Summary
Neuropeptides play many important roles in cell-cell signaling and are involved in the control of anxiety, depression, pain, reward pathways, and many other processes that are relevant to psychiatric disorders. Mass spectrometry-based peptidomics techniques can identify the precise forms of peptides that are present in a given tissue. Utilizing this technique, peptides can be identified with any post-translational modifications that may be present and the exact sequence of the peptides can be determined. Unlike radioimmunoassays, which are limited by specific antibodies and often cannot discriminate between different lengths of peptides from the same precursor, peptidomics reveals the precise sequence and allows for the identification of both known and novel peptides. The use of isotopic labels allows for quantitative peptidomics, which results in the ability to compare peptide levels between differently treated samples. These tags can be synthesized in five different isotopic forms, permitting multivariate analysis of up to five different groups of tissue extracts in a single liquid chromatography/mass spectrometry run; this is ideal for measuring changes in neuropeptides in animals subjected to drug treatments, or in comparing animal models of psychiatric disorders.
Keywords: Peptidomics, proteomics, peptidase, protease
1. Introduction
Neuropeptides play a large role in a variety of physiological processes including sleep, feeding, pain, addiction, depression, mood regulation, and many more (1, 2). Neuropeptides are vital signaling molecules in the brain, which are involved in neurotransmission. Like classical neurotransmitters, neuropeptides bind to postsynaptic receptors and elicit cellular effects. Because they are critical to brain function, peptides have become key targets in drug discovery, and in particular for the treatment of mood and anxiety disorders (1, 3). Furthermore, a clear role for neuropeptides is seen when the nervous system is stressed or diseased (4). Some examples of peptides involved in psychiatric disease include the well known substance P and neuropeptide Y. Substance P antagonists have been implicated for the clinical treatment of depression, and neuropeptide Y elicits anti-depressant like effects (4, 5). Cocaine administration leads to altered peptide levels in mouse brains, as does chronic morphine (6, 7). A multitude of data has shown a role for peptides in modulation of behavior, clearly indicating the importance for the study of peptides.
Mass spectrometry-based quantitative peptidomic techniques provide a powerful way to study peptides and peptide processing (8). Previously, radioimmunoassays were used to study peptides and their processing (9), but this technique requires antibodies for known peptides, thereby limiting the ability for discovery of novel peptides. Furthermore, most antibodies are not specific for the exact peptide sequence and can bind to peptides that are N- and/or C-terminally extended or which contains post-translational modifications. Thus, it is difficult to know which specific form of the peptide the antibody is binding to. Utilizing mass spectrometry-based quantitative peptidomics, peptides can be sequenced using tandem mass spectrometry analysis and the precise molecular form of each peptide, including any post-translational modifications, can be determined.
To perform these experiments, animals are sacrificed, brain regions are dissected out, peptides are extracted and labeled with trimethylammoniumbutyryl (TMAB) isotopic tags and analyzed using liquid chromatography and mass spectrometry. Use of the TMAB labels allows for the quantification of peptides between conditions, such as knock out versus wild type mice to compare peptide profiles between these conditions. Another utilization of this approach is to administer varying treatments to animals and evaluate whether the treatment alters levels of particular peptides. A key aspect of this technique is extraction of the peptides. If peptides are not extracted properly from the brain regions, then the TMAB labels will not bind and peptide pools will not be able to be compared. For this reason various extraction conditions have been compared and are described and evaluated in this chapter. The following protocol describes the basic method of peptide extraction, isotopic labeling, and mass spectrometry analysis.
2. Materials
2.1 Extraction of Peptides
Distilled water Milli-Q system (Millipore, Bedford, MA, USA)
0.1 M Hydrochloric Acid (Pierce, Rockford, IL, USA)
0.01 N Acetic Acid (Fisher Scientific, Fair Lawn, NJ, USA)
Acetone (Fisher Scientific, Fair Lawn, NJ, USA)
0.4 M NaH2PO4 Sigma-Aldrich, Inc. (St Louis, MO, USA)
Ultrasonic processor (W-380, Ultrasonic Inc., Farmingdale, NY, USA)
Low retention microcentrifuge tubes, Eppendorf
2.2 Peptide Labeling
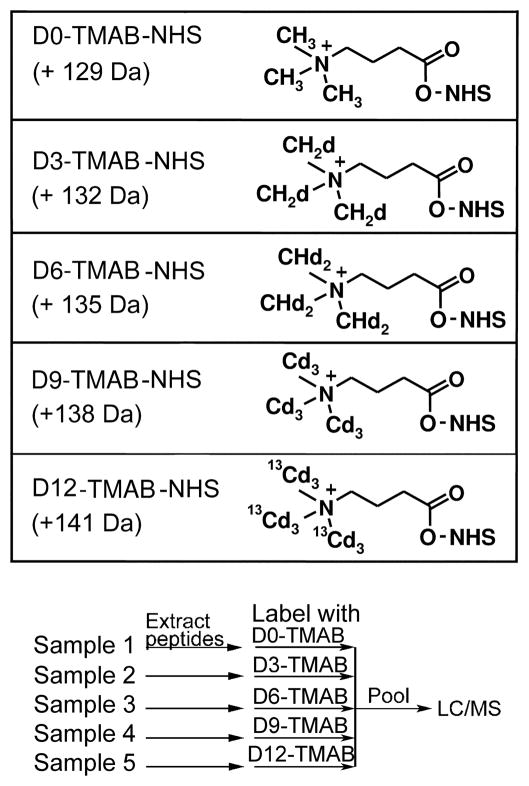
Trimethylammoniumbutyrate-N-hydroxysuccinimide (TMAB-NHS) isotopic compounds (Figure 1). The synthesis of these compounds has been previously described (10).
