Abstract
The ability to consistently detect cell-free tumor-specific DNA in peripheral blood of patients with metastatic breast cancer provides the opportunity to detect changes in tumor burden and to monitor response to treatment. We developed cMethDNA, a quantitative multiplexed methylation-specific PCR assay for a panel of ten genes, consisting of novel and known breast cancer hypermethylated markers identified by mining our previously reported study of DNA methylation patterns in breast tissue (103 cancer, 21 normal on the Illumina HumanMethylation27 Beadchip) and then validating the 10-gene panel in a TCGA breast cancer methylome database. For cMethDNA, a fixed physiological level (50 copies) of artificially constructed, standard non-human reference DNA specific for each gene is introduced into in a constant volume of serum (300 μl) prior to purification of the DNA, facilitating a sensitive, specific, robust and quantitative assay of tumor DNA, with broad dynamic range. Cancer-specific methylated DNA was detected in Training (28 normal, 24 cancer) and Test (27 normal, 33 cancer) sets of recurrent Stage 4 patient sera with a sensitivity of 91% and a specificity of 96% in the test set. In a pilot study, cMethDNA assay faithfully reflected patient response to chemotherapy (N = 29). A core methylation signature present in the primary breast cancer was retained in serum and metastatic tissues collected at autopsy 2–11 years after diagnosis of the disease. Together, our data suggest that the cMethDNA assay can detect advanced breast cancer, and monitor tumor burden and treatment response in women with metastatic breast cancer.
Keywords: breast, markers, methylation, blood, tumor
INTRODUCTION
Recent developments have shown that circulating cell-free DNA in blood offers a very convenient, noninvasive, and repeatable “liquid biopsy”, thus providing a reliable template for assessing molecular markers that reflect tumor burden. Several DNA methylation studies have suggested clinical prognostic and/or predictive utility of methylated biomarkers for predicting outcome and monitoring treatment of breast cancer. DNA methylation is one of the earliest, most robust, and frequent alteration in cancer, leading to silencing of expression of tumor suppressor genes (1). Methylation of a tumor suppressor gene allele can act as an inactivating ‘hit’, having a similar function as a genetic mutation or deletion. As examples, BRCA1 (breast, ovarian), PTEN (ovarian), HRK (prostate), APC (colorectal), and RASSF1A (multiple cancer cell types) have been found both methylated and mutated in cancer. Methylated RASSF1A and APC, identified in serum DNA from breast cancer patients, were associated with a worse outcome (2). RASSF1A and NEUROD1 were shown to be useful for monitoring the efficacy of adjuvant therapy in breast cancer patients (3, 4). Other biomarkers such as p16INK4A, CDH1, DAPK11, HIC1, RARB, CDH13, ESR1, GSTP1, have been evaluated alone or in combination in serum [reviewed in (5)]. However, a reproducible blood-based test for hypermethylated genes for diagnosis and follow-up of breast cancer has yet to be incorporated into a clinical laboratory assay. Discovery of new markers, as well as improvements in existing technologies are needed in order to provide more robust, reproducible, quantitative, sensitive and specific assays.
Here we report the identification of novel methylated breast cancer genes as well as a robust serum/plasma-specific modified QM-MSP assay called cMethDNA, which enhances the methylation signal, while maintaining the high sensitivity, specificity, reproducibility, dynamic range and quantitative advantages of the standard QM-MSP assay (6–10). We balanced several criteria to select ten biomarkers that were simultaneously, 1) highly and frequently methylated in breast tumor tissues (11), and in serum from metastatic breast cancer patients and, 2) methylated at low levels in cell-free circulating serum DNA in normal individuals. Following validation of the 10 gene panel in the TCGA (The Cancer Genome Atlas project) breast cancer methylome database, we developed and tested serum-specific prediction models, in test and training sets of sera from Stage 4 metastatic breast cancer. cMethDNA identified 91% of sera in patients with recurrent metastatic breast cancers with a specificity of 96% (AUC = 0.994, p < 0.0001) in the test set. Methylation levels reflected response to treatment, and circulating tumor DNA revealed a similar pattern of methylation as the solid tumor. We conclude that the cMethDNA assay performed on the 10 gene panel shows great potential for development as a clinical laboratory test for monitoring therapy and disease progression/recurrence.
