Abstract
Osteosarcoma (OS) is a bone malignancy that affects children and adolescents. It is a highly aggressive tumor and typically metastasizes to lungs. Despite aggressive chemotherapy and surgical treatments, the current 5 year survival rate is 60–70%. Clinically relevant models are needed to understand OS pathobiology, metastatic progression from bones to lungs, and ultimately, to develop more efficacious treatment strategies and improve survival rates in OS patients with metastasis. The main goal of this study was to develop and characterize an in vivo OS model that will allow non-invasive tracking of tumor progression in real time, and aid in studying OS pathobiology, and screening of potential therapeutic agents against OS. In this study, we have used a multi-modality approach using bioluminescent imaging, electron microscopy, micro-computed tomography, and histopathology to develop and characterize a preclinical Bioluminescent Osteosarcoma Orthotopic Mouse (BOOM) model, using 143B human OS cell line. The results of this study clearly demonstrate that the BOOM model represents the clinical disease as evidenced by a spectrum of changes associated with tumor establishment, progression and metastasis, and detection of known OS biomarkers in the primary and metastatic tumor tissue. Key novel findings of this study include: (a) multimodality approach for extensive characterization of the BOOM model using 143B human OS cell line; (b) evidence of renal metastasis in OS orthotopic model using 143B cells; (c) evidence of Runx2 expression in the metastatic lung tissue; and (d) evidence of the presence of extracellular membrane vesicles and myofibroblasts in the BOOM model.
Keywords: Extra-cellular membrane vesicles, Bioluminescent, Myofibroblasts, Orthotopic, Osteosarcoma, Preclinical
Highlights
► A multi-modality approach is an effective strategy to characterize the BOOM model. ► Evidence of pulmonary and renal metastasis in the BOOM model. ► Runx2 immunolocalization in the metastatic lung tissue of the BOOM model. ► Evidence of the presence of extracellular membrane vesicles and myofibroblasts in the BOOM model. ► The BOOM model represents the clinical disease.
1. Introduction
Osteosarcoma (OS) is the most common malignancy of bone, mainly affecting children, adolescents and young adults. According to the American Cancer Society, 750–900 new cases of OS are diagnosed annually [1]. It usually occurs near rapidly growing areas of long bones, such as distal femur, proximal tibia and humerus near metaphyseal growth plates [2]. Osteosarcoma is a highly aggressive tumor and typically metastasizes to lungs. The majority of OS patients at presentation have pulmonary micrometastases that are mostly undetectable by current diagnostic tools [3], [4]. Despite aggressive chemotherapy and surgical treatments, the current 5 year survival rate is 60–70%. To improve survival rates in patients with metastasis or relapsed OS, clinically relevant models are needed to understand OS pathobiology and develop more efficacious treatment strategies. In vitro studies using OS cell lines are greatly limited owing to biological complexity and tissue heterogeneity of the tumor. Clinical tissue samples from OS patients are suboptimal in providing important information on dynamic tumor–stroma interactions as intense chemotherapy often disrupts the cellular components of the extracellular matrix network and induces cytotoxic effects. Obtaining clinical samples from pediatric patients for research studies is a major limitation. Development and application of reliable animal models is essential to better understand the pathological and molecular basis of the disease and for preclinical screening of potential chemotherapeutic agents. Even though there are numerous studies reporting OS xenograft and allograft models using either mouse or human OS cells, [5], [6], [7] those models fail to provide information on the tumor microenvironment (TME). In this context, orthotopic models are extremely valuable as they faithfully recapitulate human disease in terms of tumor onset, progression, and metastasis, [8] and provide important information not only on tumor biology and responsiveness to chemotherapy, but also on TME. Genetically engineered models further advance our understanding of genetic basis of this aggressive disease [9], [10]. The knowledge gained is potentially significant in discovering new targets to block metastasis in the pre-clinical setting and eventually design metastasis directed therapies for effective OS management.
In recent years, bioluminescent in vivo imaging system (IVIS) has emerged as an important technology in cancer biology to track tumor progression in real time in a non-invasive manner, and in reducing the number of animals needed in in vivo studies. The bioluminescent IVIS system is a highly specific and sensitive imaging modality, and is based on the detection and quantitation of signal produced by an enzymatic reaction in which luciferin a substrate is oxidized by luciferase expressing tumor cells, in the presence of oxygen and ATP. To the best of our knowledge, there are very few reports which describe in vivo characterization of orthotopic bioluminescent OS models (143B-based) in a comprehensive manner [11], [12], [13]. Since multiple genetic risk factors are involved in the etiology of OS, it is important to develop and characterize especially those models that closely represent human disease.
In this paper, we describe the development and characterization of a bioluminescent OS orthotopic mouse (BOOM) model using human OS cell line 143B. This cell line has a mutation in p53 and K-ras genes, and when injected orthotopically in immunocompromised mice, results in a highly aggressive form of OS. The main goal is to apply the BOOM model to advance our understanding of OS pathobiology, and to aid in the preclinical screening of potential therapeutic agents that effectively inhibit OS metastasis. In conjunction with bioluminescent imaging, we have employed micro-computed tomography (μCT) to quantify intra-osseous tumor growth and changes in the bone microarchitecture, and histopathology to assess histological changes at the primary and metastatic tumor sites. Micro-CT is a powerful and sophisticated tool to monitor structural and morphological changes in bone in 3D. The 3D nature of μCT allows a detailed assessment of changes occurring in the cortical and trabecular bone. Establishment of tumor and its subsequent metastasis to distant sites like lungs and kidneys was evaluated by histochemical staining for the detection of unmineralized osteoid and for the presence of tumor biomarkers such as Ki-67, Runx2, α-smooth muscle actin and ezrin by immunohistochemistry. We have used transmission electron microscopy (TEM) to detect the presence of extracellular membrane vesicles (EMVs) and myofibroblasts in the primary tumor tissue. Extracellular membrane vesicles are nano-sized membrane bound entities which mediate intercellular communication via horizontal transfer of proteins, mRNA and miRNA [14]. Tumor derived EMVs support tumor cell proliferation, survival, invasion and metastasis either directly or indirectly by modifying stromal microenvironment. It is our hypothesis that EMVs derived from cancer cells mediate transdifferentiation of normal stromal fibroblasts or mesenchymal stem (MSCs) cells to myofibroblasts.
2. Materials and methods
2.1. Cell culture
Human OS cell line 143B was purchased from American Type Culture Collection (Manasas, Virginia) and maintained in DMEM medium supplemented with 10% fetal bovine serum, antibiotics including penicillin and streptomycin, 2 mM glutamine and 1× minimal essential medium in 5% CO2 at 37 °C as previously described [15]. Cells were grown till sub-confluence in T-75 cell culture flasks and harvested by 0.05% Trypsin/EDTA dissociation. Exponentially growing cells were used to generate luciferase expressing puromycin resistant 143B-luc cells.
2.2. Establishment of lentivirally transduced 143B cells expressing luciferase reporter gene
We engineered 143B cells to express luciferase (FUW-Luc-mCherry-puro) using a 3rd generation lentiviral vector (FUW) by following previously published methods specified for transduction techniques [16], [17]. Positive 143B-luc cells were selected in puromycin (Sigma Aldrich, St. Louis, MO, USA), expanded and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega Corp., Madison WI, USA).