Figure 1.
TMAB labels and scheme for labeling of peptides extracted from tissues. Top: The compound structures shown includes the N-hydroxysuccinimide (NHS) moiety, which is replaced by the amine (N-terminus or Lys side chain) after reaction with the peptide. The indicated mass is the net mass shift for each tag incorporated into the peptide, replacing a hydrogen on the amine. Note the quaternary amine group which provides a positive charge after labeling. This allows for the charge of the primary amine on the peptide to remain positive. The TMAB-NHS labeling reagents are synthesized as the chloride salt (not shown). Bottom: Labeling strategy for experiment comparing five different samples. After labeling, peptides are pooled prior to purification by microfiltration, desalting, and analysis on LC-MS/MS.
1.0 M NaOH Sigma-Aldrich, Inc. (St. Louis, MO, USA).
Dimethylsulfoxide Sigma-Aldrich, Inc. (St. Louis, MO, USA).
NH2OH HCl Sigma-Aldrich, Inc. (St. Louis, MO, USA).
Glycine Sigma-Aldrich, Inc. (St. Louis, MO, USA).
Hydrion pH Papers, range 8.0–9.5 (Micro Essential Laboratory, Brooklyn NY, USA).
2.3 Peptide Purification
Amicon Ultra 4 mL Ultracel 10,000 Molecular weight cutoff Centrifugal Filter Devices (Millipore, Bedford, MA, USA).
PepClean™ C-18 spin column Pierce (Rockford, IL, USA)
Acetonitrile HPLC grade Fisher Scientific (Fair Lawn, NJ)
Trifluoroacetic acid Pierce (Rockford, IL, USA)
3. Methods
3.1 Peptide Extraction
1. Sacrifice and Microwaving
Mice are sacrificed by cervical dislocation followed by decapitation. Heads are then immediately placed in a conventional microwave oven at full power for 8 seconds in order for the brain to reach an internal temperature of 80°C (11). This temperature is sufficient to inactivate proteases in the brain. Due to variability in microwave strength, the unit needs to be calibrated to ensure that the proper internal temperature of the brain is achieved. Mice can also be sacrificed by microwave irradiation, although this requires a specific piece of equipment that is not available in most laboratories (12).
2. Dissection
Following microwaving, the brain is removed from the scull and a sterile razor blade is used to dissect out brain regions of interest. The razor blade is held in coronal orientation and is used at the proper coordinates for each region, as is found in the Paxinos and Franklin mouse brain atlas (13). After removing the olfactory bulbs, the prefrontal cortex is removed by cutting at Bregma 1.94 and taking the anterior slice. Another coronal cut is made at bregma 0.00, the resulting slice of Bregma 1.94 to 0.00 contains cortex and the striatum. The cortex can be peeled off from the dorsal side and the striatum is located on both sides of the midline. The “striatum” sample includes the caudate putamen, nucleus accumbens, septum, and ventral palladium. The next coronal cut is made at Bregma −3.00, this coronal section of Bregma 0.00 to −3.00 is dissected into the hippocampus, thalamus, amygdala, and hypothalamus. Cortex can be dissected from this section as well. Lastly, the forebrain and brainstem can be removed from the rest of the brain, leaving the cerebellum to be collected.
3. Tissue Storage
Because peptides are quite sticky in character, low retention tubes need to be used during all steps of this process in order to limit the loss of peptides (see Note 1). Prior to placing brain regions into tubes for storage, the tubes need to be washed with double distilled water. As regions are dissected out they are put into the low retention tubes and are immediately placed on dry ice. At this point, tissues from multiple animals can be pooled into single tubes, if each group will consist of multiple animals. If not proceeding to the next step immediately, tissues should be stored at −70°C.
4. Sonication
Immediately prior to sonication, 5 μL of ice-cold water per μg of tissue is added to each sample in the same 2 ml low retention tube into which the samples were placed after dissection. A minimum of 200 μL needs to be used. Using an ultrasonic processor, dissected tissue pools are sonicated two times for 20 sec at 1 pulse/sec at duty cycle 3, 50% output. A small additional volume of water is used to rinse the tip of the sonicator into the tube containing the brain extracts, ensuring no peptides are left on the sonication tip. The sonicator should be washed with double distilled water between tissue extractions to prevent contamination of peptides between samples.
