Indeed, most mRNAs that normally associate with FMRP are correctly targeted to the synapse in the absence of FMRP. Thus, another RNA-binding protein may be needed for the normal active transport of the majority of FMRP targets, and FMRP may be more of a passive passenger within the RNA transport granule.
FMRP Negatively Regulates Translation
Subcellular fractionation studies originally showed that the majority of FMRP-RNA complexes are in actively translating polyribosomal fractions, particularly in synaptic preparations (Aschrafi et al. 2005; Brown et al. 2001; Corbin et al. 1997; Eberhart et al. 1996; Feng et al. 1997a, 1997b; Khandjian et al. 1995; Stefani et al. 2004; Tamanini et al. 1996; Zalfa et al. 2007). These observations, together with the knowledge that both FMRP protein and mRNA are expressed in dendrites and dendritic spines, suggested that FMRP regulates local protein synthesis at the synapse. Several independent lines of evidence support this hypothesis and show that FMRP functions to repress translation. First, purified recombinant FMRP added to rabbit reticulolysate or injected into Xenopus oocytes shows a dose-dependent suppression of mRNA translation that is abolished when FMRP-binding sequences are removed from target mRNA (Laggerbauer et al. 2001, Li et al. 2001). Second, an electrophysiological readout of synaptic protein synthesis in the hippocampus, metabotropic glutamate receptor (mGluR)-dependent long-term depression (LTD) (discussed below), is exaggerated in the absence of FMRP, consistent with increased protein synthesis (Huber et al. 2002). Third, direct measurement of the rate of protein synthesis in hippocampal slices or cortical synaptoneurosomes in vitro shows a significant elevation in the Fmr1-KO mouse (Dolen et al. 2007, Muddashetty et al. 2007, Osterweil et al. 2010) (Figure 3). Finally, similar measurements performed in the KO mouse in vivo show a global increase in brain protein synthesis (Qin et al. 2005). The fact that increased protein synthesis can be observed in the intact animal in vivo has raised the possibility that measurements of protein synthesis could serve as a biomarker of disease (Bishu et al. 2008, Bishu et al. 2009). Indeed, studies are currently underway to test this hypothesis in human patients with FX (http://www.clinicaltrials.gov).
Figure 3.
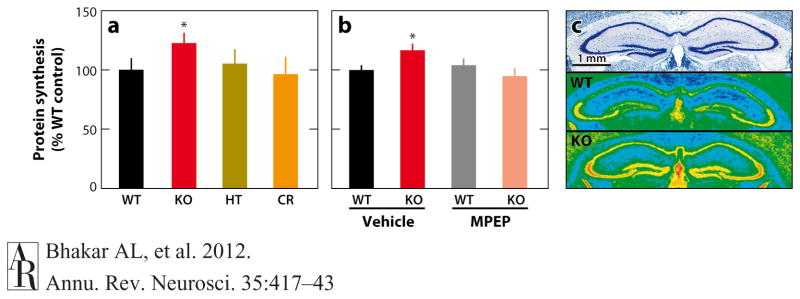
Excessive protein synthesis in the hippocampus of Fmr1-KO mice. Translation rates in the hippocampus measured by metabolic labeling in vitro (a,b) and in vivo (c) confirm that FMRP functions to negatively regulate protein synthesis in neurons. (a) Basal protein synthesis is significantly increased in Fmr1-KO hippocampal slices compared to control WT. Although there is no effect of reducing mGluR5 by 50% in Grm5 heterozygous mice (HT), crossing these mice with Fmr1-KO mice (CR) is sufficient to correct the excessive protein synthesis (modified from Dolen et al. 2007). (b) Excessive protein synthesis in Fmr1-KO hippocampal slices is restored to normal levels by acute treatment with an mGluR5 inhibitor (MPEP), demonstrating it occurs downstream of constitutive mGluR5 activity (modified from Osterweil et al. 2010). (c) Nissl-stained coronal sections (top panel) and their corresponding pseudocolored autoradiograms (middle and lower panels) show quantitative increases in translation rates throughout the hippocampus of 6-month-old Fmr1-KO mice in vivo (lower panel) compared with WT controls (middle panel). Images courtesy of C.B. Smith (Qin et al. 2005). Hot colors represent higher rates of synthesis. Abbreviations: FMRP, fragile X mental retardation protein; KO, knockout; mGluR, metabotropic glutamate receptor; MPEP, 2-methyl-6-(phenylethynyl)-pyridine; WT, wild type.
Mechanisms of Translational Regulation by FMRP
Although it is now appreciated that FMRP functions to negatively regulate protein synthesis, the mechanism by which repression is achieved remains controversial. Given that the majority of FMRP cosediments with polyribosomes, FMRP was originally suspected to repress translation by blocking elongation (Ceman et al. 2003, Feng et al. 1997a, Khandjian et al. 1996, Stefani et al. 2004, Tamanini et al. 1996). This hypothesis has received strong support in a recent study in which FMRP mRNA targets were identified following ultraviolet cross-linking (Darnell et al. 2011). The majority (66%) of mRNA binding was found within the coding sequence of the 842 transcripts cross-linked to FMRP in mouse brain polysomes. Ribosomal run-off assays on these transcripts demonstrated that FMRP is associated with transcripts on which ribosomes are stalled. These data support a model whereby FMRP dynamically represses translation in a complex consisting of target mRNAs and stalled ribosomes (Figure 4).
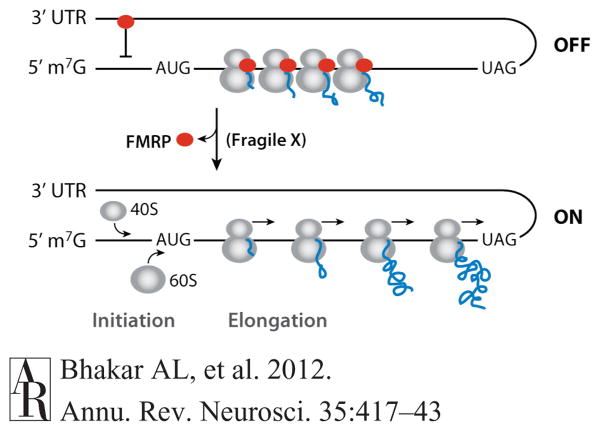
Figure 4.
FMRP regulates mRNA translation. FMRP (red ovals) can be found bound to coding regions of mRNA in association with stalled ribosomes [complexes of 40S (small gray ovals) and 60S (large gray ovals) ribosomal subunits] and bound to 3′UTRs in association with inhibitory components of the initiation machinery (indicated by an inhibitory line). Data currently suggest that FMRP normally represses translation by stalling the elongation of actively translating ribosomes and by blocking the initiation of ribosome assembly. Loss of FMRP (as in fragile X) removes both of these inhibitory associations and leads to increased protein synthesis. Curly blue lines represent ribosomally synthesized polypeptide chains that lengthen as translation proceeds. Small arrows indicate active movement. Abbreviations: AUG, initiation codon; FMRP, fragile X mental retardation protein; m7G, 7-methylguanylate cap; ON, translation on; OFF, translation off; UAG, termination codon; 3′UTR, 3 prime-end untranslated region.
However, the presence of FMRP mRNPs in p bodies, stress granules, and high-density granules has suggested that FMRP represses translation throughout many phases of translational regulation. FMRP can cosediment with the monomeric 80S ribosomes and in light mRNP complexes with BC1 (brain cytoplasmic RNA 1), CYFIP1 (cytoplasmic FMRP-interacting protein), and translation initiation factors (Centonze et al. 2008, Gabus et al. 2004, Johnson et al. 2006, Lacoux et al. 2012, Laggerbauer et al. 2001, Napoli et al. 2008, Zalfa et al. 2007). These data suggest that FMRP also represses translation at the initiation stage. In this model, FMRP represses translation by inhibiting cap-dependent initiation through interactions with CYFIP1, a eukaryotic initiation factor 4E binding protein (4E-BP). Consistent with this proposal, genetic reduction of CYFIP1 levels increases the expression of several FMRP targets (Napoli et al. 2008). The in vivo relevance of these interactions, however, has been questioned (Iacoangeli et al. 2008a,2008b; Stefani et al. 2004; Wang et al. 2005).
Mechanisms to Stall Elongation
How FMRP cooperates with the translational machinery to stall elongation or block initiation is incompletely understood. Some data have suggested that association with the microRNA (miRNA) machinery may be involved. FMRP interacts with members of the RNA-induced silencing complex (Bolduc et al. 2008, Caudy & Hannon 2004, Caudy et al. 2002, Cheever & Ceman 2009, Ishizuka et al. 2002, Jin et al. 2004, Muddashetty et al. 2011; but see Didiot et al. 2009) and several specific miRNAs (Edbauer et al. 2010, Plante et al. 2006, Xu et al. 2008, Yang et al. 2009) that function together to silence target mRNA, either by direct cleavage of transcripts or by translational repression (see, for a review, Schratt 2009). Because FMRP lacks a canonical miRNA-binding domain, it currently seems likely that this modulation occurs through protein-protein interactions between members of the RNA-induced silencing complex (e.g., Argonaute, Dicer) and FMRP, rather than direct binding to miRNAs. Still, the possibility remains that the kissing-complex structure, the putative ligand of the KH domain of FMRP, may be formed by miRNA and target mRNA together (Darnell et al. 2005, Plante et al. 2006).
Several post-translational modifications of FMRP have also been suggested to regulate translational repression. Methylation of FMRP on arginine residues can reduce FMRP binding to stem loop G-quartet structures (Stetler et al. 2006). Others have suggested that ubiquitin-proteasome degradation followed by resynthesis of FMRP may be a mechanism for transient derepression (Zhao et al. 2011), but some work has shown that FMRP synthesis increases upon stimulation prior to its degradation (Hou et al. 2006). FMRP can also be phosphorylated on a series of serine residues N terminal to the RGG box. Phosphorylation has been suggested to stall ribosomal translocation while preserving the association of FMRP with mRNA (Ceman et al. 2003, Coffee et al. 2011, Muddashetty et al. 2011). Thus, one way neural activity may gate translation is by regulating FMRP phosphorylation.