2.3. Orthotopic intratibial injections
All animal studies were performed at the Proof of Concept Laboratory (University of Kansas Cancer Center, Kansas City, KS), with the approval of the University of Kansas Institutional Animal Care and Use Committee. One million exponentially growing 143B-luc cells were injected orthotopically into Nu/Nu mice, according to the procedures previously described by Luu et al. [18] Briefly, the knee was flexed, and about 1×106 exponentially growing 143B-luc positive cells suspended in 100 μl PBS were injected in the proximal tibias of both legs, using 27 gauge needle. Tumor growth was monitored weekly for 5–6 weeks by luciferase imaging, using the Calipers IVIS system. This time point represents a reasonable end-point for evaluating osteosarcoma bone colonization, progression, and metastasis in the BOOM model using histopathology, immunohistochemistry for detection of disease specific biomarkers, electron microscopy for detection and confirmation of myofibroblasts and extra-cellular membrane vesicles, and micro-computed tomography for assessing the impact on bone architecture.
2.4. Bioluminescent imaging
Mice were prepared for imaging following intraperitoneal injection of luciferin. The mice were anesthetized by exposure to isoflurane gas anesthesia (2.5%) before placing them inside the imaging chamber. Photographic and bioluminescent images were acquired and superimposed, while still maintaining the anesthetized mice under isoflurane atmosphere. Using the Living image software, a region of interest (ROI) was manually drawn around the tumor site, and the radiance output or signal was recorded as the total flux of the ROI and expressed in photons/seconds for each animal. For ex-vivo imaging to detect luciferase positive cells at the metastatic site, lungs and kidneys were resected at necropsy from luciferin injected animals.
2.5. Detection of mCherry positive 143B cells and tumor-induced histopathological changes
For evaluating fluorescent mCherry positive 143B cells and tumor-induced histopathological changes, excised tibias (tumor-bearing), lungs and kidneys were fixed in 4% paraformaldehyde (pH 7.4). For detection of mCherry fluorescence, 5 μm sections of excised tumor tissue were examined microscopically. Slides were imaged on an Olympus IX71 inverted fluorescent microscope equipped with a xenon arc lamp and monochromatic CMOS camera. Images were acquired using SlideBook v4.2 image acquisition software (Olympus). For hematoxylin and eosin (H and E) staining and immunohistochemistry (IHC), tibias were decalcified in 10% EDTA (pH 7.5) for 2 weeks prior to sectioning and paraffin embedding. Tumor-bearing and control tissue sections were stained with H&E, for histological evaluation of primary tumors and micrometastasis in the lungs and kidneys.
2.6. Detection of unmineralized osteoid (a hallmark of OS) by Villanueva–Goldner staining
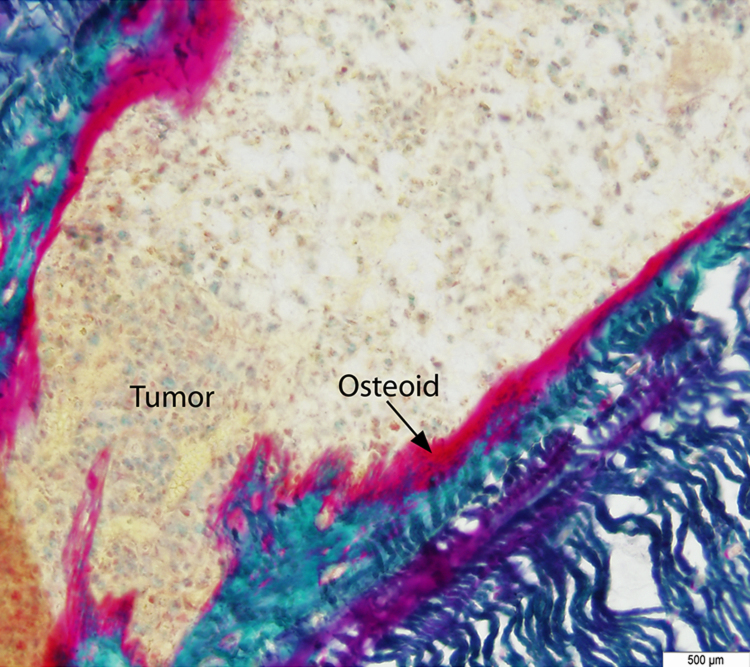
To determine osteoid content in tumor-bearing bones, excised tibias (either tumor-bearing or non-tumor-bearing) were fixed in 4% paraformaldehyde (pH 7.4) and non-decalcified sections were embedded in methyl methacrylate (MMA) according to a previously described method [19]. Serial sections of embedded samples were stained with Villanueva–Goldner stain for the detection of unmineralized osteoid [20].
2.7. Immunonohistochemistry (IHC)
Unstained tibial, lung and kidney sections were processed for immunohistochemical analysis. Primary antibodies to Ki-67 (clone MIB-1) and desmin were purchased from Dakocytomation (Carpinteria, CA). Primary antibodies to Runx2 and ezrin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Primary antibody to α-smooth muscle actin (α-SMA Sigma–Aldrich, MO) was generously provided by Dr. Smirnova, University of Kansas Medical Center. The concentrations of primary antibodies used were as follows: 1:500 for Ki-67; 1:50 for Runx2 and desmin; and 1:200 for ezrin and α-SMA. Expression of ezrin, Runx2, desmin, α-SMA and Ki-67 was detected using peroxidase conjugated secondary antibody and a peroxidase substrate (3, 5-diaminobenzidine). For Ki-67, tonsils and appendix tissue sections and for Runx2 IHC, spleen and kidney tissue sections were used as controls. For ezrin tonsil sections, and for desmin and α-SMC IHC, smooth muscle tissue sections were used as controls.
2.8. Transmission Electron Microscopy (TEM)
For TEM, tumor-bearing bones were fixed in 2.5% glutaraldehyde, post fixed in 1% osmium tetraoxide (OsO4), dehydrated, embedded in epon resin and cut into ultra-thin sections. The sections were stained with uranyl acetate and lead acetate prior to mounting on EM grids. The sections were examined and photographed using a JEM 1400 electron microscope (80 kV).
2.9. Micro-computerized tomography (μ-CT)
For μ-CT micro-computerized tomography of tibias, bones were fixed in 70% ethanol and scanned with a high-resolution μCT (μCT40; Scanco Medical, Southeastern, PA) as previously described [19]. Data were acquired at 55 KeV at 6 μm resolution and three-dimensional reconstructions of bone samples were generated using the following parameters: Sigma, 0.8; Support, 1; Threshold, 212 (cortex). Cortical thickness and tissue mineral density were calculated by integration of the value on each transverse section of a set of 160 slices chosen in the midshaft area. Tissue mineral density was derived from the linear attenuation coefficient of threshold bone through precalibration of the apparatus for the acquisition voltage chosen.
2.10. Statistical analysis
Statistical analysis was performed using Prism 5 (Graph-Pad software, La Jolla, CA). All experimental data are presented as mean±S.E.M. Student's t-test was used for determining statistical significance between control (non-tumor-bearing) versus tumor-bearing mice (saline treated) groups. A p value <0.05 was considered statistically significant.