5. Extraction of Peptides
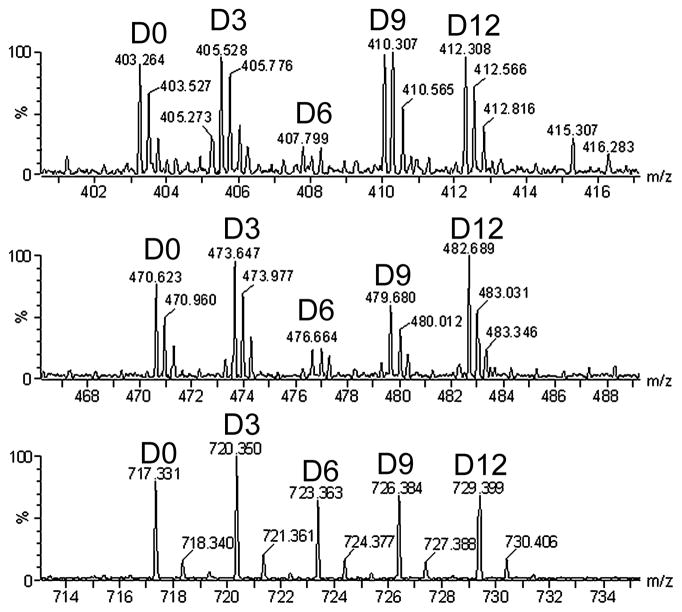
The homogenates are incubated in a 70°C water bath for 20 min and then cooled on ice for 15 min. In the representative data shown in Figure 2, different extraction protocols were used to evaluate efficacy of peptide recovery. The details of the different extraction conditions are described in the legend, Figure 2. The basic procedure used in our laboratory (and shown with the D9 label in Figure 2) involves the addition of ice-cold 0.1 M HCl to a final concentration of 10 mM HCl. Each sample was mixed and incubated on ice for 15 minutes. The homogenates were centrifuged at 13,000 g for 40 min at 4°C and the supernatants transferred to new 2 ml low-retention tubes. The extracts can be frozen and concentrated to a lower volume in a vacuum centrifuge. This may be necessary if large volumes are to be combined during the filtration step (for filtration, it is best if the total volume of all samples is less than 4.0 mL).
Figure 2.
Representative MS data. Each spectrum illustrates one peptide labeled with each of the five tags. In this experiment, different extraction conditions were tested and the sample analyzed on a Waters q-TOF mass spectrometer. Several mouse brains were homogenized by sonication, pooled, and then divided into aliquots and extracted with different conditions. After extraction, the peptides were labeled as in Figure 1, pooled, and analyzed by LC-MS/MS. The sample labeled with the D9 reagent was extracted as described in Section 3.1 using 10 mM HCl. The other samples used a generally similar procedure except for minor differences. The D0 sample extraction condition was 0.25% acetic acid, the D6 sample was extracted with water alone, and the D12 sample was extracted with 100 mM HCl. The D3 labeled sample was extracted using acidified acetone, as described (17). This procedure involved initial extraction in 10 mM HCl (as the D9 sample) followed by re-extraction of the pellet in ice-cold acetone solution (40:6:1 acetone/water/HCl). After centrifugation at 13,000 g for 30 min at 4°C, the second supernatant was lyophilized and combined with the first 10 mM HCl extract. Top panel: spectrum showing alpha-neoendorphin (YGGFLRKYPK) extracted with the five different conditions. This peptide has a 4+ charge and is labeled with 3 tags, two on lysines and one on the N-terminus. Quantitative analysis of the spectra illustrates that the peptide was poorly extracted with water alone, but extracted equally well with all other conditions. Middle panel: Dynorphin A10-17 (PKLKWDNQ). This peptide has a 3+ charge and is labeled with 3 tags. This peptide is also not efficiently extracted with water alone. Bottom panel: proenkephalin derived Met-enkephalin with oxidized Met (YGGFMox). This peptide has a charge of 1+ and has one tag at the N-terminus. This peptide is extracted equally well with all conditions used, although it is possible that there is a slight improvement with the acidified acetone relative to the other conditions (replicates would be needed to verify a small 20% change in levels).
6. Buffering
0.4 M sodium phosphate buffer (pH 9.5) is added to the peptide extracts to adjust the pH of the mixture to 9.5. These extracts can be stored at −70°C until labeling.
3.2 Labeling
Since peptides often have free amines on the N-terminus and/or on a Lys side chain, reagents that react with primary amines can be used for quantitative peptidomics studies (14). One commercially available reagent is succinic anhydride with four hydrogens or deuteriums. Another commonly used isotopic tag is 3-(2,5-dioxopyrrolidin-1-yloxycarbonyl)propyl trimethylammonium chloride (15), which is the N-hydroxysuccinimide (NHS) ester of trimethylammoniumbutyrate (TMAB). The TMAB reagents can be easily synthesized from gamma-aminobutyric acid and iodomethane containing various numbers of hydrogen and deuterium (10). We find that the TMAB reagent is superior to succinic anhydride for several reasons. First, for accurate quantification, it is important that the heavy and light forms of the peptides co-elute from HPLC columns. We have found that peptides labeled with the TMAB reagent containing 9 deuteriums co-elute with peptides labeled with the reagent containing 9 hydrogens, whereas peptides labeled with succinic acid containing four deuteriums do not precisely co-elute with peptides labeled with succinic acid containing four hydrogens, (16). Second, because the TMAB reagent contains a positive charge (a quaternary amine) the positive charge of the N-terminus and any Lys residues of the peptide will be maintained when labeled with TMAB. With succinic anhydride the positive charge of the free amines is converted to a negative charge. If there are no other positive charges in the peptide sequence, then the peptide will not be detected with mass spectrometry performed in positive ion mode. Third, succinic anhydride-labeled peptides tend to show weaker signals than TMAB-labeled peptides (16). Lastly, five different isotopic forms of the TMAB labels can be produced. This provides the ability to perform multivariate analysis of different tissue extracts. The five forms differ by a mass difference of 3 Da, the D)-TMAB-NHS contains all hydrogens and no deuteriums, the D3-TMAB-NHS has 3 deuteriums, the D6-TMAB-NHS has 6 deuteriums, the D9-TMAB-NHS has 9 deuteriums, and the fifth label has nine deuteriums and three atoms of 13C, thereby making its mass 3 Da higher than the D9 label and is therefore referred to as the D12-TMAB-NHS (10). Structures of the reagents are shown in Figure 1 along with the masses added to the peptide by the addition of the TMAB group to an amine and loss of one hydrogen atom. It should be pointed out that some peptides have N-terminal modifications, such as acetylation, and no Lys residues in their sequence. In this case there are no free amines, and therefore the tags would not have anywhere to attach. These peptides would not be quantifiable using the following approach (see Note 2).