Synaptic Regulation of Protein Synthesis
Although FMRP is expressed throughout the neuron, it has attracted particular attention as a regulator of protein synthesis at excitatory synapses. Because exaggerated protein synthesis is believed to be pathogenic in FX and possibly in other disorders associated with autism (Kelleher & Bear 2008, Darnell 2011), the question of how synaptic activity can trigger FMRP-regulated mRNA translation is of particular interest. Conversely, because neuronal protein synthesis has a fundamental role in synaptic plasticity and information storage (Kandel 2001), understanding how FMRP functions at the synapse has also become a high priority in basic neurobiology.
Interest in synaptically localized protein synthesis originated with the discovery that polyribosomes accumulate at the base of many dendritic spines that are postsynaptic to glutamatergic excitatory synapses (Steward & Levy 1982). These synaptic polyribosomes seemed to provide an ideal substrate for the structural changes that support long-term synaptic modifications, such as long-term potentiation (LTP) and LTD, that store memories. Consistent with this proposal, the transitions from early to late phases of LTP and LTD require new protein synthesis independent of transcription (Cracco et al. 2005, Huber et al. 2000, Kang & Schuman 1996). Furthermore, these modifications can be maintained by new translation in isolated dendrites, implicating pre-existing dendritically localized mRNA. Thus, glutamate release at individual synapses appears to stimulate local protein synthesis to maintain long-lasting synaptic change.
Translational Control at Glutamatergic Synapses
An understanding of the molecular mechanisms by which synaptic activity regulates local protein synthesis is beginning to emerge. Two types of postsynaptic glutamate receptors have been implicated: the calcium-permeable N-methyl-d-aspartate ionotropic receptors (NMDARs) and the Gq-coupled (group 1) mGluR1 and mGluR5. The mGluRs have complementary expression patterns: mGluR5 expression is highest in the forebrain and mGluR1 expression is highest in the cerebellum (Shigemoto et al. 1993). NMDARs are also widely expressed throughout the brain and stimulate the release of brain-derived neurotrophic factor, a ligand for TrkB receptors, which can contribute to synaptic protein synthesis (Kang & Schuman 1996, Schratt 2009).
Of particular interest in the context of FX is protein synthesis stimulated by activation of Gp1 mGluRs. Weiler & Greenough (1993) provided the first evidence that Gp1 mGluR agonists stimulate protein synthesis in biochemical preparations enriched for cortical synapses. It is now understood that Gp1 mGluRs couple to the synaptic translation machinery at synapses in many parts of the brain and that many functional consequences of Gp1 mGluR activation depend on new protein synthesis (see Krueger & Bear 2011 for a review).
Two intracellular signaling cascades have been proposed to couple mGluRs and other synaptic receptors to the translational machinery: (a) the mammalian target of rapamycin (mTOR) pathway and (b) the extracellular signal–regulated kinase (ERK) pathway. Both mTOR and ERK pathways can stimulate cap-dependent translation by regulating components of initiation. Initiation is the step during which the small ribosome subunit is recruited to the 5′ end of mRNA and scans toward the start codon to assemble into the complete ribosome (see Gebauer & Hentze 2004 for a review).
One key regulatory step in initiation is the recognition of the 5′ mRNA cap by eIF4E (Supplemental Figure 1), which leads to assembly of the eIF4F complex and recruitment of the small ribosomal subunit (Richter & Sonenberg 2005). A family of 4E-BPs inhibits this process by binding to eIF4E. This inhibition is relieved by phosphorylation of 4E-BPs by both mTOR and ERK or, in postnatal mammalian brain, by deamination (Bidinosti et al. 2010). The mTOR pathway can also facilitate initiation through phosphorylation of p70 ribosomal protein S6 kinases (S6Ks), leading to ribosomal protein S6 phosphorylation and phosphorylation of eIF4B. Similarly, the ERK pathway can facilitate initiation by phosphorylation of S6 and eIF4B through activation of p90 ribosomal protein S6 kinases (RSKs); however, it can also lead to phosphorylation of eIF4E through activation of MNK. Phosphorylation of eIF4B stimulates the eIF4F complex activity by potentiating the RNA-helicase activity of eIF4A. Phosphorylation of eIF4E generally decreases eIF4E affinity for the cap, however, and may function to reduce overall translation rates. Some researchers have hypothesized that this mechanism may allow for increases in the translation of a specific subset of mRNAs (Costa-Mattioli et al. 2009). This is likely to be one mechanism whereby specific pools of mRNAs are selected for translation (a topic we discuss below).
Another major regulatory step in initiation is the formation of the ternary complex (eIF2, Met-tRNA, and GTP) required to complete the 43S ribosomal complex. Phosphorylation of eIF2 inhibits the GDP/GTP exchange required to reconstitute a functional ternary complex, causing a decrease in general translation and an impairment in some forms of late-phase LTP and long-term memory (Costa-Mattioli et al. 2009). Curiously, however, eIF2 phosphorylation can also stimulate translation of a subset of mRNAs that contain short upstream open reading frames. Initiation can also be regulated at the mRNA 3′ end by CPEB (cytoplasmic polyadenylation element–binding protein), an RNA-binding protein that inhibits poly(A) tail addition and formation of the eIF4F complex. CPEB, similar to FMRP, is commonly found to repress the translation of dendritically transported mRNAs (Costa-Mattioli et al. 2009). How synaptic activity couples to eIF2 phosphorylation or CPEB regulation has yet to be fully explained.
Although initiation is usually the rate-limiting step in translation, in some instances excitatory synaptic stimulation can regulate the elongation phase of translation. Both mGluR5 and NMDAR, via activation of calcium/calmodulin-dependent eEF2 kinase, can increase phosphorylation of eEF2. Phospho-eEF2 stalls general elongation but allows translation of a subset of mRNAs (Scheetz et al. 2000), including those that encode the proteins Arc and MAP-1B (Park et al. 2008). Arc and MAP-1B are well-characterized targets of translation repression by FMRP. Below, we return to the question of how mGluRs couple specifically to FMRP-regulated protein synthesis.
The Mglur Theory of Fragile X
As mentioned above, it is now appreciated that Gp1 mGluRs couple to the translational machinery at many synapses in the brain. The mGluR theory of FX posits that many psychiatric and neurological aspects of FX are due to exaggerated downstream consequences of mGluR1/5 activation (Bear et al. 2004). The origins of this theory have been reviewed recently elsewhere (Krueger & Bear 2011). Briefly, Huber et al. (2000) showed that one protein synthesis-dependent consequence of Gp1 mGluR activation in the CA1 region of the hippocampus is a form of LTD, later shown to be expressed by internalization of AMPA-type glutamate receptors (Snyder et al. 2001). The early finding that FMRP can be synthesized in response to mGluR activation (Weiler et al. 1997) led to the study of LTD in the Fmr1-KO mouse (Huber et al. 2002). The prediction at that time was that absence of FMRP would result in impaired LTD, given the hypothesis that FMRP was one of the proteins synthesized to stabilize LTD. Instead, LTD was found to be exaggerated, suggesting that FMRP serves as a brake on mGluR-stimulated protein synthesis. As reviewed above, strong consensus now indicates that FMRP is a translational suppressor in vivo. The mGluR theory arose from the recognition that exaggerated consequences of mGluR activation at synapses throughout the nervous system could potentially provide a thread to connect seemingly unrelated FX phenotypes.
In the intervening decade, researchers have accumulated evidence that strongly supports the mGluR theory. The assumption that FMRP regulates varied responses triggered by mGluR-stimulated protein synthesis has been well validated (Auerbach & Bear 2010, Chuang et al. 2005, Dolen et al. 2007, Hou et al. 2006, Huber et al. 2002, Koekkoek et al. 2005, Lu et al. 2004, Muddashetty et al. 2007, Nosyreva & Huber 2006, Park et al. 2008, Ronesi & Huber 2008, Todd et al. 2003, Waung & Huber 2009, Westmark & Malter 2007, Zalfa et al. 2007, Zhang & Alger 2010, Zhao et al. 2005). Moreover, as summarized in Table 1 and reviewed in greater detail elsewhere (Dolen et al. 2010, Krueger & Bear 2011), the important prediction that FX phenotypes can be corrected by reducing mGluR5 activity has been confirmed using both pharmacological and genetic approaches in evolutionarily distant animal models (Aschrafi et al. 2005; Bolduc et al. 2008; Chang et al. 2008; Choi et al. 2010, 2011; Chuang et al. 2005; de Vrij et al. 2008; Dolen et al. 2007; Hays et al. 2011; Koekkoek et al. 2005; Levenga et al. 2011; Liu et al. 2011; Malter et al. 2010; McBride et al. 2005; Meredith et al. 2011; Min et al. 2009; Nakamoto et al. 2007; Osterweil et al. 2010; Pan & Broadie 2007; Pan et al. 2008; Repicky & Broadie 2009; Su et al. 2011; Suvrathan et al. 2010; Tauber et al. 2011; Thomas et al. 2011, 2012; Tucker et al. 2006; Veloz et al. 2012; Yan et al. 2005). A way of conceptualizing the constellation of findings is that FX is a disorder of excess—an excess that develops as Gp1 mGluR-dependent signaling cascades operate unchecked and that can be corrected by intervening at the first step in the cascade, the mGluR. The evolutionarily conserved relationship of Gp1 mGluRs and FMRP has provided a strong rationale for studies in human FX (see review by Hagerman et al. 2012).