MATERIALS AND METHODS
Patients and Sample Collections
Whole blood and tissue were collected prospectively from women with Stage 4 metastatic breast carcinoma following disease recurrence after prior therapy [Training set: J0214, NCT00080665 (12)], and J0425, NCT00274768 collected at JH 2004–2008; Test set: J0524, TBCRC 005 collected 2004–2012], as well as from healthy controls (randomly divided into training and test sets: TBCRC 005). To evaluate concordance between sera and tissue, tissue were obtained from a subset of cancer patients in the training set, along with additional samples collected at diagnosis of de novo metastatic disease (TBCRC 013 collected at participating institutions from 11/2009–4/2012). All the samples were collected with appropriate approval from the various institutional review boards. Supplementary Methods has additional details. Patient characteristics are summarized in Table 1 and Supplementary Table 1.
Table 1.
Patient characteristics
| Patient characteristics - Metastatic Breast Cancer | Recurrent | De Novo | |
|---|---|---|---|
|
| |||
| A. Stage IV serum | Training set | Test set | TBCRC 013 |
| Patient characteristics | n = 24 | n = | n = 18 |
| Race | |||
| Caucasian | 17 | 23 | 13* |
| Black | 6 | 10 | 2 |
| Other | 1 | 0 | 3 |
| ECOG performance status | |||
| 0 | 17 | 22 | 7 |
| 1 | 7 | 9 | 11 |
| 2 | 0 | 2 | 0 |
| Location of disease: | |||
| Visceral | 4 | 5 | 4 |
| Non-visceral | 11 | 8 | 7 |
| Both | 19 | 20 | 7 |
| Receptor status: | |||
| ER/PR-positive, HER2-negative | 12 | 17 | 9 |
| HER2-positive | 6 | 10 | 8 |
| Triple-negative (ER,PR,HER2 negative) | 6 | 6 | 1 |
| No. treated with prior chemotherapy for metastatic | 11 | 10 | 0 |
| No. prior chemotherapy regimens for metastatic disease: | |||
| 0 | 13 | 23 | 18 |
| 1 | 3 | 1 | 0 |
| 2 | 6 | 4 | 0 |
| 3 | 1 | 3 | 0 |
| ≥ 4 | 1 | 2 | 0 |
| Age: | |||
| Median | 53.5 | 57.0 | 54 |
| Range | 28–73 | 29–78 | 21–73 |
|
| |||
| B. Normal serum | Training set | Test set | |
| Patient Characteristics | n = 28 | n = | |
| Race: | |||
| Caucasian | 16 | 12 | |
| Black | 11 | 15 | |
| Other | 1 | 0 | |
| Age: | |||
| Median | 42.5 | 40.0 | |
| Range | 21–59 | 20–63 | |
Includes 1 Hispanic, 12 non-Hispanic
Purification of Cell-free Circulating DNA
DNA extraction from serum was tested using three different serum DNA purification kits: QiaAmp MinElute Virus Spin Kit (ME; Qiagen), QiaAmp UltraSens Virus Kit (US; Qiagen), and Quick-gDNA MiniPrep (ZR; Zymo Research, Orange, CA). The MinElute method was then selected for use in the study because of superior performance. External recombinant gene-specific standards (STDgene; 50 copies per gene for up to 12 genes) and carrier DNA/RNA [250 ng salmon sperm (Life Technologies, Grand Island, NY), 250 ng tRNA (Roche Applied Science, Indianapolis, IN), and 5.6 μg “Carrier RNA” (Qiagen, Valencia, CA)] were added to each serum sample (300 μl), and cell-free DNA was extracted per manufacturer’s instructions. Extracted DNA was then modified with sodium bisulfite, cleaned and eluted in 15 μl of water according to the EZ DNA Methylation Kit protocol (Zymo Research). Supplementary Methods has detailed protocols.
Extraction of tissue DNA
Tissue DNA was extracted overnight at 56°C in buffer containing TNES (10 mM Tris, 150 mM NaCl, 2 mM EDTA, 0.5% SDS) and proteinase K (30 μg), heat inactivated 10 min at 90°C, then treated with sodium bisulfite as described previously (11, 13).