3. Results
3.1. Non-invasive tracking of tumor growth in real time following injection of 143B-luc cells in Nu/Nu mice
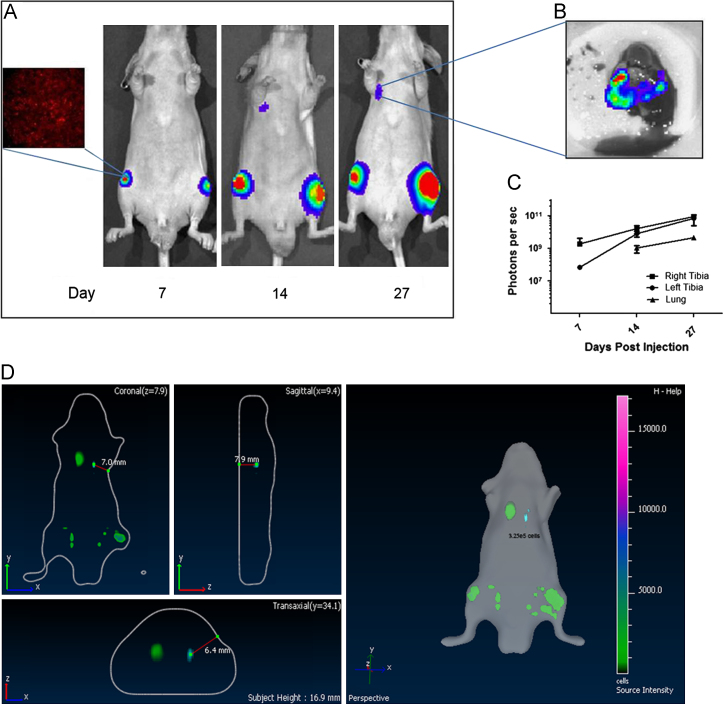
In this study, we developed an orthotopic OS mouse model using human OS cell line 143B expressing luciferase (143B-luc). Quantification of luciferase activity in serial dilutions of lentivirally transduced 143B-luc cells revealed that 0.3×105 cells was the threshold number of cells to detect luminescence, in vitro. For 143B-luc cells, total flux/cell was determined to be 2165 p/s/cell. Following intra-tibial injection, the 143B-luc cells generated tumors in the medullary cavities of the injected bones that subsequently metastasized to lungs within two weeks (Fig. 1A–D), as detected by bioluminescent imaging (BLI). Using the BLI approach, tumor burden was 100 % in the BOOM model in 2 separate experimental trials (Table 1). Ex-vivo bioluminescence confirmed the presence of 143B-luc cells in the lungs (Fig. 1B). The kinetics of tumor progression in real time revealed an increase in signal intensity i.e. the number of photons/second (Fig. 1C).
Fig. 1.
Bioluminescent imaging of intra-tibial growth of 143B-luc cells. Mice were injected with 143B-luc cells intra-tibially and tumor growth was monitored weekly by luciferase imaging (A). Within 2 weeks, mice developed spontaneous metastasis to lungs (A). A representative image of ex-vivo bioluminescent imaging of lung metastatic lesions is shown (B). Kinetics of 143B-luc generated tumor in real time is shown in C. Detection of bioluminescent signal in 3D views (coronal, transaxial and saggital) is shown in D.
Table 1.
Tumor burden in the BOOM model as detected by bioluminescent imaging (BLI) and histology.
| Experiment # | Tumors detected in the BOOM model |
||||||
| BLI (# of mice with detectable tumors) | Histology (# of slides with detectable tumors) |
||||||
| Bone | Lungs |
Kidneys |
|||||
| 1 | 7 | 7/7* (*tail-vein injection-1) | 7/7** (** tail-vein injections-2) | 3/5 | |||
| 2 | 9 | 9/9 | 7/9 | 0/4 | |||
| Tumor burden (%) | 100% (16/16) | 16/16 (100%) | 14/16 (88%) | 3/9 (33%) | |||
3.2. Histopathological characterization of tumors derived from 143B-luc cells in the BOOM model
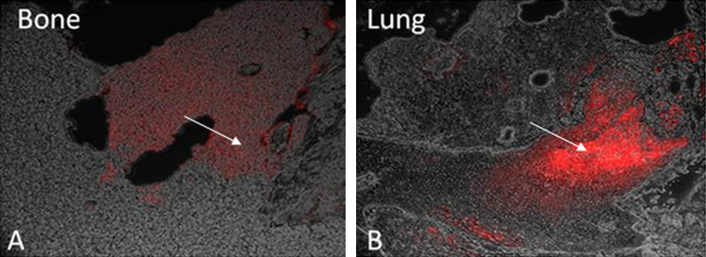
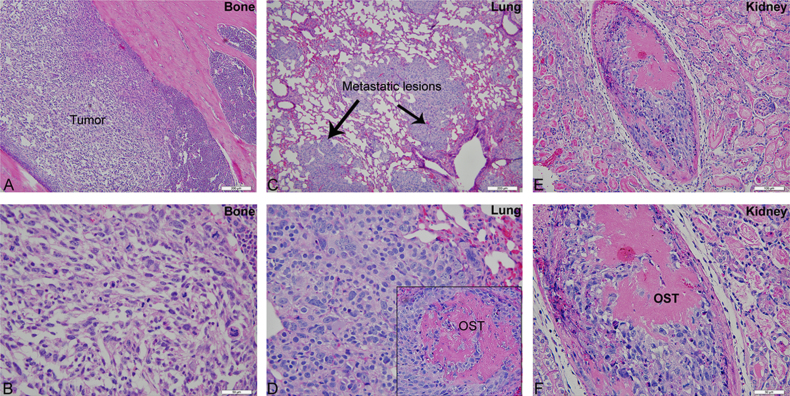
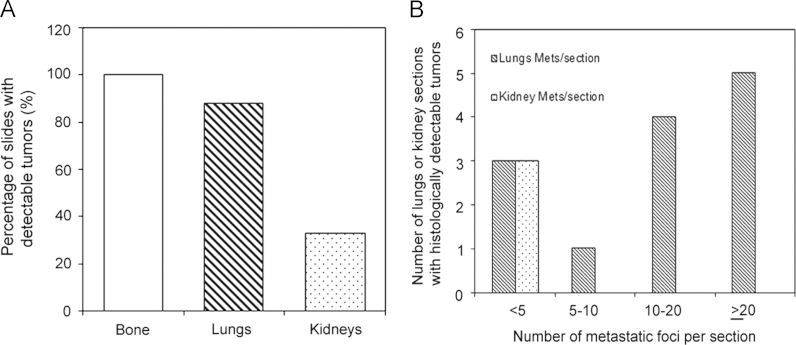
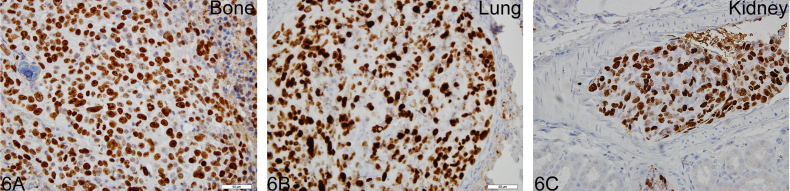
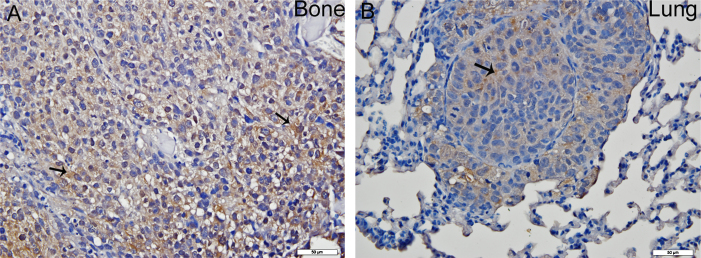
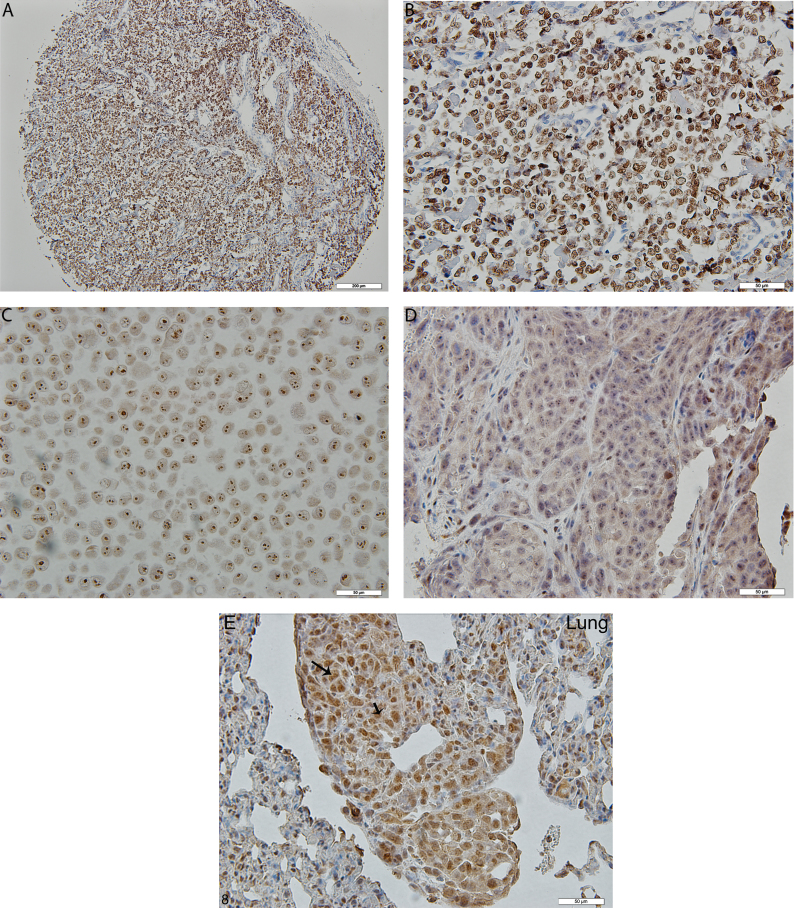
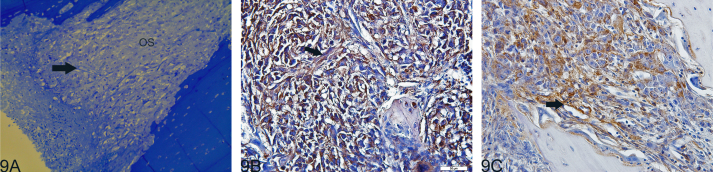
Histological examination revealed the presence of fluorescent mCherry positive 143B-luc cells in paraffin embedded sections of lungs and bones (Fig. 2). Hematoxylin and eosin stained sections revealed the presence of intramedullary tumor composed of highly anaplastic polygonal and spindle shaped OS cells in primary bone and metastatic lesions of tumor-bearing mice (Fig. 3A–F). The tumor cells had pleomorphic, and hyperchromatic nuclei. Actively dividing OS cells in different mitotic stages were detected in the primary and metastatic tumors of tibial, lung and kidney sections (Fig. 3A–F). Metastatic tumor emboli were also observed within small and medium sized angiolymphatic vessels in kidneys. Areas with geographic necrosis were also detected in the tumor-bearing tissue. Evidence of osteoid formation was observed in some tumors (Fig. 3E and F). Histopathologial evaluation of slides revealed the presence of tumors in tibial, lung and kidney sections. We detected and confirmed the presence of tumor foci in 16/16 (100%) tibial, 14/16 (88%) lung, and 3/9 (33.3%) kidney sections, at the end of 2 experimental pilot trials (Fig. 4a). The number of metastatic foci per lung section ranged from 1–50 (Fig. 4b). Metastatic lesions in kidneys were detectable in fewer number of slides examined. Average tumor burden profile in the BOOM model from 2 trials, as evaluated histologically is shown in Table 1 and Fig. 4. Goldner staining on