The number of labels used and the number of replicates depends on the experimental questions being addressed. For example, it is possible to use all five isotopic tags to directly compare the levels of various peptides five separate samples, as shown in Figure 1, bottom panel. Representative data comparing five different extraction conditions is shown in Figure 2. If comparing only two conditions, such as wild-type (WT) and knock-out (KO) mice, only two isotopic tags need to be used. However, all five tags could be used by including three replicates of one condition and two of another. This provides several experimental advantages over using two tags. First, a higher n value can be achieved without having to perform additional runs. Also, because the resulting analysis provides relative values, statistical analysis of the data requires the comparison of the relative level of peptides in WT vs WT groups. Comparing WT vs WT also allows evaluation of the variability between control animals. If five isotopic tags are used, multiple WT animals can be evaluated in the same run. If only two labels are used, it is important to perform a duplicate analysis in which the tags are switched so that WT and KO are labeled with the opposite tags from those used in the previous run. This additional run eliminates the possibility that changes in peptide levels are due to particular TMAB tags rather than the conditions themselves. If the five tags are used for five different samples, as shown in Figures 1 and 2, it is important to replicate the results with additional groups in which the order of tags is switched, which provides replicates as well as a control for labeling efficiency.
1. Labels
Depending on how many samples are to be compared, two to five labels may be used in an experiment. 5 mg of TMAB-NHS label is used per brain region. The label is dissolved in DMSO at a concentration of 400 μg TMAB per μl of DMSO. The mixture needs to be mixed until the label has fully dissolved (see Note 3).
2. Label Volume Calculation
To determine the volume of labeling solution added per round of labeling, divide the total volume of DMSO used to dissolve TMAB by seven. This is done because seven rounds of labeling will be carried out. For example, if 5 mg label was used and dissolved in 12.5 μl of DMSO, then 1.8 μl will be added during each round of labeling.
3. Labeling
Before starting the labeling process, the pH of each sample is checked by blotting <1 μL of each sample onto pH paper (range = 8 to 9.5). If the pH is below 9.5, 1.0 M NaOH is used to bring the pH up to the desired 9.5. Once the pH is adjusted, the first round of labeling begins. At the start of each round, one seventh of the label volume is added to each sample and mixed using a vortex mixer. This is repeated with each label being added to its respective sample. Samples are allowed to incubate at room temperature for 10 minutes. After this incubation period, the pH is verified and adjusted to 9.5 as was done at the start of labeling. Samples are incubated for another ten minutes at room temperature before the next round of label is added. This process is repeated six more times to ensure all peptides are properly labeled. It is important that the pH of the sample is at 9.5 before label is added to the sample. After the final addition of TMAB-NHS reagent, the samples should be incubated at room temperature for another 10–30 minutes. The entire labeling procedure requires approximately four hours.
4. Quenching
After the labeling and prior to combining the samples for filtration, any unreacted TMAB-NHS labels need to be quenched with 2.5 M glycine in water. For this, 10 μl of the glycine solution is added per 5 mg of TMAB-NHS reagent used. The samples are then incubated at room temperature for 40 minutes.
3.3 Peptide Purification
1. Pool and Filter
After labeling, all samples that are going into the same run (i.e. samples labeled with different tags) are pooled together, as per the scheme in Figure 1. Microfiltration is performed with Amicon Ultra 4 mL Ultracel filters. These filters have a 10,000 nominal molecular weight limit and therefore trap most proteins and allow only peptides and small proteins through the pores. Before placing samples on filters, the filters need to be prepared by washing off any glycerol that may be on the filter from the manufacturer (see Note 4). To do this, approximately 2 mL of distilled water is placed onto the filter and the filter is placed into a 15 mL falcon tube (do not use the tube that comes with the filters for this wash). The filters are then placed into a centrifuge and spun until all of the water passes through the filter into the collection tube. The filters are then placed into the collection tubes that are provided from the manufacturer and pooled samples are loaded onto the clean filter. The samples are then spun according to usage guidelines in a refrigerated centrifuge kept at 4°C. The flow through is used for further steps and analysis.
2. Hydroxylamine Step
It is important that peptides are labeled only on lysines and N-termini. To ensure this, hydroxylamine (NH2OH) is used to hydrolyze any tyrosine labeling. 2.0 M hydroxylamine in DMSO solution is used for this step. 3 μl of the hydroxylamine solution is used for every 10 mg of TMAB label in the entire filtrate. For example, if 5 mg of each label was used for each sample and the run includes samples with all 5 tags, then the total in the filtrate is 25 mg label. In this case 7.5 μl of hydroxylamine solution would be needed. To start, the pH of the filtrate is adjusted to 9.0 using 1.0 M NaOH. Then, because this is done in three rounds, 1/3 of the total volume of hydroxylamine is added to the filtrate, vortex mixed, and incubated at room temperature for 10 minutes. The pH is adjusted back to 9.0 and the second aliquot of hydroxylamine is added. After another 10 minutes of incubation, the final round of pH and hydroxylamine is performed. After 10 more minutes at room temperature, these filtrates can be stored at −70°C until desalting.