Table 1. Phenotypes corrected by mGluR1/5 inhibition in animal models of FX*.
| Animal model | Fragile X phenotype (versus WT) | mGluR1/5 manipulation | Reference(s) |
|---|---|---|---|
| Mouse | Exaggerated mGluR-LTD |
Grm5+/- cross Lithium |
Dolen et al. 2007 Choi et al. 2011 |
| Mouse | Increased AMPA receptor internalization | MPEP | Nakamoto et al. 2007 |
| Mouse | Impaired spontaneous EPSCs in juvenile hippocampus | MPEP | Meredith et al. 2011 |
| Mouse | Increased protein synthesis |
Grm5+/- cross MPEP (mGluR5 NAM) Lithium |
Dolen et al. 2007 Osterweil et al. 2010 Liu et al. 2011 |
| Mouse | Decreased number of mRNA granules in whole brain | MPEP | Aschrafi et al. 2005 |
| Mouse | Increased glycogen synthase kinase-3 activity | MPEP, Lithium |
Min et al. 2009 Gross et al. 2010 |
| Mouse | Increased beta amyloid | MPEP | Malter et al. 2010 |
| Mouse | Increased dendritic spine/filopodia density |
Grm5+/- cross Fenobam (mGluR5 NAM) MPEP AFQ056 (mGluR5 NAM) |
Dolen et al. 2007 de Vrij et al. 2008 Su et al. 2011 Levenga et al. 2011 |
| Mouse | Altered visual cortical plasticity | Grm5+/- cross | Dolen et al. 2007 |
| Mouse | Exaggerated inhibitory avoidance extinction | Grm5+/- cross | Dolen et al. 2007 |
| Mouse | Impaired eyelid conditioning | MPEP | Koekkoek et al. 2005 |
| Mouse | Decreased initial performance on rotorod | MPEP | Thomas et al. 2012 |
| Mouse | Associative motor-learning deficit | Fenobam | Veloz et al. 2012 |
| Mouse | Increased audiogenic seizure |
Grm5+/- cross MPEP Lithium JNJ16259685 (mGluR1NAM) |
Dolen et al. 2007 Thomas et al. 2012, Yan et al. 2005 Min et al. 2009 Thomas et al. 2012 |
| Mouse | Prolonged epileptiform discharges in hippocampus | MPEP | Chuang et al. 2005 |
| Mouse | Increased persistent activity states in neocortex | MPEP, Grm5+/- cross | Hays et al. 2011 |
| Mouse | Increased open-field activity | MPEP Lithium Grm1 +/- cross |
Min et al. 2009, Yan et al. 2005 Thomas et al. 2011 |
| Mouse | Defective prepulse inhibition of acoustic startle | MPEP AFQ056 |
de Vrij et al. 2008 Levenga et al. 2011 |
| Mouse | Abnormal social interaction with unfamiliar mouse | Grm5+/- cross | Thomas et al. 2011 |
| Mouse | Increased marble burying (repetitive behavior) | JNJ16259685, MPEP | Thomas et al. 2012 |
| Mouse | Impaired presynaptic function in amygdala | MPEP | Suvrathan et al. 2010 |
| Mouse | Avoidance behavior deficit | Fenobam | Veloz et al. 2012 |
| Mouse | Pubertal increase in body weight | Grm5+/- cross | Dolen et al. 2007 |
| Zebrafish | Abnormal axon branching | MPEP | Tucker et al. 2006 |
| Zebrafish | Craniofacial abnormalities | MPEP | Tucker et al. 2006 |
| Zebrafish | Reduced number of trigeminal neurons | MPEP | Tucker et al. 2006 |
| Fly | Increased synaptic transmission | dmGluR-A null cross | Repicky & Broadie 2009 |
| Fly | Increased NMJ axon arborization | dmGluR-A null cross, MPEP | Pan et al. 2008 |
| Fly | Increased NMJ presynaptic vesicle density | dmGluR-A null cross | Pan et al. 2008 |
| Fly | Mushroom-body structural abnormalities | MPEP, lithium | McBride et al. 2005, Pan et al. 2008 |
| Fly | Age-dependent cognitive decline | MPEP, lithium | Choi et al. 2010 |
| Fly | Altered regulation of ionotropic glutamate receptor subtypes | dmGluR-A null cross | Pan & Broadie 2007 |
| Fly | Decreased courtship/social learning | MPEP, lithium | McBride et al. 2005, Tauber et al. 2011 |
| Fly | Decreased olfactory memory | MPEP | Bolduc et al. 2008 |
| Fly | Increased embryonic lethality on glutamate enriched diet | MPEP | Chang et al. 2008 |
| Fly | Increased roll-over (righting) time | dmGluR-A null cross | Pan et al. 2008 |
Abbreviations: AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; EPSC, excitatory postsynaptic currents; FX, fragile X; dmGluR, drosophila mGluR; LTD, long-term depression; mGluR, metabotropic glutamate receptor; MPEP, 2-methyl-6-(phenylethynyl)-pyridine; mRNA, messenger RNA; NAM, negative allosteric modulator; NMJ, neuromuscular junction; WT, wild type.
However, given when and where FMRP is normally expressed during development, it is clear that FX is a result of more than just altered mGluR signaling. Furthermore, because FMRP regulates signaling initiated by other neuronal receptors (Lee et al. 2011, Volk et al. 2007), reduction of Gp1 mGluR signaling seems unlikely to have a therapeutic benefit across all cognitive and somatic domains of what is a complex and pervasive neurodevelopmental disorder. Accordingly, efforts are under way to identify the aspects of FX pathophysiology that may not be related to mGluR function or that may arise before birth (Desai et al. 2006; Dolen et al. 2007; Suvrathan et al. 2010; Tauber et al. 2011; Thomas et al. 2011, 2012). Such knowledge is important in guiding therapy, both by defining the limits of what to expect from mGluR-based approaches and by suggesting additional therapeutic targets (see Fragile X Mental Retardation Protein and Neurogenesis, sidebar below).
How Mglur4 Couples to Fmrp-Regulated Protein Synthesis
Although the mGluR theory of FX has been well validated, it remains poorly understood how mGluR5 couples to protein synthesis and how this process is altered in the absence of FMRP to disrupt synaptic function. In addition to providing additional insight into FX pathophysiology and suggesting new therapeutic targets, investigating this question promises to shed light on long-standing but unresolved questions concerning how protein synthesis stabilizes LTD, LTP, and memory.
mGluR5 Signaling Pathways
Gp1 mGluRs were originally discovered on the basis of their ability to stimulate phospholipase C, the hydrolysis of phosphoinositides (PI), and the release of calcium from intracellular stores (Dudek et al. 1989, Nicoletti et al. 1988, Pin et al. 1992, Schoepp & Conn 1993). One phosphoinositide product, DAG (diacyl-glycerol), subsequently activates protein kinase C and protein kinase D (Krueger et al. 2010). This canonical signaling cascade does not appear to be critically involved in FMRP-regulated protein synthesis, however, as mGluR-LTD is insensitive to Ca2+ chelators and inhibitors of phospholipase C (Fitzjohn et al. 2001, Gallagher et al. 2004, Huber et al. 2001). Rather, signaling via the mTOR and ERK pathways is crucial for LTD and mGluR coupling to protein synthesis (Figure 5).
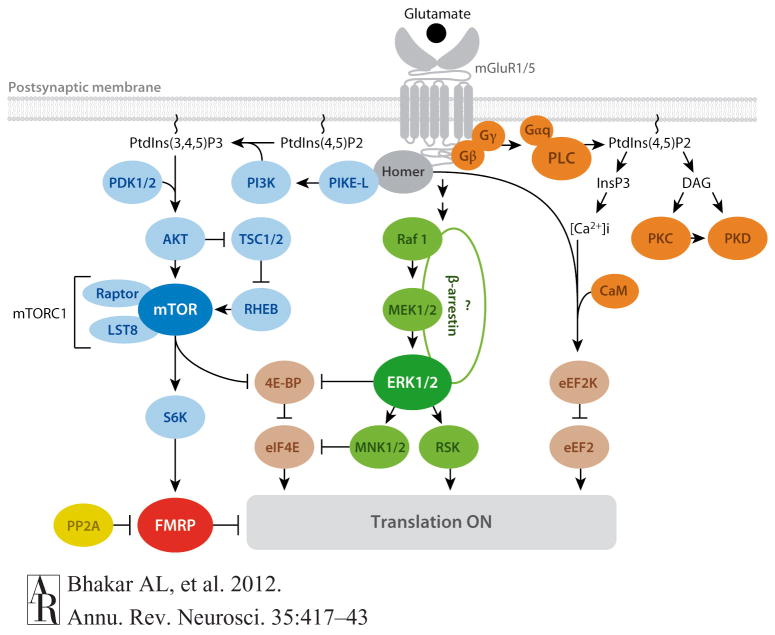
Figure 5.
mGluR1/5 signaling pathways relevant to protein synthesis. Glutamate binding to Gp1 mGluRs activates three main pathways that couple the receptors to translational regulation: (a) the PLC/calcium-calmodulin pathway (orange ovals), (b) the mTOR pathway (blue ovals), and (c) the ERK pathway (green ovals). See main text for details. Key translational regulatory components implicated in these pathways are shown in brown. mGluR1/5 may also inhibit FMRP (red oval) function to regulate translation through a fourth pathway requiring stimulation of PP2A (yellow oval). Question marks indicate undetermined associations. Arrows indicate a positive consequence on downstream components; perpendicular lines indicate an inhibitory consequence. Abbreviations: [Ca2+]i, calcium release from intracellular stores; CaM, calmodulin; ERK, extracellular signal–regulated kinase; FMRP, fragile X mental retardation protein; (Gαq, Gβ, Gγ), heterotrimeric G proteins; InsP3, inositol-1,4,5-triphosphate (InsP3); mGluR, metabotropic glutamate receptor; mTOR, mammalian target of rapamycin; PtdIns, phosphoinositides; PLC, phospholipase C; PP2A, protein phosphatase 2A; Raptor, regulatory-associated protein of mTOR.
To activate the mTOR pathway, mGluR5 couples to Homer, a postsynaptic-density scaffolding protein that recruits the GTPase, PIKE-L, forming an mGluR-Homer-PIKE complex (Ahn & Ye 2005). PIKE directly enhances the lipid kinase activity of PI3K (phosphoinositide 3-kinase), leading to the phosphorylation of PIP2 (phosphatidylinositol-4,5-bisphosphate). PIP3 together with PDK (phosphoinositide-dependent kinase) activates the serine/threonine kinase Akt. Akt, in turn, can activate mTOR by direct phosphorylation and indirectly through inhibition of the tumor-suppressor complex, composed of TSC1 and TSC2 (for review see Han & Sahin 2011). The TSC1/2 complex has GAP activity against the small GTP-binding protein RHEB. When free of TSC1/2, RHEB activates mTOR within a rapamycin-sensitive protein complex called mTORC1. Activation of mTORC1 is best known for stimulating cap-dependent translation through its main effector proteins, namely the 4E-BPs and S6Ks (see above section).