non-decalcified tibial sections indicated the presence of more osteoid in the tumor tissue compared to control, thereby indicating the presence of active osteoid secreting osteosarcoma cells (Fig. 5). High proliferative activity of OS lesions was confirmed by the increased number of Ki-67 positively stained cells, as shown in Fig. 6. Expression of ezrin, an OS metastasis biomarker (Fig. 7A and B), and Runx2 (Fig. 8A–E), a differential tumor modulator was detected in all OS cell from primary and metastatic lesions, by IHC. Light microscopy revealed the presence of fibroblasts in and around the tumor tissue (Fig. 9A). Expression of α-SMA and desmin was detected in the primary tumor tissue of the BOOM model (Fig. 9B and C).
Fig. 2.
Photomicrograph illustration showing the presence of mCherry positive 143B-luc cells (as indicated by an arrow) in the bone (primary tumor site) (A) and lung (metastatic site) tissue (B).
Fig. 3.
Hematoxylin and Eosin staining showing primary tumor in the tibia (A and B) and OS metastatic lesions in lungs (C and D) and kidney (E and F). The inset shows the presence of osteoid (pink color) in the tumor. The reader is recommended to zoom to 200% to view tumor cells in different mitotic stages. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Graphic illustrations showing tumor burden profile in the BOOM model from two rounds of experiments. Bone, lung and kidney sections were examined histogically for the detection of tumors (A). Number of metastatic lesions per lung or kidney sections were also examined (B).
Fig. 5.
Photomicrograph illustration showing the presence of osteoid (pink color) in the primary tumor as detected by Goldner staining. The osteoid (as indicated by an arrow) is deposited by rapidly proliferating osteosarcoma cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Photomicrograph illustration showing Ki-67 (a proliferation marker) immunostaining in actively dividing osteosarcoma cells in tibia (A), lungs (B) and kidneys (c). Arrows indicate mitotic figures where Ki-67 is maximal.
Fig. 7.
Photomicrograph illustration showing ezrin immunostaining in osteosarcoma tumor tissue in tibia and lungs. Panels A and B are treated with anti-ezrin primary antibody. Brown staining indicates ezrin expression (also indicated by arrows). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Photomicrograph illustration showing immunlocalization and expression of Runx2 in osteosarcoma tissue microarray, 143B human OS cells and tumor tissue of the BOOM model. Panels A (10×), B (40×), C and D demonstrate positive staining in osteoblastic osteosarcoma core sample of bone cancer tissue microarrays, 143B human OS cell line in vitro, and in the tumor tissue isolated from the BOOM model, respectively. Panel E (40X) shows positive immunostaining for Runx2 in metastatic OS lesions in the lung tissue.
Fig. 9.
Photomicrograph illustration showing the presence of myofibroblast like cells in the primary tumor tissue of the BOOM model as identified by light microscopy (Panel A) and α-SMA (Panel B), and desmin (Panel C) immunostaining.
3.3. Detection of Extracellular Membrane Vesicles (EMVs) and myofibroblasts in the tumor tissue of the BOOM model by Electron Microscopy (EM)
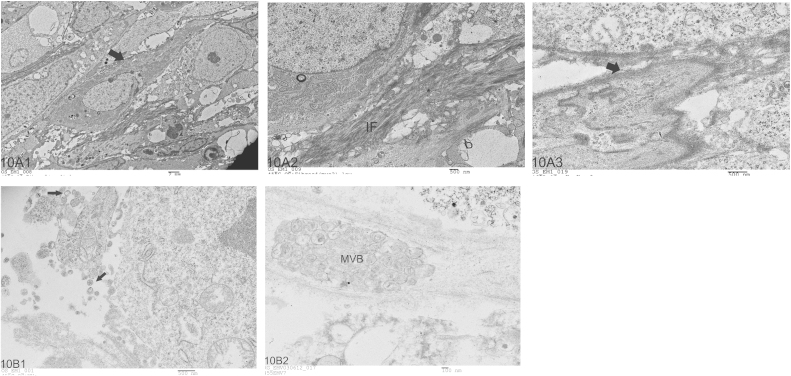
Electron microscopic examination of the primary tumor tissue from the BOOM model revealed the presence of myofibroblasts in and around the tumor tissue, and were identified based on their cellular content and distinct morphological features such as presence of abundant Rough Endoplasmic Reticulum (RER), peripheral distribution of filaments, fibronectin fibrils and fibronexus (cell-to-matrix junction) (Fig. 10A). Extracellular matrix vesicles which were about 50–200 nm in diameter were detected in the extracellular matrix of tumor, in the vicinity of the cells from which they are derived (Fig. 10B). Multivesicular bodies were detected in the tumor tissue (Fig. 10B2).