3. Desalting
Desalting is carried out with a PepClean™ C-18 spin column (Pierce). First, the resin from two C-18 colums are combined in one. This is done due to the large amount of peptides that will be loaded onto the column. Once the resin is combined, the procedure described in the manufacturers’ instructions is followed. Briefly, the column is first activated several times with acetonitrile solution and is then equilibrated several times with acetonitrile and trifluoroacetic acid solution. The sample, with added sample buffer, is loaded on top of the resin bed (400 μl aliquots at a time). The resin with bound sample is repeatedly washed. Lastly, the peptides are eluted with 80 μl of 70% acetonitrile and 0.1% trifluoroacetic acid in water. These eluents are frozen on dry ice, concentrated to 10–20 μl in a vacuum centrifuge, and stored at −70°C until mass spectrometry (MS) analysis.
3.4. Liquid chromatography and mass spectrometry (LC-MS/MS)
To detect and quantify the largest number of natural peptides, best results are obtained by chromatography on a reverse phase column with direct electrospray ionization mass spectrometry on a quadrupole time-of-flight (q-TOF) instrument. Matrix-assisted laser desorption ionization is less effective than electrospray ionization instruments because the laser causes decomposition of the TMAB tag and the loss of the trimethylamine group containing the isotopic label. The extent of the decomposition depends on the intensity of the laser, and can represent the majority of the signal. Because the trimethylamine group contains the isotopic-tag, the signal resulting from the decomposed peptide cannot be quantified. The trimethylamine label is also largely removed by the collision-induced dissociation used for MS/MS sequencing (described below).
Many different LC systems have been used with success. When small amounts of sample are used, such as when analyzing individual mouse brain regions, nanospray is optimal. If larger sample amounts are available, then nanospray is not critical and microspray can be used. Typical protocols for both microspray and nanospray LC/MS analysis on q-TOF mass spectrometers are described below:
3.4.1. Nanospray LC/MS using a Waters Synapt G1 Q-TOF mass spectrometer
Frozen samples are thawed and briefly centrifuged in a microfuge to remove particulates.
An aliquot (typically 2–5 μl) is injected onto a Symmetry C18 trapping column (5 μm particles, 180 μm i.d.× 20 mm, Waters, USA).
The material is desalted online for 10 min and the trapped peptides are separated by elution with a water/acetonitrile 0.1% formic acid gradient through a BEH 130 - C18 column (1.7 μm particles, 100 μm i.d. x 100 mm, Waters, USA), at a flow rate of 1 μl/min.
Data are acquired in data-dependent mode and selected peptides dissociated by collisions with argon.
3.4.2. Microspray LC/MS using an Agilent 6520 Q-TOF mass spectrometer
Frozen samples are thawed and briefly centrifuged in a microfuge to remove particulates. The supernatant is dried in a SpeedVac and reconstituted with 20 μl of 0.1% TFA.
An aliquot (2–5 μl) is injected onto a ZORBAX 300 SB-C18 column (150 X 0.5 mm, 5 μm particles, Agilent, USA) using a 1200 series capillary HPLC system
Peptides are separated and eluted with a water/acetonitrile 0.1% formic acid gradient (75 min stop time) at a flow rate of 5 μl/min.
The MS/MS data is acquired in data-dependent mode using nitrogen as collision gas.
3.4.3. Effect of collision energy on peptide identification by MS/MS
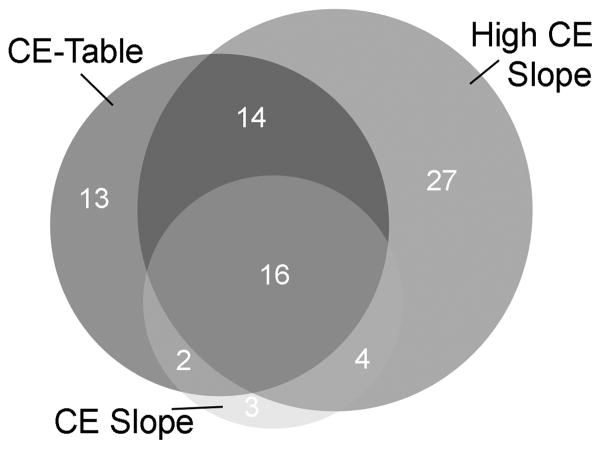
The identification of peptides requires optimal fragmentation of peptides by collision-induced dissociation. Depending on the length, charge, and amino acid sequence, peptides can require different energies to produce sufficient fragmentation to allow for the unequivocal identification. Moreover, the presence of the TMAB group on the peptides adds additional complexity to the fragmentation because this group partially fragments upon collision-induced dissociation, eliminating the trimethylamine group through neutral loss. As a result, higher collision energies are needed to ensure adequate fragmentation of the peptide bonds of the TMAB-labeled peptides. For example, we recently compared the same sample run under three different conditions for selecting the collision energy. When analyzed using energy values that are standard for analysis of un-tagged peptides (3V/100 m/z + off set 2V), only 25–45 peptides could be identified after Mascot searches of the MS/MS data (Figure 3). In contrast, when another aliquot of the sample was analyzed using a higher collision energy (3.9V/100 m/z + off set 2.9V), 61 peptides could be identified from the Mascot searches (Figure 3). These 61 peptides represent 77% of all peptides identified in all three runs. Therefore, while multiple runs using different collision energies will identify the most peptides, if only one run is possible, it is optimal to use higher collision energy for the TMAB-labeled peptides than typically used for unlabeled peptides.