The ERK cascade, as with all mitogen-activated protein kinase (MAPK) cascades, typically involves sequential activation of a small GTPase (Ras), a MAPK kinase kinase (Raf), and a MAPK kinase (MEK), to activate ERK. How mGluR5 couples to Ras or other downstream components of the ERK cascade is not fully understood. ERK activation is required for both mGluR LTD (Gallagher et al. 2004) and mGluR5 activation of FMRP-regulated mRNA translation (Osterweil et al. 2010).
Recent work on related G-protein-coupled receptors (GPCRs) suggest that mGluR5 may couple to the ERK cascade through β-arrestins. β-arrestins are scaffold proteins that are typically recruited to the receptor tails following serine/threonine phosphorylation by GPCR kinases (GRKs)—a response that is best understood for terminating the receptor's G-protein signaling (Ferguson 2001, Premont et al. 1995). However, more recent work has shown that β-arrestin binding to GPCRs may also serve to regulate mRNA translation by providing a scaffold for Raf, MEK, ERK, and MNK (DeWire et al. 2008).
Interestingly, FMRP appears to be a component of the signaling pathway that couples mGluR5 activation to protein synthesis. As mentioned above, dephosphorylation shifts FMRP from stalled to active polyribosomes (Ceman et al. 2003, Muddashetty et al. 2011), motivating a few groups to identify FMRP phosphatases and kinases that lie downstream of mGluR5. S6K1 can phosphorylate FMRP on a conserved serine residue required for mRNA binding and PP2A can remove this phosphorylation (Mao et al. 2005, Narayanan et al. 2007, Narayanan et al. 2008, Wang et al. 2010a). Both enzymes are activated in response to mGluR5 stimulation, and one model proposes that activation of PP2A rapidly dephosphorylates FMRP to enable translation, followed by delayed translation suppression caused by S6K phosphorylation of FMRP downstream of mTOR (Santoro et al. 2011).
Regulation of mGluR5-dependent protein synthesis exclusively via FMRP is unlikely, however. In the Fmr1-KO mouse, which lacks FMRP, the excessive basal protein synthesis (and many other phenotypes) are rescued by inhibiting mGluR5 (Figure 3). If loss of FMRP completely uncoupled mGluR5 from protein synthesis regulation, there would be no effect of inhibiting mGluR5 on protein synthesis in the Fmr1 KO. Therefore, mGluR5 stimulation of protein synthesis must occur via additional pathway(s) that are independent of FMRP (Figure 6).
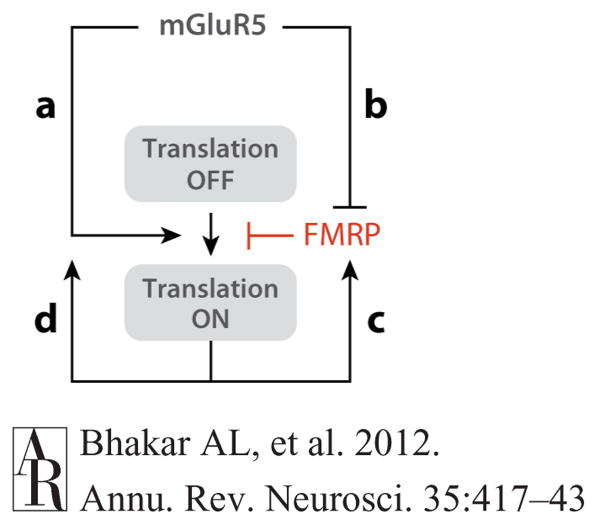
Figure 6.
Schema for coupling mGluR5 to FMRP-regulated protein synthesis. Several lines of evidence suggest that mGluR5 couples to FMRP-regulated protein synthesis through multiple pathways. (a) Activation of mGluR5 directly stimulates mRNA translation through the ERK signaling pathway. (b) Additionally, activation of mGluR5 can trigger dephosphorylation of FMRP by PP2A, which derepresses translation. (c) FMRP is rapidly synthesized in response to mGluR5 activation, providing a negative-feedback loop to turn off protein synthesis. (d) Several FMRP target proteins are known components of mGluR5 signaling pathways, suggesting that positive feedback may occur, particularly in the context of FX. Abbreviations: ERK, extracellular signal–regulated kinase; FMRP, fragile X mental retardation protein; FX, fragile X; mGluR, metabotropic glutamate receptor; PP2A, protein phosphatase 2A.
Altered Signaling in the Absence of FMRP
Because both ERK and mTOR pathways can be activated by mGluR5 (Antion et al. 2008, Banko et al. 2006, Ferraguti et al. 1999, Gallagher et al. 2004, Hou et al. 2006, Ronesi & Huber 2008, Sharma et al. 2010) and both regulate protein synthesis, these two pathways have been most studied in the context of FX. One hypothesis has been that alterations in mGluR5 signaling through ERK or mTOR may be responsible for the excessive protein synthesis and exaggerated LTD in the Fmr1-KO mice. Consistent with the notion of altered signaling, mGluR5 receptors are less tightly associated with synaptic plasma membrane and Homer (Giuffrida et al. 2005), and they are unable to activate the mTOR pathway in Fmr1-KO mice (Ronesi & Huber 2008). Other reports suggest a basal increase in ERK activity (Hou et al. 2006), an aberrant mGluR-induced inactivation of ERK (Kim et al. 2008), and a basal increase in AKT/mTOR signaling that occludes further activation by mGluR stimulation (Gross et al. 2010, Sharma et al. 2010).
Although mGluR5 signaling is evidently altered in FX, it has not been shown that these alterations are responsible for the excessive protein synthesis that is believed to be the core pathogenic mechanism in FX. Indeed, one recent study examined ERK and mTOR pathways under the same experimental conditions that reveal excessive protein synthesis and exaggerated LTD and found no evidence for altered signaling (Osterweil et al. 2010). Protein synthesis rates could be restored to WT levels by acute partial inhibition of mGluR5 or ERK activity (but not mTOR), however, indicating that increased protein synthesis in FX occurs downstream of constitutive mGluR5/ERK activity (Osterweil et al. 2010). These data suggest that the excessive basal protein synthesis in Fmr1-KO mice is due to hypersensitivity of the translation machinery to normal mGluR signals (ERK, in particular), rather than to hyperactivity of the mGluR signaling pathways (Figure 6). If this model is correct, altered intracellular signaling in FX should be viewed as a consequence, rather than a cause, of the increased protein synthesis in this disease.
ERK and mTOR May Regulate Separate Pools of mRNA
Disentangling the contributions of ERK and mTOR signaling pathways to the protein synthesis required for mGluR-LTD has been difficult, but recent studies of a mouse model of tuberous sclerosis complex (TSC) have been illuminating. TSC is another single-gene disorder characterized by intellectual disability, seizures, and autism and is caused by heterozygous loss of function of either the TSC1 or TSC2 gene. The protein products of these genes form the TSC1/2 complex that normally represses mTOR signaling via inhibition of RHEB, as discussed above. Thus, TSC is caused by excessive mTOR signaling. If the excessive protein synthesis in FX were driven by the mTOR signaling pathway, one would expect TSC mutations to have similar effects on mGluR-dependent LTD. Very recently, three groups examined this hypothesis in the CA1 region of hippocampus using different but complementary animal models of TSC (Auerbach et al. 2011, Bateup et al. 2011, Chevere-Torres et al. 2012). The surprising result is that mouse Tsc mutants with excessive mTOR activity show impaired mGluR-LTD and basal protein synthesis, the exact opposite of what is observed in the Fmr1-KO. Moreover, synaptic, biochemical, and cognitive deficits in the Tsc2+/− mouse model were corrected by treatment with a positive allosteric modulator of mGluR5 as well as by introducing the FX mutation into the Tsc2+/− animals (Auerbach et al. 2011). These findings indicate that elevated mTOR signaling is not a proximal cause of FX pathophysiology.
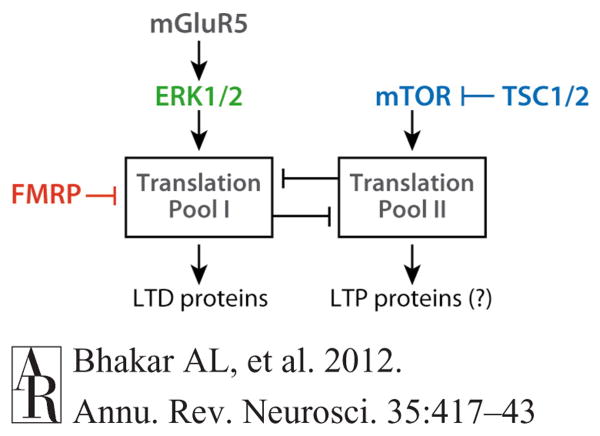
The recent findings in Tsc mutants suggest that excessive mTOR signaling suppresses the synthesis of proteins required for LTD (Auerbach et al. 2011). One simple hypothesis is that elevated mTOR causes hyperphosphorylation of FMRP via activation of S6K1 (Figure 5), resulting in translational suppression of the FMRP-target mRNAs that gate LTD. However, this explanation is not easily reconciled with the observation that excess LTD in the Fmr1-KO mice (lacking FMRP) is rescued by crossing them with the Tsc2+/− mice. An alternative model is that mTOR stimulates translation of a pool of mRNA (call it Pool II) that competes with a second, ERK- and FMRP-regulated pool (Pool I) for access to the translation machinery (Figure 7) (see also Bear et al. 2004).
Figure 7.