Fig. 10.
Transmission Electron Micrograph composite showing representative images of myofibroblasts and EMVs in the primary tissue of the BOOM model. The ultrastructural features that are characteristic of myofibroblasts include presence of RER, peripheral distribution of filaments, and a prominent fibronexus junction (Panel A). Panel B shows representative images (B1) of EMVs and multivesicular body (B2) in the tumor tissue of the BOOM model.
3.4. Characterization of cancer induced bone disease in OS by μ-CT
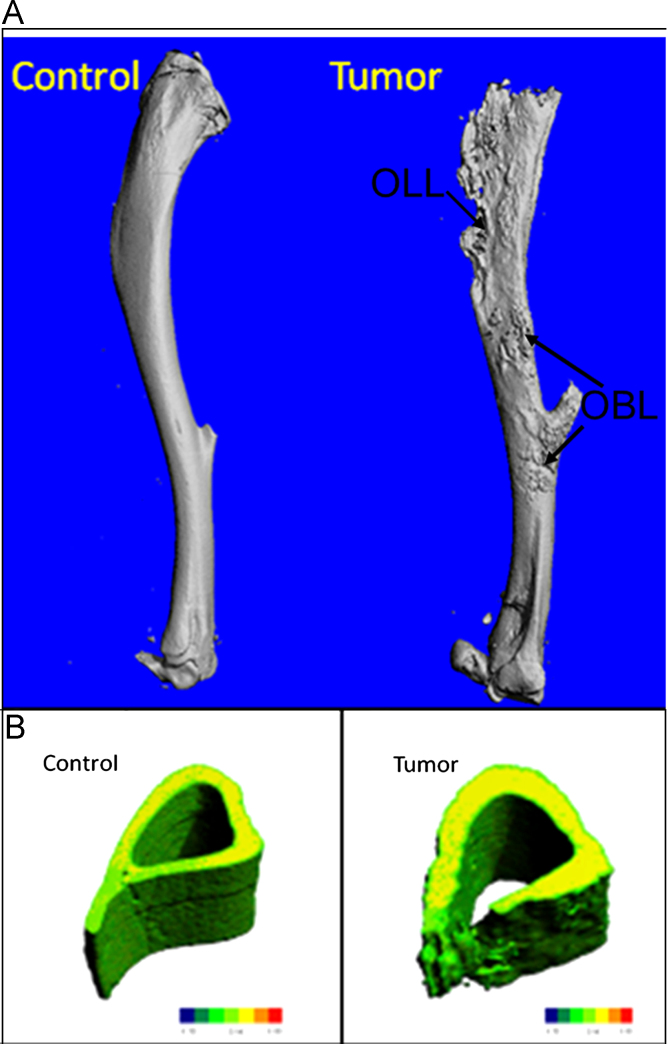
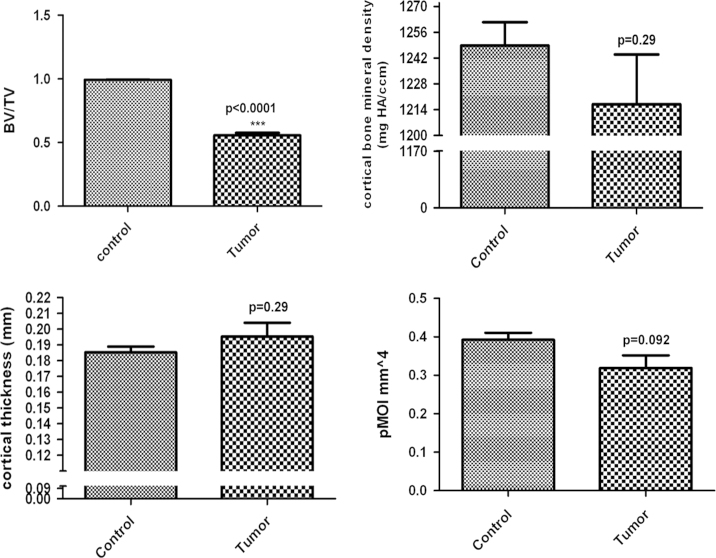
Our μ-CT results demonstrate that 143B-luc cells in the BOOM model produced both osteolytic and osteoblastic lesions in the tibias of tumor-bearing mice (Fig. 11A). These lesions were absent in (vehicle injected) control tibia. Micro-CT analyses revealed changes in the bone architecture due to cortical penetration of tumor in tumor-bearing tibia vs. control tibia (Fig. 11B). Bone parameters such BV/TV was significantly decreased (p<0.0001) in the tumor-bearing tibia vs. control, while no significant changes were observed in the cortical thickness and cortical bone mineral density (Fig. 12). Polar moment of inertia (pMOI) which is an indicator of bone strength was reduced in tumor-bearing tibia vs. non-tumor-bearing control, but the difference was not significance (p=0.09, n≥3) (Fig. 12).
Fig. 11.
Photomicrograph illustration showing representative 3D reconstructions of μCT scans of control (non-tumor-bearing) vs. orthograft tumor (tumor-bearing). Osteoblastic (OBL)/osteolytic (OLL) lesions are indicated by an arrow (Panel A). The lower panel (B) shows representative 3D μCT–generated sections of tibias showing increased cortical thickening and destruction of cortical bone in tumor-bearing mice versus control.
Fig. 12.
Graphic illustration comparing cortical bone parameters i.e. cortical fraction (BV/TV) (A), cortical bone mineral density (B), cortical thickness (C), and polar moment of inertia (pMOI) (D) between control (non-tumor-bearing) and tumor-bearing bone (n≥3).
4. Discussion
In this study, we report the development of a preclinical Bioluminescent Osteosarcoma Orthotopic Mouse (BOOM) model and its characterization by bioluminescent imaging, μ-CT and histopathology, using 143B, an aggressive human OS cell line. We have used bioluminescent imaging for easy detection of luciferase-tagged cancer cells in real-time, and histopathological techniques like light microscopy and IHC for morphological and histopathological characterization i.e. tumor establishment at the primary site and progression or metastasis to distant sites including lungs and kidneys. Finally, the impact of intraosseous tumor growth on local bone microenvironment and architecture in the BOOM model was assessed using μ-CT, and detection of osteoid by Goldner's staining.
To our best knowledge, a multi-modality approach to systematically characterize an OS in vivo orthotopic model, using an aggressive OS cell line has not been previously reported. The advantage of using multi-modality approach i.e. BLI, histopathology, immunohistochemistry, and electron microscopy allowed us to detect tumor cells and disease progression in real time, detect OS-specific biomarkers, and successfully detect the presence of myofibroblasts and extra-cellular membrane vesicles in the osteosarcoma tissue.
The primary tumor generated by 143B cells in the BOOM model revealed histological features similar to those of clinical OS as detected by light microscopy, IHC and Goldner staining. Histological detection of newly formed osteoid in our studies indicates that the 143B cells in the BOOM model proliferate rapidly, secreting osteoid, and form osteoblastic lesions similar to neoplastic OS cells in the clinical disease [21], [22]. Bone involvement of the experimental OS was also confirmed by μ-CT which revealed the presence of both osteoblastic and osteolytic lesions in tumor-bearing bones.