Figure 3.
Comparison of collision energy effect on the numbers of peptides identified by Mascot analysis of MS/MS data. A sample of mouse brain peptides was run on the same Agilent LC/MS Q-TOF instrument under identical conditions except for variations in the collision energy. The green circle (CE Slope) represents peptides run under standard energy values: 3V/100m/z +2V offset. The red circle (High CE Slope) represents a higher collision energy version of the above: 3.9V/100m/z +2.9V offset. The blue circle (CE Table) represents a higher collision energy using parameters selected from a table: m/z 300–1500, Z = 1, CE 9-69V; Z = 2, CE 9-69V; Z = 3, CE 9-54V; Z >3, CE 9-54V. The number of peptides identified by Mascot searches for each dataset is indicated; those peptides found in multiple searches are shown in the overlap of the circles, while the peptides found uniquely to each dataset are in the area with no overlap.
3.5 Data analysis
Relative levels of peptides between different groups are compared by measuring peak heights within mass spectra, using the appropriate software program (for example, Mass Hunter qualitative analysis software is used for viewing data obtained on a Agilent instrument, and MassLynx for analysis of data obtained on Waters intruments). Spectra are scanned in this program and peaks sets are selected based on co-elution and separation from one another. Peptides separate along the x-axis, which is the mass to charge ratio of each peptide, with a typical range of 300–1700. The majority of peptides elute from the LC step over a 20–30 minute period. Some examples of peak sets are shown in Figure 2. Each of the spectra is a snapshot of 1 sec of the mass spec run and is zoomed in to the m/z range at which the peptides are seen. The top panel shows a neuropeptide from prodynorphin, named alpha-neoendorphin with the sequence YGGFLRKYPK. Based on the sequence we know that the charge should be 4. But we can also see the charge of the peptide by comparing the 13C-containing peaks to the monoisotopic peak in the spectra. This calculation considers that the monoisotopic peak is at m/z 403.264 and the peak containing one atom of 13C is 403.527, therefore the difference is 0.25 m/z or 1/4, meaning the charge is 4. The m/z difference between the monoisotopic peak of the first and second peak set is ~2.25 m/z (405.528-403.264). This means that since m/z=2.25 and z=4, then the mass difference is 4 x 2.25, or 9 Da. Since we know that these peak sets are labeled with the D0 and D3, which has a 3 Da difference, to account for the 9 Da difference, the peptide therefore must be labeled with 3 isotopic tags. The mass of this peptide without isotopic tags or protons can be calculated from the following formula.
where m/z is the observed mass to charge value for the monoisotopic peak,
z is the charge state,
c is the mass of the TMAB tag (128.118 for the D0 TMAB, 137.170 for the D9 TMAB),
T is the number of tags incorporated,
1.008 is the mass of a proton,
and (z-T) is the calculation of the number of protons (i.e. the difference between the charge and the number of tags). This last part of the equation is essential because the TMAB tags add a positive charge due to the quaternary amine group and therefore the charge state is not equal to the number of protons.
Typically, the mass of the unmodified peptide is taken as the average of the values determined from all of the isotopic tags. Using these calculations for the example in the top panel of Figure 2, the average of the values from all of the tags results in a mass of 1227.71 for this peptide. We can then check databases of known neuropeptides and see that alpha-neoendorphin has an expected mass of 1227.68 Da. In addition, alpha-neoendorphin is predicted to incorporate 3 TMAB isotopic tags (one on the N-terminus and two on Lys residues), and to have a charge of 4+ (one for each TMAB tag plus one for the proton on the Arg residue). While this identification of the peptide is highly likely based on the similarities between the predicted and observed mass, number of incorporated tags, and charge state, unequivocal identification requires MS/MS sequencing, which is discussed in the next section.
Quantification of the results is performed by measuring peak intensity. Typically, the height of the monoisotopic peak and the peak containing 1 atom of 13C are averaged so that the peak intensity is based on multiple points. Figure 2 top panel shows that alpha-neoendorphin abundance is different between the 5 extraction conditions. Whereas the height of the peaks that are D0, D3, D9, and D12 labeled are relatively equal, the D6 labeled peak is much lower than the rest. This example illustrates that the water alone extraction condition (which was labeled with the D6 TMAB) did not extract this peptide as well as the other conditions. Figure 2 middle panel shows an example with another peptide derived from prodynorphin, dynorphinA10-17 (PKLKWDNQ) with a charge of 3+ and 3 tags. This peptide was also not extracted as well with water alone. Figure 2 bottom panel shows the spectra for a proenkephalin derived peptide named met-enkephalin with an oxidized Met (YGGFMox). This peptide has a charge of 1+ with 1 tag on the N-terminus. In this example, all conditions are relatively equal, suggesting that this neuropeptide is not extracted preferentially with any of the conditions.
It should be noted that when five labels are used, there are sometimes spectra which contain overlapping peaks in which the signals with multiple 13C atoms extend into the range of the next TMAB tag. In these cases, the isotopic distribution of the peptide must be calculated and the contribution from the 13C-containing peaks subtracted as described (10).