The two-pool hypothesis. A model to account for the opposing mGluR5 responses detected in the Tsc2+/− and Fmr1-KO mice proposes that activation of mGluR5 stimulates the translation of a pool of mRNAs (Pool I), through ERK- and FMRP-dependent pathways, that are in competition for the translational machinery with a second pool of mRNAs (Pool II) that are regulated by mTOR activation. Current data suggest that mRNAs translated in Pool I may comprise the proteins required to stabilize LTD (LTD proteins), whereas mRNAs within Pool II stabilize LTP (LTP proteins). Consistent with this proposal, derepression of Pool I in FX causes excessive LTD, whereas derepression of Pool II in TSC causes enhanced LTP. Arrows indicate a positive consequence on downstream components; perpendicular lines indicate an inhibitory consequence. Abbreviations: ERK, extracellular signal–regulated kinase; FMRP, fragile X mental retardation protein; FX, fragile X; KO, knockout; LTD, long-term synaptic depression; LTP, long-term synaptic potentiation; mGluR, metabotropic glutamate receptor; mTOR, mammalian target of rapamycin; TSC, tuberous sclerosis complex.
As mentioned above, there is considerable precedent for a “push-pull” regulation of translation by different pools of mRNA. Inhibition of what is often called general translation enables certain types of specific translation of mRNAs that can include FMRP targets. Although this can occur via multiple mechanisms, to illustrate consider regulation of translation via the elongation factor eEF2. Phosphorylation of eEF2 by eEF2 kinase occurs in response to mGluR5 activation and promotes translation of specific transcripts in Pool I (including those for the FMRP targets Arc and MAP1b) by inhibiting translation of Pool II transcripts (Park et al. 2008). Conversely, activation of the mTOR pathway causes inhibitory phosphorylation of the eEF2 kinase (via S6 kinase), which stimulates translation of Pool II and thereby inhibits translation of Pool I (Costa-Mattioli et al. 2009, Herbert & Proud 2007).
Two distinct effects on protein synthesis–dependent synaptic plasticity have been reported in Tsc2 mutants with increased mTOR activity: (a) The persistence of late-phase LTP is increased, presumably by increasing translation of the proteins required to make synapses stronger (Ehninger et al. 2008), and (b) mGluR-LTD is inhibited by eliminating the protein synthesis required to make synapses weaker (Auerbach et al. 2011, Bateup et al. 2011). It is tempting to speculate that Pool II includes LTP proteins regulated by mTOR signaling and that Pool I comprises LTD proteins regulated by mGluR5, ERK, and FMRP. According to this idea, derepression of Pool I in FX causes excessive LTD, whereas derepression of Pool II in TSC causes enhanced LTP.
Such simple models are useful if they generate hypotheses and stimulate experiments. If this conjecture is correct, for example, proteomic comparison of Tsc2+/− and Fmr1-KO hippocampus may be a fruitful path to discover the elusive plasticity gating proteins. Of course, the regulation of plasticity-related protein synthesis is unlikely to be this simple. For example, the model suggests that LTP may be impaired in FX owing to repression of Pool II translation. Although there are some reports of deficient LTP in the hippocampus of Fmr1-KO mice (Hu et al. 2008, Lauterborn et al. 2007, Lee et al. 2011, Meredith & Mansvelder 2010, Shang et al. 2009), many have found no difference in LTP threshold or long-term maintenance (Auerbach & Bear 2010, Godfraind et al. 1996, Zhang et al. 2009). Another element of the model that requires further clarification is how activity couples to the mTOR pathway. A recent study showed that inhibition of the mTOR pathway derepresses translation of the Pool I mRNA Kv4.2, but that this occurs via dephosphorylation of FMRP downstream of NMDA receptors instead of mGluRs (Lee et al. 2011). Other studies have shown mTOR is activated by Gp1 mGluR activation and is required for LTD (Hou & Klann 2004; but see Auerbach et al. 2011). One thing is certain: Intracellular signaling is complicated. Clarity will require that experiments be performed on the same synapses, prepared in the same way, and from animals that are at the same age.
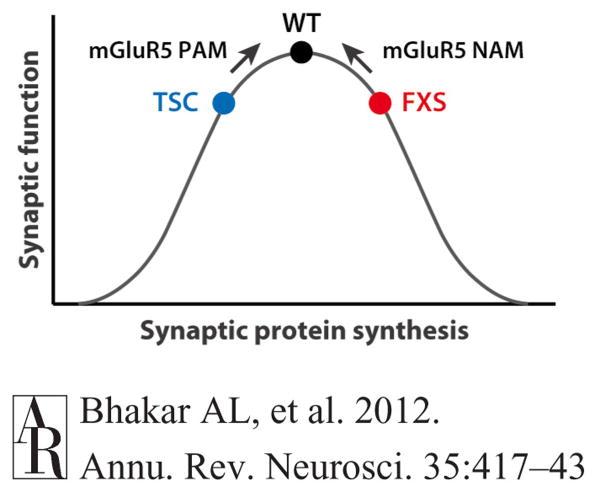
These caveats notwithstanding, under identical experimental conditions, littermate mice carrying the Fmr1 mutation, the Tsc2 mutation, and both mutations show augmented, impaired, and WT levels, respectively, of mGluR-dependent LTD and protein synthesis (Auerbach et al. 2011). Of particular interest, both single mutants showed deficits in context-discrimination memory that were erased in the double mutants. These findings support the ideas that proper synaptic function requires an optimal level of mGluR-regulated protein synthesis and that deviations in either direction can yield similar behavioral disturbances that can include cognitive impairment (Figure 8).
Figure 8.
Mutations causing monogenic autism define an axis of synaptic pathophysiology. Recent data suggest that proper synaptic function requires an optimal level of mGluR-regulated protein synthesis and that deviations in either direction can produce similar impairments in cognitive function (Auerbach et al. 2011). Two types of monogenic autism, TSC and FXS, lie on opposite ends of this spectrum and, correspondingly, show reduced and increased protein synthesis rates, and respond to opposite alterations in mGluR5 activation (PAM and NAM, respectively). Abbreviations: FXS, fragile X syndrome; mGluR, metabotropic glutamate receptor; NAM, negative allosteric modulator; PAM, positive allosteric modulator; TSC, tuberous sclerosis complex; WT, wild type.
Pathogenic Proteins
Evidence suggests that synaptically controlled protein synthesis must be maintained in a normal range to ensure proper synaptic (and cognitive) function, and that important aspects of FX are a consequence of altered protein expression. Several-hundred mRNAs have been implicated as targets of FMRP (Darnell et al. 2011). Among these are the proteins that disrupt synaptic function in FX, and it is of great interest to identify those that are pathogenic.
Given the reversal of FX phenotypes by reducing mGluR1/5 stimulation, one way to prioritize the list of pathogenic proteins may be to determine which of the identified direct targets show (a) altered protein expression profiles in the Fmr1-KO mice, (b) translation under normal circumstances in response to mGluR1/5 activation, and (c) a contribution to the functional responses to activated mGluR5, e.g., mGluR LTD. For example, the plasticity protein Arc is an identified FMRP mRNA target, upregulated in the Fmr1-KO mouse and synthesized at the synapse in response to mGluR5 activation (Auerbach et al. 2011, Park et al. 2008, Waung et al. 2008). Similarly, the amyloid precursor protein (APP) and the brain-specific tyrosine phosphatase STEP are FMRP mRNA targets, synthesized in response to mGluR5 (Westmark & Malter 2007, Westmark et al. 2009, Zhang et al. 2008), and both APP cleavage products and STEP protein are overexpressed in the Fmr1-KO mouse (Goebel-Goody et al. 2011). Arc and STEP are both considered to be LTD proteins, involved in regulating AMPA-receptor membrane trafficking. The cleavage product of APP, β-amyloid, also triggers AMPA receptor internalization and LTD (Hsieh et al. 2006). Of particular interest, removing a single allele of APP in the Fmr1 KO partially or completely corrects audiogenic seizure, anxiety, and mGluR LTD phenotypes (Westmark et al. 2011). Another FMRP target of interest is metalloproteinase 9 (MMP-9), also overexpressed in the Fmr1-KO downstream of mGluR5. MMP-9 is a secreted extracellular endopeptidase that, similar to Gp1 mGluR agonists (Vanderklish & Edelman 2002), elongates and thins dendritic spines (Michaluk et al. 2011). Treatment with the tetracycline analogue minocycline (among other actions) inhibits MMP-9 and corrects the spine phenotype in the Fmr1-KO mouse (Bilousova et al. 2009). Moreover, both minocycline and genetic reduction of MMP rescue circuit disruptions in the dfmr1-null fly model of FX (Siller & Broadie 2011).
Additional downstream consequences of altered synaptic protein expression may be dysregulation of the signaling components that normally control protein synthesis. For example, both the catalytic subunit of PI3K (p110b) and the PI3K enhancer PIKE-L are FMRP mRNA targets, translated in response to mGluR activation and elevated in the Fmr1-KO mice (Gross et al. 2010, Sharma et al. 2010). Indeed, 62% of the genes composing the mGluR5 postsynaptic proteome (Croning et al. 2009) are direct FMRP targets (Darnell et al. 2011). These findings fit with data showing abnormal mGluR5 signaling in FX.
The list of pathogenic proteins is sure to expand as additional research is conducted. Particularly interesting are those that can be targeted with small-molecule therapeutics. In addition to those mentioned above, interesting prospects include p21-activated kinase (Hayashi et al. 2007) and glycogen synthase kinase-3 (Mines & Jope 2011).
The overlap of FMRP targets and genes implicated in autism is intriguing. One-quarter of the SFARI database of autism risk genes (http://gene.sfari.org) are FMRP targets. Among these are NLGN3, NRXN1, SHANK3, PTEN, TSC2, and NF1, all of which encode proteins that control synaptic structure or protein synthesis. Rare mutations of these genes all cause autism (Zoghbi & Bear 2012). These findings reinforce the belief that the study of FX, the most common known genetic cause of autism, provides insight into the molecular pathophysiology of autism and associated intellectual disability of unknown etiology. The hope is that treatments developed for FX will be useful for treating autism of diverging etiologies, with the important caveat that it will be critical to understand where an individual is on the spectrum of altered synaptic protein synthesis to devise an appropriate therapy (Auerbach et al. 2011).