DNA Methylation Array
Details are presented in Supplementary Methods.
The cMethDNA assay
cMethDNA was performed in two sequential PCR reactions (Fig. 1A). PCR reaction #1 (Multiplex Reaction): DNA was isolated from 300 μl serum containing 50 copies of each STDgene DNA, bisulfite treated, and amplified with a cocktail of external DNA-specific primer pairs specific for each of the 10 genes. Reaction #1 was then diluted 1:500–1:50,000. PCR reaction #2 (Real-time qMSP): Diluted amplicons (4 μl) from Reaction #1 were amplified by real-time PCR, with the TARGETgene and reference STDgene in the same well. Patient samples were purified, assayed in duplicate, and results averaged. Detailed information is available in Supplemental Methods.
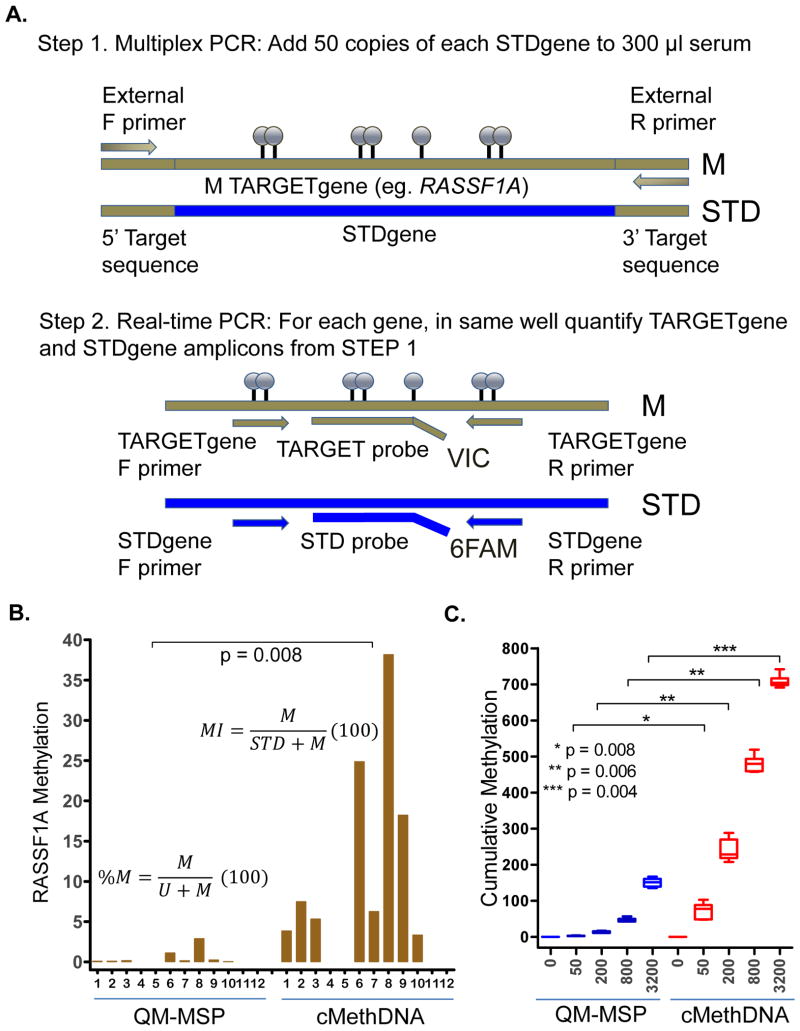
Figure 1. The cMethDNA Assay.