3.6. Identification of peptides by MS/MS sequencing
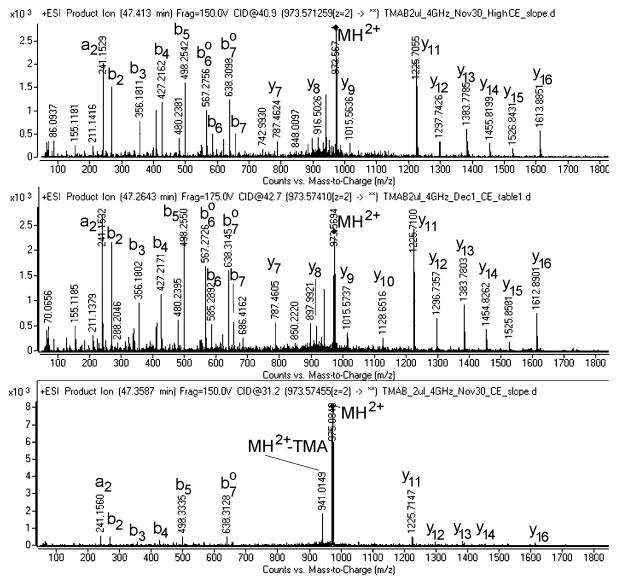
Interpretation of MS/MS data is typically performed by computer-assisted searching of databases consisting of proteins or translated cDNA. There are a number of programs available for database searching. For peptides labeled with the TMAB reagents we have found the optimal program to be Mascot. This program currently has four of the five TMAB labels included as options (the missing one is the D12-TMAB). Most importantly, the Mascot program considers the neutral loss of TMA from the peptides during collision-induced dissociation; this causes the loss of 59 Da from peptides labeled with one D0-TMAB tag, 62 Da from peptides labeled with one D3-TMAB tag, 65 Da from peptides labeled with one D6-TMAB tag, and 68 Da from peptides labeled with one D9-TMAB. For example, the peptide little SAAS incorporates a single TMAB tag on the N-terminus, and for the D6-TMAB-labeled peptide, all of the b ions are 65 Da lighter than if the intact TMAB was attached (Figure 4). In this example, the precursor ion (M+H)2+ also shows loss of the TMA moiety. Note the impact of collision energy on the fragmentation pattern, with “normal” collision energy parameters for unlabeled peptides (“CE-Slope) producing only minimal fragmentation while the two higher energy LC/MS runs showed substantially more fragmentation of the peptide (Figure 4).
Figure 4.
Representative MS/MS spectra showing peptide identification. All three MS/MS spectra are from the same peptide (named little SAAS; sequence SLSAASAPLVETSTPLRL) labeled with the D6-TMAB tag. Top panel: High CE Slope (collision energy 40.9V). Middle panel: CE Table (collision energy 42.7V). Lower panel: CE Slope (collision energy 31.2V).
Mascot searches need to be followed by manual interpretation to eliminate false positives. Several criteria are used to accept or decline the peptides identified by Mascot:
The isotopic form of TMAB matched by Mascot is the correct one based on analysis of the peak set. While this may seem obvious, Mascot does not consider the peak set and know which of the individual peaks correspond to the D0, D3, D6, D9, or S12 forms. Therefore, if using the five isotopic forms of TMAB, there is a 1 in 5 chance that a false positive labeled with one tag is correct. If two tags, there is a 1 in 25 chance that a false positive has the correct tags, and for three tags a 1 in 125 chance that a false positive has the correct tags. Therefore, correlation of the isotopic TMAB form in the observed peak set with the predicted Mascot match is a simple and necessary step, and most false positives will fail this test.
The number of tags incorporated into the peptide matches the number of free amines (N-terminus and side chains of Lys). If multiple tags are incorporated, all should be the same isotopic form on a particular peptide. As mentioned above in point #1, for peptides labeled with multiple tags the false positives will rarely have consistent forms of the tags, while real positives will all be the same isotopic form.
The Mascot score is either the top score of all potential peptides, or the other peptides with comparable scores can be excluded by the above criteria, leaving only one peptide that matches all criteria.
The majority (>80%) of the major MS/MS fragment ions match predicted a, b, or y ions, internal ions, or precursor ions with loss of trimethylamine.
The mass accuracy of the fragment ions is within the accepted specification for the q-TOF instrument used for the analysis. For older q-TOF instruments, this would mean within 50 parts per million; for newer instruments this can be within 10 parts per million or better.
A minimum of 5 fragment ions match b or y ions. For small peptides, this criterion can be a problem.
The charge state should be reasonable based on the peptide sequence. For most peptides, this means that the charge state equals the number of TMAB tags plus the number of Arg and His residues, although His residues are not always positively charged and peptides detected with two different charge states usually indicates the presence of a His residue. On occasion, some peptides will pick up an additional proton to give a charge state one higher than the maximum predicted from the number of amines, although this is usually a minor form relative to the ion with the correct charge state.
Acknowledgments
The development of the techniques described in this chapter was supported by National Institutes of Health grants DA-04494 (L.D.F.). Some of the mass spectrometry was performed in the Dalton Mass Spectrometry Laboratory at the Institute of Chemistry, University of Campinas, Brazil, supported by FAPESP, INCT Bioanalitica and CNPq.