Concluding Remarks
Interest in FX has burgeoned in recent years. It is now appreciated to be a disease of the synapse, amenable to potentially disease-altering therapeutic interventions and relevant to understanding the pathophysiology of autism and intellectual disability more broadly. We appear to be close to fulfilling the promise of molecular medicine in FX (Krueger & Bear 2011). We have gone from identification of the gene to the discovery and validation of novel therapeutic targets, and there is good reason for optimism that new therapies will emerge that can greatly enhance the quality of life for affected individuals and their families (see Figure 1).
This field has grown so large that it is impossible to cover adequately all the developments given the space limitations of this review. We have chosen to focus on synaptic control of protein synthesis because it appears to be proximal to the biology of FMRP and the pathogenesis of the disease in multiple animal models. In addition to targeting synaptic protein synthesis, other approaches also show promise, for example, changing the balance of excitation to inhibition by enhancing GABA signaling (Hampson et al. 2011, Rooms & Kooy 2011). Whether different approaches will converge on the same pathophysiological processes or whether they will target distinct aspects of the disease remains to be determined. Regardless, understanding how synaptic transmission differs in FX holds the key to developing new therapies. Furthermore, the study of FX has greatly enriched our understanding of the neurobiology of synaptic transmission.
Supplementary Material
Fragile X Mental Retardation Protein An Neurogenesis.
The metabotropic glutamate receptor (mGluR) theory has contributed to a paradigm shift in the way fragile X (FX) and other genetic disorders of brain development are viewed medically. The data now indicate that a constellation of seemingly unrelated and complex symptoms could be a consequence of altered cerebral metabolism—synaptic protein synthesis in the case of FX—that can be substantially improved by therapies begun after symptom onset, possibly even in adulthood. It is important to recognize, however, that FMR1 is normally expressed early in embryogenesis (Devys et al. 1993, Hinds et al. 1993) and that full-mutation FX patients fail to express FMRP very early in gestation (Willemsen et al. 1996). FMRP is required for proper prenatal neurogenesis and neuronal differentiation (Callan et al. 2010, Castren et al. 2005, Eadie et al. 2009, Tervonen et al. 2009). Thus, the FX brain is different at birth.
However, neurogenesis occurs throughout life in the dentate gyrus of the hippocampus. Remarkably, hippocampus-dependent memory impairments have been rescued by re-expression of FMRP in adult neural stem cells in an Fmr1 knockout (KO) mouse line (Guo et al. 2011). Moreover, these defects can be reversed in adults by treatment with an inhibitor of glycogen synthase kinase 3 (GSK3) (Guo et al. 2012). GSK3 activity is elevated in the Fmr1 KO downstream of mGluR5 (Yuskaitis et al. 2009), suggesting that the mGluR theory may also be relevant to this aspect of FX pathophysiology.
Acknowledgments
The authors thank Dr. Emily Osterweil for help with the manuscript and the Howard Hughes Medical Institute, FRAXA Research Foundation, U.S. Department of Defense, National Institute of Child Health and Human Development, and National Institute of Mental Health for support.
Footnotes
Disclosure Statement: M.B. has a financial interest in Seaside Therapeutics, Inc.
Contributor Information
Asha L. Bhakar, Email: abhakar@mit.edu.
Gül Dölen, Email: gul@stanford.edu.
Mark F. Bear, Email: mbear@mit.edu.
References
- Ahn JY, Ye K. PIKE GTPase signaling and function. Int J Biol Sci. 2005;1:44–50. doi: 10.7150/ijbs.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–8. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–55. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–59. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Antion MD, Hou L, Wong H, Hoeffer CA, Klann E. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol Cell Biol. 2008;28:2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Cunningham BA, Edelman GM, Vanderklish PW. The fragile X mental retardation protein and group I metabotropic glutamate receptors regulate levels of mRNA granules in brain. Proc Natl Acad Sci USA. 2005;102:2180–85. doi: 10.1073/pnas.0409803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley C, Wilkinson K, Reines D, Warren S. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–66. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Auerbach BD, Bear MF. Loss of the fragile X mental retardation protein decouples metabotropic glutamate receptor dependent priming of long-term potentiation from protein synthesis. J Neurophysiol. 2010;104:1047–51. doi: 10.1152/jn.00449.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–73. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31:8862–69. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–77. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidinosti M, Ran I, Sanchez-Carbente MR, Martineau Y, Gingras AC, et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilousova TV, Dansie L, Ngo M, Aye J, Charles JR, et al. Minocycline promotes dendritic spine maturation and improves behavioural performance in the fragile X mouse model. J Med Genet. 2009;46:94–102. doi: 10.1136/jmg.2008.061796. [DOI] [PubMed] [Google Scholar]
- Bishu S, Schmidt KC, Burlin T, Channing M, Conant S, et al. Regional rates of cerebral protein synthesis measured with L-[1-11C]leucine and PET in conscious, young adult men: normal values, variability, and reproducibility. J Cereb Blood Flow Metab. 2008;28:1502–13. doi: 10.1038/jcbfm.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishu S, Schmidt KC, Burlin TV, Channing MA, Horowitz L, et al. Propofol anesthesia does not alter regional rates of cerebral protein synthesis measured with L-[1-(11)C]leucine and PET in healthy male subjects. J Cereb Blood Flow Metab. 2009;29:1035–47. doi: 10.1038/jcbfm.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum Mol Genet. 2010;19:1314–23. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–45. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Callan MA, Cabernard C, Heck J, Luois S, Doe CQ, Zarnescu DC. Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Hum Mol Genet. 2010;19:3068–79. doi: 10.1093/hmg/ddq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren M, Tervonen T, Karkkainen V, Heinonen S, Castren E, et al. Altered differentiation of neural stem cells in fragile X syndrome. Proc Natl Acad Sci USA. 2005;102:17834–39. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Hannon GJ. Induction and biochemical purification of RNA-induced silencing complex from Drosophila S2 cells. Methods Mol Biol. 2004;265:59–72. doi: 10.1385/1-59259-775-0:059. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–96. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Centonze D, Rossi S, Mercaldo V, Napoli I, Ciotti MT, et al. Abnormal striatal GABA transmission in the mouse model for the fragile X syndrome. Biol Psychiatry. 2008;63:963–73. doi: 10.1016/j.biopsych.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Chang S, Bray SM, Li Z, Zarnescu DC, He C, et al. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–63. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. RNA. 2009;15:362–66. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–17. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Chevere-Torres I, Kaphzan H, Bhattacharya A, Kang A, Maki JM, et al. Metabotropic glutamate receptor-dependent long-term depression is impaired due to elevated ERK signaling in the DeltaRG mouse model of tuberous sclerosis complex. Neurobiol Dis. 2012;45(3):1101–10. doi: 10.1016/j.nbd.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CH, McBride SM, Schoenfeld BP, Liebelt DA, Ferreiro D, et al. Age-dependent cognitive impairment in a Drosophila fragile X model and its pharmacological rescue. Biogerontology. 2010;11:347–62. doi: 10.1007/s10522-009-9259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CH, Schoenfeld BP, Bell AJ, Hinchey P, Kollaros M, et al. Pharmacological reversal of synaptic plasticity deficits in the mouse model of fragile X syndrome by group II mGluR antagonist or lithium treatment. Brain Res. 2011;1380:106–19. doi: 10.1016/j.brainres.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SC, Zhao W, Bauchwitz R, Yan Q, Bianchi R, Wong RK. Prolonged epileptiform discharges induced by altered group I metabotropic glutamate receptor-mediated synaptic responses in hippocampal slices of a fragile X mouse model. J Neurosci. 2005;25:8048–55. doi: 10.1523/JNEUROSCI.1777-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Keith K, Albizua I, Malone T, Mowrey J, et al. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet. 2009;85:503–14. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee RL, Jr, Williamson AJ, Adkins CM, Gray MC, Page TL, Broadie K. In vivo neuronal function of the fragile X mental retardation protein is regulated by phosphorylation. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, et al. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 2010;152A:2512–20. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–72. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracco JB, Serrano P, Moskowitz SI, Bergold PJ, Sacktor TC. Protein synthesis-dependent LTP in isolated dendrites of CA1 pyramidal cells. Hippocampus. 2005;15:551–56. doi: 10.1002/hipo.20078. [DOI] [PubMed] [Google Scholar]
- Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the Genes to Cognition database. Nucleic Acids Res. 2009;37:D846–51. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC. Defects in translational regulation contributing to human cognitive and behavioral disease. Curr Opin Genet Dev. 2011;21:465–73. doi: 10.1016/j.gde.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, et al. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–18. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- De Diego Otero Y, Severijnen LA, van Cappellen G, Schrier M, Oostra B, Willemsen R. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol Cell Biol. 2002;22:8332–41. doi: 10.1128/MCB.22.23.8332-8341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrij FM, Levenga J, van der Linde HC, Koekkoek SK, De Zeeuw CI, et al. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol Dis. 2008;31:127–32. doi: 10.1016/j.nbd.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai NS, Casimiro TM, Gruber SM, Vanderklish PW. Early postnatal plasticity in neocortex of Fmr1 knockout mice. J Neurophysiol. 2006;96:1734–45. doi: 10.1152/jn.00221.2006. [DOI] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–40. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- DeWire SM, Kim J, Whalen EJ, Ahn S, Chen M, Lefkowitz RJ. Beta-arrestin-mediated signaling regulates protein synthesis. J Biol Chem. 2008;283:10611–20. doi: 10.1074/jbc.M710515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–39. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell. 2009;20:428–37. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010;127:78–93. doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–62. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek SM, Bowen WD, Bear MF. Postnatal changes in glutamate stimulated phosphoinositide turnover in rat neocortical synaptoneurosomes. Brain Res Dev Brain Res. 1989;47:123–28. doi: 10.1016/0165-3806(89)90114-4. [DOI] [PubMed] [Google Scholar]