Human OS cell line 143B is derived from TE85 (ras wild type) OS cell line. The metastatic ability of 143B cells is due to the activation of K-ras [18]. The propensity of ras-activated tumor cells to lungs is known [23] but the underlying mechanisms that facilitate lung metastasis are not clear. While pulmonary metastasis is a common feature associated with OS, we neither know the frequency of ras mutations nor its association with pulmonary metastasis in clinical OS. Metastatic OS in the kidney was detected in the BOOM model. Renal metastasis is rare in clinical OS, but there are a few studies which report that renal metastasis develops subsequent to pulmonary metastasis [24], [25], [26], [27], [28], [29]. Renal metastasis of OS is correlated with aggressive cancer and poor survival [30] and usually detected after death in about 10–12% OS autopsies [31], [32].To our best knowledge, this is the first study that reports renal metastasis in OS orthotopic model using 143B cells. It is not clear whether the renal metastasis as observed in few animals in our study is cell-line specific or due to immune-incompetence, and/or the chosen animal.
Ezrin is one of most consistent metastasis biomarker of OS and its role in metastasis is discussed in a number of investigative studies [33], [34], [35]. Ezrin is a protein that links cytoskeleton proteins like actin to the cell membrane. It plays an important role in cell–cell interaction and signal transduction [36]. In pediatric OS patients, increased ezrin expression is correlated with poor prognosis [37], [38] and down regulation of ezrin expression in murine OS model resulted in inhibition of pulmonary metastasis [33]. The expression of ezrin in the tumor tissue of bones and lungs observed in our study supports the clinical relevance of the BOOM model.
A key pathophysiological feature of OS is the cumulative defect in osteoblastic differentiation. Runx2 is a transcription factor which regulates osteogenic differentiation and bone formation. The role of Runx2 in OS is a subject of intense debate. While some studies report that OS cells express lower levels of Runx2 and suggest that Runx2 insufficiency contributes towards osteosarcomagenesis and progression, [39] others report high incidence of Runx2 genomic amplification, [40] and increased Runx2 expression (mRNA and protein) in OS biopsy samples [41], [42]. Increased Runx2 expression also correlates with poor response to neoadjuvant chemotherapy in OS [41]. Presence of increased expression of Runx2 in the tumor tissue obtained from the BOOM model confirmed our observation of increased expression of Runx2 in the osteoblastic type of OS in the bone cancer tissue microarrays.
The presence of myofibroblasts in OS is not surprising as previous studies have reported the presence of myofibroblasts in bone tumors [43], [44], [45], [46]. To our best knowledge, this is the first study that reports the presence of myofibroblasts in an in vivo OS experimental model, as identified by immunoexpression of α-SMA and characteristic ultrastructural features by TEM. Myofibroblasts of the reactive tumor stroma mediate ECM remodeling mainly by active synthesis of extracellular matrix (ECM) components like collagen type I, III, fibronectin, and enzymes like MMPs, urokinase plasminogen activator and fibronectin activation protein [47], [48], [49], [50]. Expression of α-SMA in OS cells as seen in this study and others suggest that (a) highly aggressive OS cells may tend to become de-differentiated and pleomorphic [51], and (b) tumor cells may fuse with stromal cells such as myofibroblasts from tumor microenvironment [52]. Previous studies suggest that expression of α-SMA in OS tissue may be of prognostic significance [51], [52], [53]. Previous studies report that the expression of desmin in OS is highly variable (absent to minimal). This could be attributed to heterogeneity of tumor stroma [54]. Evidence of strongly positive staining for desmin expression in our OS tissue could be associated with K-ras mutational background of 143B cells as previous studies with soft tissue sarcomas have reported that expression of Kras in satellite cells induced expression of myogenic proteins such as Myo D and desmin [55].
The tumor microenvironment is enriched with EMVs that are derived not only by proliferating tumor cells, but also from infiltrating macrophages and neutrophils, and support tumor cell survival, growth, invasion and metastasis. Tumor derived EMVs also have the potential to modulate surrounding stromal cells, and facilitate rapid phenotype adjustments for maintaining cellular homeostasis during physiological and pathological states. In particular, a recent study has demonstrated that exosomes or EMVs derived from mesothelioma cancer cells can stimulate differentiation of fibroblasts to α-smooth muscle actin expressing myofibroblasts [56]. Acquisition of myofibroblast phenotype in the stromal microenvironment is associated with increased tumor growth, angiogenesis and metastasis [56], [57]. In our study, we report the presence of both EMVs and myofibroblasts in OS tissue, isolated from the primary tumors of the BOOM model.
A preclinical BOOM model like the one described in this paper and others [11], [12], [58] will continue to shed light on OS pathobiology and tumor microenvironment in primary and metastatic sites. These models are also invaluable in conducting chemopreventive, chemotherapeutic and genetic (gene silencing or RNA interference or gene therapy) studies especially when clinical trials are challenging because of limited number of samples and the patient category (pediatric). Our goal is to expand the use of the preclinical BOOM model and obtain mechanistic information underlying tumorigenesis; matrix remodeling, dynamic cross-talk between tumor–stroma; and metastasis, and screen potential antineoplastic or tumor modulating agents. Multi-modality application of the preclinical BOOM model will help to identify new therapeutically relevant targets for drug development against OS. We anticipate this strategy will provide a powerful first step before further evaluation using naturally occurring in canine OS models (comparative oncology approach) and then testing in pediatric patients.
Our model has provided a powerful tool for following the progression of cancer cells in real time. Also, for future studies the BOOM model provides a platform for collecting information on threshold-dose of luciferase-tagged OS cell lines to generate tumors orthotopically, cellular, molecular, histological and μ-CT changes dynamically. One of the limitations of the BOOM model is that it does not allow a “complete” evaluation of host immune responses to OS. The lymphocyte mediated immune response especially the T-cell response is lost in the immunocompromised mouse model [59]. Despite the above discussed limitations, there are clear scientific benefits that can be derived from the BOOM model. For example, as stated previously, the frequency of ras mutations in clinical OS is unknown, but successful metastasis of 143B cells to lungs and kidneys from tibia i.e. the primary site demonstrates the reliability of this model to investigate the mechanism(s) underlying tumor progression and metastasis. In conclusion, we have successfully developed and characterized a bioluminescent orthotopic model that can be used to study OS pathobiology, and to evaluate potential therapeutic agents. Key novel findings of this study include: (a) multimodality approach for extensive characterization of the BOOM model using 143B human OS cell line; (b) evidence of in vivo Ki-67 staining of 143B cells; (c) evidence of renal metastasis in OS orthotopic model using 143B cells, (d) evidence of Runx2 expression in the metastatic lung tissue, (e) evidence of osteoblastic and osteolytic lesions with destruction of cortical bone due to increasing tumor burden; and (f) evidence of the presence of extracellular membrane vesicles and myofibroblasts in the tumor tissue from the BOOM model, suggesting a possible role of tumor-stroma intercellular communication in osteosarcoma progression.
Acknowledgment
We thank Ms. Marsha Danley for helping with Ki-67, Runx2, desmin immunohistochemistry and the Department of Pathology for allowing us to use their digital microscope. We appreciate the generosity of Dr. Irina Smirnova for providing α-SMA antibody. We thank Professor Shrikant Anant for helpful suggestions and feedback. We thank Drs. Peter Van Veldhuizen, Raymond Perez, and Debra Sullivan for their support. Funding sources for this study include: Division of Hematology and Oncology, School of Health Professions (SHP), and The University of Kansas Cancer Center (KUCC) research support to Rama Garimella, PhD, MS, M.Sc.