Footnotes
Avoiding contaminants is important. Small molecules and polymers can substantially interfere with the MS analysis. Clean water is essential. Some brands of microfuge tubes and filtration devices have polymeric contaminants that appear as polyethylene glycol-related compounds on MS; these contaminant signals usually overwhelm the signals from the tissue-derived peptides. Using low-retention mirofuge tubes and pipette tips is important, as some peptides bind regular tubes and tips and are lost during transfers and labeling steps. Whenever possible, rinse and dry tubes and tips with ultrapure deionized water before use.
While the quantitative peptidomics technique is able to identify hundreds of peptides and their specific processing forms from a single LC-MS/MS run, the method does not detect every peptide in the sample. For example, peptides lacking an N-terminal free amine (due to post translational modification such as acetylation or pyroglutamylation) that also lack an internal lysine residue are not labeled by the TMAB reagent. These peptides are therefore present as single peaks, and no information on relative levels can be obtained using this method. In other cases, intrinsic factors can cause low ionization efficiency of certain peptides during mass spectrometry. The mass/charge ratios used in these studies (typically 300–1700 m/z) excludes very small peptides as well as large peptides that contain few positive charges so that the m/z is greater than 1700. Finally, the dynamic range of peptide levels in biological samples varies beyond the dynamic range of the mass spectrometry equipment and low abundance peptides are not detectable above the background.
Prepare labeling solutions as well as glycine and hydroxylamine solutions fresh each day. It is extremely important to prepare all solutions (HCl, NaOH, glycine, phosphate buffer and desalting solutions) with ultrapure deionized water to avoid contamination from small organic molecules that can interfere with mass spectrometry.
It is extremely important to clean Centricon filters before filtering peptides. These filters often contain glycerol which needs to be washed off before filtering to avoid sample contamination which will substantially interfere with mass spectrometry. Make sure that filters are covered with ultrapure deionized water, and then run the water through the filters at the designated centrifugal speed. Be sure to empty any remaining water before loading extracts for filtering.
References
- 1.Hokfelt T, Bartfai T, Bloom F. Neuropeptides: opportunities for drug discovery. Lancet Neurol. 2003;2:463–472. doi: 10.1016/s1474-4422(03)00482-4. [DOI] [PubMed] [Google Scholar]
- 2.Strand FL. Neuropeptides: general characteristics and neuropharmaceutical potential in treating CNS disorders, Progress in drug research. Fortschritte der Arzneimittelforschung. 2003;61:1–37. doi: 10.1007/978-3-0348-8049-7_1. [DOI] [PubMed] [Google Scholar]
- 3.Rotzinger S, Lovejoy DA, Tan LA. Behavioral effects of neuropeptides in rodent models of depression and anxiety. Peptides. 31:736–756. doi: 10.1016/j.peptides.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Hokfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M. Neuropeptides--an overview. Neuropharmacology. 2000;39:1337–1356. doi: 10.1016/s0028-3908(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 5.Ishida H, Shirayama Y, Iwata M, Katayama S, Yamamoto A, Kawahara R, Nakagome K. Infusion of neuropeptide Y into CA3 region of hippocampus produces antidepressant-like effect via Y1 receptor. Hippocampus. 2007;17:271–280. doi: 10.1002/hipo.20264. [DOI] [PubMed] [Google Scholar]
- 6.Che FY, Vathy I, Fricker LD. Quantitative peptidomics in mice: effect of cocaine treatment. J Mol Neurosci. 2006;28:265–275. doi: 10.1385/JMN:28:3:265. [DOI] [PubMed] [Google Scholar]
- 7.Decaillot FM, Che FY, Fricker LD, Devi LA. Peptidomics of Cpefat/fat mouse hypothalamus and striatum: effect of chronic morphine administration. J Mol Neurosci. 2006;28:277–284. doi: 10.1385/JMN:28:3:277. [DOI] [PubMed] [Google Scholar]
- 8.Fricker LD, Lim J, Pan H, Che FY. Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 9.Gray TS, Morley JE. Neuropeptide Y: anatomical distribution and possible function in mammalian nervous system. Life sciences. 1986;38:389–401. doi: 10.1016/0024-3205(86)90061-5. [DOI] [PubMed] [Google Scholar]
- 10.Morano C, Zhang X, Fricker LD. Multiple isotopic labels for quantitative mass spectrometry. Analytical chemistry. 2008;80:9298–9309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Svensson M, Skold K, Svenningsson P, Andren PE. Peptidomics-based discovery of novel neuropeptides. Journal of proteome research. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 13.Paxinos G, Franklin KB. The Mouse Brain in Stereotaxic Coordinates. Academic Press; San Diego: 2001. [Google Scholar]
- 14.Julka S, Regnier F. Quantification in proteomics through stable isotope coding: a review. Journal of proteome research. 2004;3:350–363. doi: 10.1021/pr0340734. [DOI] [PubMed] [Google Scholar]
- 15.Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE. Controlling deuterium isotope effects in comparative proteomics. Analytical chemistry. 2002;74:3662–3669. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- 16.Che FY, Fricker LD. Quantitative peptidomics of mouse pituitary: comparison of different stable isotopic tags. J Mass Spectrom. 2005;40:238–249. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- 17.Lee JE, Atkins N, Jr, Hatcher NG, Zamdborg L, Gillette MU, Sweedler JV, Kelleher NL. Endogenous peptide discovery of the rat circadian clock: a focused study of the suprachiasmatic nucleus by ultrahigh performance tandem mass spectrometry. Mol Cell Proteomics. 2010;9:285–297. doi: 10.1074/mcp.M900362-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]