- Dutch-Belgian Fragile X Consort. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Eadie BD, Zhang WN, Boehme F, Gil-Mohapel J, Kainer L, et al. Fmr1 knockout mice show reduced anxiety and alterations in neurogenesis that are specific to the ventral dentate gyrus. Neurobiol Dis. 2009;36:361–73. doi: 10.1016/j.nbd.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–91. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–84. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843–48. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahling M, Mrowka R, Steege A, Kirschner KM, Benko E, et al. Translational regulation of the human achaete-scute homologue-1 by fragile X mental retardation protein. J Biol Chem. 2009;284:4255–66. doi: 10.1074/jbc.M807354200. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997a;1:109–18. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997b;17:1539–47. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SS. Evolving concepts in G protein–coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53:1–24. [PubMed] [Google Scholar]
- Ferraguti F, Baldani-Guerra B, Corsi M, Nakanishi S, Corti C. Activation of the extracellular signal-regulated kinase 2 by metabotropic glutamate receptors. Eur J Neurosci. 1999;11:2073–82. doi: 10.1046/j.1460-9568.1999.00626.x. [DOI] [PubMed] [Google Scholar]
- Fitzjohn SM, Palmer MJ, May JE, Neeson A, Morris SA, Collingridge GL. A characterisation of long-term depression induced by metabotropic glutamate receptor activation in the rat hippocampus in vitro. J Physiol. 2001;537:421–30. doi: 10.1111/j.1469-7793.2001.00421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–58. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J Neurosci. 2004;24:10579–83. doi: 10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–37. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SM, Daly CA, Bear MF, Huber KM. Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. J Neurosci. 2004;24:4859–64. doi: 10.1523/JNEUROSCI.5407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrida R, Musumeci S, D'Antoni S, Bonaccorso CM, Giuffrida-Stella AM, et al. A reduced number of metabotropic glutamate subtype 5 receptors are associated with constitutive homer proteins in a mouse model of fragile X syndrome. J Neurosci. 2005;25:8908–16. doi: 10.1523/JNEUROSCI.0932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfraind JM, Reyniers E, De Boulle K, D'Hooge R, De Deyn PP, et al. Long-term potentiation in the hippocampus of fragile X knockout mice. Am J Med Genet. 1996;64:246–51. doi: 10.1002/(SICI)1096-8628(19960809)64:2<246::AID-AJMG2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Goebel-Goody SM, Baum M, Paspalas CD, Fernandez SM, Carty NC, et al. Therapeutic implications for striatal-enriched protein tyrosine phosphatase (STEP) in neuropsychiatric disorders. Pharmacol Rev. 2011;64:65–87. doi: 10.1124/pr.110.003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Nakamoto M, Yao X, Chan CB, Yim SY, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–38. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Allan AM, Zong R, Zhang L, Johnson EB, et al. Ablation of FMRP in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17:559–65. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Murthy AC, Zhang L, Johnson EB, Schaller EG, et al. Inhibition of GSK3β improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–91. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman R, Lauterborn J, Au J, Berry-Kravis E. Fragile x syndrome and targeted treatment trials. Results Probl Cell Differ. 2012;54:297–335. doi: 10.1007/978-3-642-21649-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson DR, Adusei DC, Pacey LK. The neurochemical basis for the treatment of autism spectrum disorders and Fragile X Syndrome. Biochem Pharmacol. 2011;81:1078–86. doi: 10.1016/j.bcp.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Han JM, Sahin M. TSC1/TSC2 signaling in the CNS. FEBS Lett. 2011;585:973–80. doi: 10.1016/j.febslet.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton DD, Sideris J, Skinner M, Mankowski J, Bailey DB, Jr, et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A. 2006;140A:1804–13. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Rao BS, Seo JS, Choi HS, Dolan BM, et al. Inhibition of p21-activated kinase rescues symptoms of fragile X syndrome in mice. Proc Natl Acad Sci USA. 2007;104:11489–94. doi: 10.1073/pnas.0705003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SA, Huber KM, Gibson JR. Altered neocortical rhythmic activity states in Fmr1 KO mice are due to enhanced mGluR5 signaling and involve changes in excitatory circuitry. J Neurosci. 2011;31:14223–34. doi: 10.1523/JNEUROSCI.3157-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert TP, Proud CG. Regulation of translation elogation and the cotranslational protein target pathway. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2007. pp. 601–24. [Google Scholar]
- Hinds HL, Ashley CT, Sutcliffe JS, Nelson DL, Warren ST, et al. Tissue-specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–54. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–61. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H, Boehm J, Sato C, Iwatsubo T, Tomita T, et al. AMPAR removal underlies Aβ-induced synaptic depression and dendritic spine loss. Neuron. 2006;52:831–43. doi: 10.1016/j.neuron.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Qin Y, Bochorishvili G, Zhu Y, van Aelst L, Zhu JJ. Ras signaling mechanisms underlying impaired GluR1-dependent plasticity associated with fragile X syndrome. J Neurosci. 2008;28:7847–62. doi: 10.1523/JNEUROSCI.1496-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–50. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–57. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis-dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86:321–5. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, et al. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008a;105:734–39. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, et al. Reply to Bagni: on BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008b;105:E29. doi: 10.1073/pnas.0803737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–17. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, et al. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–43. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–38. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–6. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Abrams MT, Chen W, Reiss AL. Genotype, molecular phenotype, and cognitive phenotype: correlations in fragile X syndrome. Am J Med Genet. 1999;83:286–95. [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Fortin A, Thibodeau A, Tremblay S, Cote F, et al. A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum Mol Genet. 1995;4:783–89. doi: 10.1093/hmg/4.5.783. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–90. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol Cell Biol. 2009;29:214–28. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Markham JA, Weiler IJ, Greenough WT. Aberrant early-phase ERK inactivation impedes neuronal function in fragile X syndrome. Proc Natl Acad Sci USA. 2008;105:4429–34. doi: 10.1073/pnas.0800257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koekkoek SK, Yamaguchi K, Milojkovic BA, Dortland BR, Ruigrok TJ, et al. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron. 2005;47:339–52. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–96. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–29. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DD, Osterweil EK, Bear MF. Activation of mGluR5 induces rapid and long-lasting protein kinase D phosphorylation in hippocampal neurons. J Mol Neurosci. 2010;42:1–8. doi: 10.1007/s12031-010-9338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoux C, Di Marino D, Pilo Boyl P, Zalfa F, Yan B, et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–38. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lauterborn JC, Rex CS, Kramar E, Chen LY, Pandyarajan V, et al. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J Neurosci. 2007;27:10685–94. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Ge WP, Huang W, He Y, Wang GX, et al. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron. 2011;72:630–42. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenga J, de Vrij FM, Oostra BA, Willemsen R. Potential therapeutic interventions for fragile X syndrome. Trends Mol Med. 2010;16:516–27. doi: 10.1016/j.molmed.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenga J, Hayashi S, de Vrij FM, Koekkoek SK, van der Linde HC, et al. AFQ056, a new mGluR5 antagonist for treatment of fragile X syndrome. Neurobiol Dis. 2011;42:311–17. doi: 10.1016/j.nbd.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZH, Huang T, Smith CB. Lithium reverses increased rates of cerebral protein synthesis in a mouse model of fragile X syndrome. Neurobiol Dis. 2011;45:1145–52. doi: 10.1016/j.nbd.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch DZ, Huggins R, Petrovic V, Slater H. Expansion of the CGG repeat in fragile X in the FMR1 gene depends on the sex of the offspring. Am J Hum Genet. 1995;57:1408–13. [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O'Donnell WT, et al. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci USA. 2004;101:15201–6. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet. 1995;10:483–85. doi: 10.1038/ng0895-483. [DOI] [PubMed] [Google Scholar]
- Maghsoodi B, Poon MM, Nam CI, Aoto J, Ting P, Chen L. Retinoic acid regulates RARα-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc Natl Acad Sci USA. 2008;105:16015–20. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter JS, Ray BC, Westmark PR, Westmark CJ. Fragile X syndrome and Alzheimer's disease: another story about APP and beta-amyloid. Curr Alzheimer Res. 2010;7:200–6. doi: 10.2174/156720510791050957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Yang L, Arora A, Choe ES, Zhang G, et al. Role of protein phosphatase 2A in mGluR5-regulated MEK/ERK phosphorylation in neurons. J Biol Chem. 2005;280:12602–10. doi: 10.1074/jbc.M411709200. [DOI] [PubMed] [Google Scholar]
- Martin JP, Bell J. A pedigree of mental defect showing sex-linkage. J Neurol Psychiatry. 1943;6:154–57. doi: 10.1136/jnnp.6.3-4.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–17. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–64. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Meredith RM, de Jong R, Mansvelder HD. Functional rescue of excitatory synaptic transmission in the developing hippocampus in Fmr1-KO mouse. Neurobiol Dis. 2011;41:104–10. doi: 10.1016/j.nbd.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Meredith RM, Mansvelder HD. STDP and mental retardation: dysregulation of dendritic excitability in fragile X syndrome. Front Synaptic Neurosci. 2010;2:10. doi: 10.3389/fnsyn.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaluk P, Wawrzyniak M, Alot P, Szczot M, Wyrembek P, et al. Influence of matrix metalloproteinase MMP-9 on dendritic spine morphology. J Cell Sci. 2011;124:3369–80. doi: 10.1242/jcs.090852. [DOI] [PubMed] [Google Scholar]