References
- 1.Abe M., Akeno N., Ohida S., Horiuchi N. Inhibitory effects of 1,25-dihydroxyvitamin D3 and 9-cis-retinoic acid on parathyroid hormone-related protein expression by oral cancer cells (HSC-3) The Journal of Endocrinology. 1998;156:349–357. doi: 10.1677/joe.0.1560349. [DOI] [PubMed] [Google Scholar]
- 2.Meyers P.A., Gorlick R. Osteosarcoma. Pediatric Clinics of North America. 1997;44:973–989. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- 3.Link M.P. Adjuvant therapy in the treatment of osteosarcoma. Important Advances in Oncology. 1986:193–207. [PubMed] [Google Scholar]
- 4.Ward W.G., Mikaelian K., Dorey F., Mirra J.M., Sassoon A., Holmes E.C. Pulmonary metastases of stage IIB extremity osteosarcoma and subsequent pulmonary metastases. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 1994;12:1849–1858. doi: 10.1200/JCO.1994.12.9.1849. [DOI] [PubMed] [Google Scholar]
- 5.Dass C.R., Ek E.T., Choong P.F. Human xenograft osteosarcoma models with spontaneous metastasis in mice: clinical relevance and applicability for drug testing. Journal of Cancer Research and Clinical Oncology. 2007;133:193–198. doi: 10.1007/s00432-006-0157-x. [DOI] [PubMed] [Google Scholar]
- 6.Dass C.R., Ek E.T., Contreras K.G., Choong P.F. A novel orthotopic murine model provides insights into cellular and molecular characteristics contributing to human osteosarcoma. Clinical & Experimental Metastasis. 2006;23:367–380. doi: 10.1007/s10585-006-9046-6. [DOI] [PubMed] [Google Scholar]
- 7.Houghton P.J., Morton C.L., Tucker C., Payne D., Favours E., Cole C. The pediatric preclinical testing program: description of models and early testing results. Pediatric Blood & Cancer. 2007;49:928–940. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 8.Khanna C., Prehn J., Yeung C., Caylor J., Tsokos M., Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clinical & Experimental Metastasis. 2000;18:261–271. doi: 10.1023/a:1006767007547. [DOI] [PubMed] [Google Scholar]
- 9.Walkley C.R., Qudsi R., Sankaran V.G., Perry J.A., Gostissa M., Roth S.I. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes & Development. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman S.D., Calo E., Landman A.S., Danielian P.S., Miller E.S., West J.C. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miretti S., Roato I., Taulli R., Ponzetto C., Cilli M., Olivero M. A mouse model of pulmonary metastasis from spontaneous osteosarcoma monitored in vivo by Luciferase imaging. PloS One. 2008;3:e1828. doi: 10.1371/journal.pone.0001828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sottnik J.L., Duval D.L., Ehrhart E.J., Thamm D.H. An orthotopic, postsurgical model of luciferase transfected murine osteosarcoma with spontaneous metastasis. Clinical & Experimental Metastasis. 2010;27:151–160. doi: 10.1007/s10585-010-9318-z. [DOI] [PubMed] [Google Scholar]
- 13.Noh K., Kim K.O., Patel N.R., Staples J.R., Minematsu H., Nair K. Targeting inflammatory kinase as an adjuvant treatment for osteosarcomas. The Journal of Bone and Joint Surgery American Volume. 2011;93:723–732. doi: 10.2106/JBJS.J.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson H.C., Mulhall D., Garimella R. Role of extracellular membrane vesicles in the pathogenesis of various diseases, including cancer, renal diseases, atherosclerosis, and arthritis. Laboratory Investigation. 2010;90:1549–1557. doi: 10.1038/labinvest.2010.152. [DOI] [PubMed] [Google Scholar]
- 15.Thompson L., Wang S., Tawfik O., Templeton K., Tancabelic J., Pinson D. Effect of 25-hydroxyvitamin D(3) and 1 alpha,25 dihydroxyvitamin D(3) on differentiation and apoptosis of human osteosarcoma cell lines. Journal of Orthopaedic Research : Official Publication of the Orthopaedic Research Society. 2011 doi: 10.1002/jor.21585. [DOI] [PubMed] [Google Scholar]
- 16.Kimbrel E.A., Davis T.N., Bradner J.E., Kung A.L. In vivo pharmacodynamic imaging of proteasome inhibition. Molecular Imaging. 2009;8:140–147. [PubMed] [Google Scholar]
- 17.Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 18.Luu H.H., Kang Q., Park J.K., Si W., Luo Q., Jiang W. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clinical & Experimental Metastasis. 2005;22:319–329. doi: 10.1007/s10585-005-0365-9. [DOI] [PubMed] [Google Scholar]
- 19.Rowe P.S., Matsumoto N., Jo O.D., Shih R.N., Oconnor J., Roudier M.P. Correction of the mineralization defect in hyp mice treated with protease inhibitors CA074 and pepstatin. Bone. 2006;39:773–786. doi: 10.1016/j.bone.2006.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva A.R., Mehr L.A. Modifications of the Goldner and Gomori one-step trichrome stains for plastic-embedded thin sections of bone. The American Journal of Medical Technology. 1977;43:536–538. [PubMed] [Google Scholar]
- 21.Miller S.L., Hoffer F.A. Malignant and benign bone tumors. Radiologic Clinics of North America. 2001;39:673–699. doi: 10.1016/s0033-8389(05)70305-5. [DOI] [PubMed] [Google Scholar]
- 22.Gross M., Stevens K. Sunburst periosteal reaction in osteogenic sarcoma. Pediatric Radiology. 2005;35:647–648. doi: 10.1007/s00247-005-1442-6. [DOI] [PubMed] [Google Scholar]
- 23.Tome Y., Tsuchiya H., Hayashi K., Yamauchi K., Sugimoto N., Kanaya F. In vivo gene transfer between interacting human osteosarcoma cell lines is associated with acquisition of enhanced metastatic potential. Journal of Cellular Biochemistry. 2009;108:362–367. doi: 10.1002/jcb.22259. [DOI] [PubMed] [Google Scholar]
- 24.Dash H., Little J.R., Zaino R., Colao D.J., Chaurushiya P., Schoolwerth A.C. Metastatic periosteal osteosarcoma causing cardiac and renal failure. The American Journal of Medicine. 1983;75:145–149. doi: 10.1016/0002-9343(83)91178-6. [DOI] [PubMed] [Google Scholar]
- 25.Nelson J.A., Clark R.E., Palubinskas A.J. Osteogenic sarcoma with calcified renal metastasis. The British Journal of Radiology. 1971;44:802–804. doi: 10.1259/0007-1285-44-526-802. [DOI] [PubMed] [Google Scholar]
- 26.Watson R.C., Cubilla A.L. Osteogenic sarcoma metastatic to the kidney. Clinical Bulletin. 1975;5:114–115. [PubMed] [Google Scholar]
- 27.Ayres R., Curry N.S., Gordon L., Bradford B.F. Renal metastases from osteogenic sarcoma. Urologic Radiology. 1985;7:39–41. doi: 10.1007/BF02926848. [DOI] [PubMed] [Google Scholar]
- 28.Raby W.N., Kopplin P., Weitzman S. Metastatic osteosarcoma of the kidney presenting as renal hemorrhage. Journal of Pediatric Hematology/Oncology. 1996;18:321–322. doi: 10.1097/00043426-199608000-00020. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein C., Ambos M.A., Bosniak M.A. Multiple ossified metastases to the kidney from osteogenic sarcoma. AJR American Journal of Roentgenology. 1977;128:148–149. doi: 10.2214/ajr.128.1.148. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto L.H., Mendes W., Pecora M., Andrade R.G., Begnani M.D., de Camargo B. Bilateral renal metastases from osteosarcoma: a case report and review of the literature. Journal of Pediatric Hematology/Oncology. 2006;28:618–621. doi: 10.1097/01.mph.0000212961.73844.fc. [DOI] [PubMed] [Google Scholar]
- 31.Jeffree G.M., Price C.H., Sissons H.A. The metastatic patterns of osteosarcoma. British Journal of Cancer. 1975;32:87–107. doi: 10.1038/bjc.1975.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna R.J., Schwinn C.P., Higinbotham N.L. Osteogenic sarcoma in children. CA: a Cancer Journal for clinicians. 1966;16:26–28. doi: 10.3322/canjclin.16.1.26. [DOI] [PubMed] [Google Scholar]
- 33.Khanna C., Wan X., Bose S., Cassaday R., Olomu O., Mendoza A. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nature Medicine. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 34.Khanna C. Novel targets with potential therapeutic applications in osteosarcoma. Current Oncology Reports. 2008;10:350–358. doi: 10.1007/s11912-008-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan X., Kim S.Y., Guenther L.M., Mendoza A., Briggs J., Yeung C. Beta4 integrin promotes osteosarcoma metastasis and interacts with ezrin. Oncogene. 2009;28:3401–3411. doi: 10.1038/onc.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter K.W. Ezrin, a key component in tumor metastasis. Trends In Molecular Medicine. 2004;10:201–204. doi: 10.1016/j.molmed.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Park H.R., Jung W.W., Bacchini P., Bertoni F., Kim Y.W., Park Y.K. Ezrin in osteosarcoma: comparison between conventional high-grade and central low-grade osteosarcoma. Pathology Research And Practice. 2006;202:509–515. doi: 10.1016/j.prp.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Ogino W., Takeshima Y., Mori T., Yanai T., Hayakawa A., Akisue T. High level of ezrin mRNA expression in an osteosarcoma biopsy sample with lung metastasis. Journal of Pediatric Hematology/Oncology. 2007;29:435–439. doi: 10.1097/MPH.0b013e3180640d18. [DOI] [PubMed] [Google Scholar]
- 39.Thomas D.M., Johnson S.A., Sims N.A., Trivett M.K., Slavin J.L., Rubin B.P. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. The Journal of Cell Biology. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X.Y., Lu Y., Zhao Y.J., Jaeweon K., Kang J., Xiao-Nan L. Cell cycle regulator gene CDC5L, a potential target for 6p12-p21 amplicon in osteosarcoma. Molecular Cancer Research : MCR. 2008;6:937–946. doi: 10.1158/1541-7786.MCR-07-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadikovic B., Thorner P., Chilton-Macneill S., Martin J.W., Cervigne N.K., Squire J. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202. doi: 10.1186/1471-2407-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Won K.Y., Park H.R., Park Y.K. Prognostic implication of immunohistochemical Runx2 expression in osteosarcoma. Tumori. 2009;95:311–316. doi: 10.1177/030089160909500307. [DOI] [PubMed] [Google Scholar]
- 43.Reddick R.L., Michelitch H.J., Levine A.M., Triche T.J. Osteogenic sarcoma: a study of the ultrastructure. Cancer. 1980;45:64–71. doi: 10.1002/1097-0142(19800101)45:1<64::aid-cncr2820450112>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Komiya S. Electron microscopy of bone tumors--osteosarcoma, chondrosarcoma, giant cell tumor of bone. Nihon Seikeigeka Gakkai zasshi. 1982;56:635–657. [PubMed] [Google Scholar]
- 45.Vuletin J.C. Myofibroblasts in parosteal osteogenic sarcoma. Archives of Pathology & Laboratory Medicine. 1977;101:272. [PubMed] [Google Scholar]
- 46.Martinez-Tello F.J., Navas-Palacios J.J. The ultrastructure of conventional, parosteal, and periosteal osteosarcomas. Cancer. 1982;50:949–961. doi: 10.1002/1097-0142(19820901)50:5<949::aid-cncr2820500525>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 47.Seemayer T.A., Schurch W., Lagace R. Myofibroblasts in human pathology. Human Pathology. 1981;12:491–492. doi: 10.1016/s0046-8177(81)80061-5. [DOI] [PubMed] [Google Scholar]
- 48.Brown L.F., Dubin D., Lavigne L., Logan B., Dvorak H.F., Van de Water L. Macrophages and fibroblasts express embryonic fibronectins during cutaneous wound healing. The American Journal of Pathology. 1993;142:793–801. [PMC free article] [PubMed] [Google Scholar]
- 49.Brown L.F., Lanir N., McDonagh J., Tognazzi K., Dvorak A.M., Dvorak H.F. Fibroblast migration in fibrin gel matrices. The American Journal of Pathology. 1993;142:273–283. [PMC free article] [PubMed] [Google Scholar]
- 50.Jodele S., Blavier L., Yoon J.M., DeClerck Y.A. Modifying the soil to affect the seed: role of stromal-derived matrix metalloproteinases in cancer progression. Cancer Metastasis Reviews. 2006;25:35–43. doi: 10.1007/s10555-006-7887-8. [DOI] [PubMed] [Google Scholar]
- 51.Hemingway F., Kashima T.G., Mahendra G., Dhongre A., Hogendoorn P.C., Mertens F. Smooth muscle actin expression in primary bone tumours. Virchows Archiv : An International Journal of Pathology. 2012;460:525–534. doi: 10.1007/s00428-012-1235-x. [DOI] [PubMed] [Google Scholar]
- 52.Yu L., Guo W., Zhao S., Wang F., Xu Y. Fusion between cancer cells and myofibroblasts is involved in osteosarcoma. Oncology Letters. 2011;2:1083–1087. doi: 10.3892/ol.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salas S., Bartoli C., Deville J.L., Gaudart J., Fina F., Calisti A. Ezrin and alpha-smooth muscle actin are immunohistochemical prognostic markers in conventional osteosarcomas. Virchows Archiv : An International Journal of Pathology. 2007;451:999–1007. doi: 10.1007/s00428-007-0474-8. [DOI] [PubMed] [Google Scholar]
- 54.Skalli O., Schurch W., Seemayer T., Lagace R., Montandon D., Pittet B. Myofibroblasts from diverse pathologic settings are heterogeneous in their content of actin isoforms and intermediate filament proteins. Laboratory Investigation; A Journal of Technical Methods and Pathology. 1989;60:275–285. [PubMed] [Google Scholar]
- 55.Hettmer S., Liu J., Miller C.M., Lindsay M.C., Sparks C.A., Guertin D.A. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20002–20007. doi: 10.1073/pnas.1111733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webber J., Steadman R., Mason M.D., Tabi Z., Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Research. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 57.Fuyuhiro Y., Yashiro M., Noda S., Matsuoka J., Hasegawa T., Kato Y. Cancer-associated orthotopic myofibroblasts stimulates the motility of gastric carcinoma cells. Cancer Science. 2012;103:797–805. doi: 10.1111/j.1349-7006.2012.02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noh K., Kim K.O., Patel N.R., Staples J.R., Minematsu H., Nair K. Targeting inflammatory kinase as an adjuvant treatment for osteosarcomas. The Journal of Bone and Joint Surgery American Volume. 2011;93:723–732. doi: 10.2106/JBJS.J.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richmond A., Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Disease Models & Mechanisms. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]