- Min WW, Yuskaitis CJ, Yan Q, Sikorski C, Chen S, et al. Elevated glycogen synthase kinase-3 activity in Fragile X mice: key metabolic regulator with evidence for treatment potential. Neuropharmacology. 2009;56:463–72. doi: 10.1016/j.neuropharm.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mines MA, Jope RS. Glycogen synthase kinase-3: a promising therapeutic target for fragile x syndrome. Front Mol Neurosci. 2011;4:35. doi: 10.3389/fnmol.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–48. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–88. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Nalavadi V, Epstein MP, Narayanan U, Bassell GJ, Warren ST. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc Natl Acad Sci USA. 2007;104:15537–42. doi: 10.1073/pnas.0707484104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–54. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–57. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, et al. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–82. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti F, Wroblewski JT, Fadda E, Costa E. Pertussis toxin inhibits signal transduction at a specific metabolotropic glutamate receptor in primary cultures of cerebellar granule cells. Neuropharmacology. 1988;27:551–56. doi: 10.1016/0028-3908(88)90174-8. [DOI] [PubMed] [Google Scholar]
- Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–95. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–27. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Broadie KS. Drosophila fragile X mental retardation protein and metabotropic glutamate receptor A convergently regulate the synaptic ratio of ionotropic glutamate receptor subclasses. J Neurosci. 2007;27:12378–89. doi: 10.1523/JNEUROSCI.2970-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Woodruff E, 3rd, Liang P, Broadie K. Mechanistic relationships between Drosophila fragile X mental retardation protein and metabotropic glutamate receptor A signaling. Mol Cell Neurosci. 2008;37:747–60. doi: 10.1016/j.mcn.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–22. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pin JP, Waeber C, Prezeau L, Bockaert J, Heinemann SF. Alternative splicing generates metabotropic glutamate receptors inducing different patterns of calcium release in Xenopus oocytes. Proc Natl Acad Sci USA. 1992;89:10331–35. doi: 10.1073/pnas.89.21.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, et al. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont RT, Inglese J, Lefkowitz RJ. Protein kinases that phosphorylate activated G protein–coupled receptors. FASEB J. 1995;9:175–82. doi: 10.1096/fasebj.9.2.7781920. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–95. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Dant CC. The behavioral neurogenetics of fragile X syndrome: analyzing gene-brain-behavior relationships in child developmental psychopathologies. Dev Psychopathol. 2003;15:927–68. doi: 10.1017/s0954579403000464. [DOI] [PubMed] [Google Scholar]
- Repicky S, Broadie K. Metabotropic glutamate receptor-mediated use-dependent down-regulation of synaptic excitability involves the fragile X mental retardation protein. J Neurophysiol. 2009;101:672–87. doi: 10.1152/jn.90953.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–47. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooms L, Kooy RF. Advances in understanding fragile X syndrome and related disorders. Curr Opin Pediatr. 2011;23:601–6. doi: 10.1097/MOP.0b013e32834c7f1a. [DOI] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile x syndrome: a twenty-year perspective. Annu Rev Pathol. 2011;7:219–45. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–16. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Schoepp DD, Conn PJ. Metabotropic glutamate receptors in brain function and pathology. Trends Pharmacol Sci. 1993;14:13–20. doi: 10.1016/0165-6147(93)90107-u. [DOI] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–49. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Shang Y, Wang H, Mercaldo V, Li X, Chen T, Zhuo M. Fragile X mental retardation protein is required for chemically-induced long-term potentiation of the hippocampus in adult mice. J Neurochem. 2009;111:635–46. doi: 10.1111/j.1471-4159.2009.06314.x. [DOI] [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993;163:53–57. doi: 10.1016/0304-3940(93)90227-c. [DOI] [PubMed] [Google Scholar]
- Siller SS, Broadie K. Neural circuit architecture defects in a Drosophila model of Fragile X syndrome are alleviated by minocycline treatment and genetic removal of matrix metalloproteinase. Dis Model Mech. 2011;4:673–85. doi: 10.1242/dmm.008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–98. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Snyder EM, Philpot BD, Huber KM, Dong X, Fallon JR, Bear MF. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat Neurosci. 2001;4:1079–85. doi: 10.1038/nn746. [DOI] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–76. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler A, Winograd C, Sayegh J, Cheever A, Patton E, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein FMRP. Hum Mol Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- Steward O, Bakker CE, Willems PJ, Oostra BA. No evidence for disruption of normal patterns of mRNA localization in dendrites or dendritic transport of recently synthesized mRNA in FMR1 knockout mice, a model for human fragile-X mental retardation syndrome. Neuroreport. 1998;9:477–81. doi: 10.1097/00001756-199802160-00022. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–91. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Fan HX, Jiang T, Sun WW, Den WY, et al. Early continuous inhibition of group 1 mGlu signaling partially rescues dendritic spine abnormalities in the Fmr1 knockout mouse model for fragile X syndrome. Psychopharmacology. 2011;215:291–300. doi: 10.1007/s00213-010-2130-2. [DOI] [PubMed] [Google Scholar]
- Suvrathan A, Hoeffer CA, Wong H, Klann E, Chattarji S. Characterization and reversal of synaptic defects in the amygdala in a mouse model of fragile X syndrome. Proc Natl Acad Sci. 2010;107:11591–96. doi: 10.1073/pnas.1002262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanini F, Meijer N, Verheij C, Willems PJ, Galjaard H, et al. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–13. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- Tauber JM, Vanlandingham PA, Zhang B. Elevated levels of the vesicular monoamine transporter and a novel repetitive behavior in the Drosophila model of fragile X syndrome. PLoS ONE. 2011;6:e27100. doi: 10.1371/journal.pone.0027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervonen TA, Louhivuori V, Sun X, Hokkanen ME, Kratochwil CF, et al. Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol Dis. 2009;33:250–59. doi: 10.1016/j.nbd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Thomas AM, Bui N, Graham D, Perkins JR, Yuva-Paylor LA, Paylor R. Genetic reduction of group 1 metabotropic glutamate receptors alters select behaviors in a mouse model for fragile X syndrome. Behav Brain Res. 2011;223:310–21. doi: 10.1016/j.bbr.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AM, Bui N, Perkins JR, Yuva-Paylor LA, Paylor R. Group I metabotropic glutamate receptor antagonists alter select behaviors in a mouse model for fragile X syndrome. Psychopharmacology. 2012;219:47–58. doi: 10.1007/s00213-011-2375-4. [DOI] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci USA. 2003;100:14374–78. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker B, Richards RI, Lardelli M. Contribution of mGluR and Fmr1 functional pathways to neurite morphogenesis, craniofacial development and fragile X syndrome. Hum Mol Genet. 2006;15:3446–58. doi: 10.1093/hmg/ddl422. [DOI] [PubMed] [Google Scholar]
- Vanderklish PW, Edelman GM. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2002;99:1639–44. doi: 10.1073/pnas.032681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloz MF, Buijsen R, Willemsen R, Cupido A, Bosman LW, et al. The effect of an mGluR5 inhibitor on procedural memory and avoidance discrimination impairments in Fmr1 KO mice. Genes Brain Behav. 2012 doi: 10.1111/j.1601-183X.2011.00763.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–14. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Volk LJ, Pfeiffer BE, Gibson JR, Huber KM. Multiple Gq-coupled receptors converge on a common protein synthesis-dependent long-term depression that is affected in fragile X syndrome mental retardation. J Neurosci. 2007;27:11624–34. doi: 10.1523/JNEUROSCI.2266-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, et al. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol. 2005;171:811–21. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kim SS, Zhuo M. Roles of fragile X mental retardation protein in dopaminergic stimulation-induced synapse-associated protein synthesis and subsequent alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-4-propionate (AMPA) receptor internalization. J Biol Chem. 2010a;285:21888–901. doi: 10.1074/jbc.M110.116293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Berry-Kravis E, Hagerman RJ. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics. 2010b;7:264–74. doi: 10.1016/j.nurt.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, fragile X. Curr Opin Neurobiol. 2009;19:319–26. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Greenough WT. Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc Natl Acad Sci USA. 1993;90:7168–71. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmark CJ, Westmark PR, Malter JS. MPEP reduces seizure severity in Fmr-1 KO mice over expressing human Aβ. Int J Clin Exp Pathol. 2009;3:56–68. [PMC free article] [PubMed] [Google Scholar]
- Westmark CJ, Westmark PR, O'Riordan KJ, Ray BC, Hervey CM, et al. βPP/Aβ levels in Fmr1KO mice. PLoS ONE. 2011;6:e26549. doi: 10.1371/journal.pone.0026549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Oosterwijk JC, Los FJ, Galjaard H, Oostra BA. Prenatal diagnosis of fragile X syndrome. Lancet. 1996;348:967–68. doi: 10.1016/s0140-6736(05)65388-3. [DOI] [PubMed] [Google Scholar]
- Xu XL, Li Y, Wang F, Gao FB. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci. 2008;28:11883–89. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan QJ, Rammal M, Tranfaglia M, Bauchwitz RP. Suppression of two major Fragile X syndrome mouse model phenotypes by the mGluR5 antagonist MPEP. Neuropharmacology. 2005;49:1053–66. doi: 10.1016/j.neuropharm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, Wang J, Wen S, et al. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuskaitis CJ, Mines MA, King MK, Sweatt JD, Miller CA, Jope RS. Lithium ameliorates altered glycogen synthase kinase-3 and behavior in a mouse model of fragile X syndrome. Biochem Pharmacol. 2009;79:632–46. doi: 10.1016/j.bcp.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–87. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hou L, Klann E, Nelson DL. Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models. J Neurophysiol. 2009;101:2572–80. doi: 10.1152/jn.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Alger BE. Enhanced endocannabinoid signaling elevates neuronal excitability in fragile X syndrome. J Neurosci. 2010;30:5724–29. doi: 10.1523/JNEUROSCI.0795-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Venkitaramani DV, Gladding CM, Kurup P, Molnar E, et al. The tyrosine phosphatase STEP mediates AMPA receptor endocytosis after metabotropic glutamate receptor stimulation. J Neurosci. 2008;28:10561–66. doi: 10.1523/JNEUROSCI.2666-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Toyoda H, Ko SW, Ding HK, Wu LJ, Zhuo M. Deficits in Trace fear memory and long-term potentiation in a mouse model for fragile X syndrome. J Neurosci. 2005;25:7385–92. doi: 10.1523/JNEUROSCI.1520-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Chuang SC, Bianchi R, Wong RK. Dual regulation of fragile X mental retardation protein by group I metabotropic glutamate receptors controls translation-dependent epileptogenesis in the hippocampus. J Neurosci. 2011;31:725–34. doi: 10.1523/JNEUROSCI.2915-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.