A) Reference DNA (50 copies of each gene-specific standard; STD) was spiked into 300 μl serum and total DNA was purified. Nested PCR was performed where the first PCR reaction (STEP 1) contained one pair of external primers per gene (forward and reverse) that co-amplifies DNA from the gene of interest (TARGETgene) and the gene-specific standard (STDgene). In the second PCR reaction (STEP 2), amplicons of Step 1 are assayed by absolute quantitative real-time PCR with specific sets of primers (forward and reverse) and hydrolysis probes (in two colors) recognizing methylated TARGETgene or reference STDgene. B) RASSF1A methylation in QM-MSP and cMethDNA methods was compared by testing the same aliquot of multiplexed DNA from twelve metastatic breast cancer patient sera. Significantly higher methylation values are seen by the cMethDNA assay compared to the QM-MSP method (p = 0.008; Wilcoxon signed-rank test). C) Cumulative methylation as assessed by QM-MSP and cMethDNA methods was compared by testing 6 replicate sets of normal serum spiked with increasing number of copies of MDA-MB-453 cell line DNA (X-axis; permuted one-sided t test p-values).
Calculation of Methylation
For an individual serum sample, cMethDNA calculations were as follows: Methylation Index (MI) = [Methylated TARGETgene copies/(Methylated TARGETgene + STDgene) copies](100); and cumulative methylation index (CMI) = the sum of all MI values within the gene panel. Serum samples were assayed in duplicate and then results were averaged. For an individual sample, QM-MSP calculations: % Methylation (%M) = [Methylated TARGETgene copies/(Methylated TARGETgene + Unmethylated TARGETgene) copies](100); CMI = the sum of all %M values within the panel.
Statistical Analysis
cMethDNA data analyses were performed using GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA), SAS software (v 9.2, SAS Institute Inc., Cary, NC), or with R version 2.15.2 (2012-10-26). Statistical tests were two-sided and considered statistically significant at p < 0.05 unless otherwise stated. Distributions of cMethDNA data between independent groups were described using box plots and difference was tested using nonparametric Mann-Whitney test. The Wilcoxon signed-rank test was performed when comparing two related samples (e.g., measurements on the same subjects). Receiver Operating Characteristic (ROC) analyses were used to characterize performance and define laboratory thresholds. The performance of the 10-gene panel was characterized through estimating the area under the ROC curve (AUC), sensitivity, specificity, classification accuracy and likelihood ratio along with the 95% confidence intervals. Details of statistical analyses can be found in Supplementary Methods.
RESULTS
Whole genome methylation array
For the selection of the 10 gene panel used in the cMethDNA assay, we relied heavily on our previously published whole genome analysis of breast tissue DNA (N = 103 tumors, N = 21 normal breast samples, Infinium Human Methylation27K Beadchip) (11), along with data generated in a new array analysis of DNA methylation in serum [N = 6 recurrent metastatic breast cancer sera, N = 5 normal sera, N = 4 leukocyte pools (5 normal individuals per pool)]. For identifying candidate markers in primary breast tissue, we serially selected: 1) 8376/27,578 probes with SD > 0.100 between tumor tissues and probe detection p-values < 0.0001, 2) 2674/8376 probes having at least 1.5-fold higher mean methylation in tumors than in normal breast organoids, and, 3) 1752/2674 probes with β-methylation < 0.15 in normal breast tissue adjacent to tumor (N = 6, laser microdissected). We further filtered this pool of candidate markers on the basis of data generated in sera, choosing, 4) 212/1752 probes ≥ 2.0-fold more methylated in cancer serum than in normal serum, and 198/212 probes with β-methylation < 0.15 in individual serum samples.
The final 10-gene marker panel was chosen by careful inspection of the 198 probes, prioritizing probes that combined very low methylation in normal samples with frequent and high levels of methylation in both ER+ and ER-negative cancers. Eighteen of these probes were present among a set of 100-candidate recurrence markers identified previously in the tissue DNA methylation array (11). Among these were seven novel markers (AKR1B1, COL6A2, GPX7, HIST1H3C, HOXB4, RASGRF2, and TM6SF1) and two known ones, ARHGEF7, and TMEFF2. RASSF1 was selected to complete the 10-gene marker panel (Supplementary Table 2; Supplementary Figures 1A and 2).
The 10-gene methylation panel was verified in silico for sensitivity and specificity in The Cancer Genome Atlas Project databases (TCGA, N = 316 breast cancer, N = 27 normal breast samples, BRCA; Supplementary Figure 1B), and subsequently verified in sera using the cMethDNA assay. The 10-probe test panel outperformed 97.9% of 100,000 iterations of randomly created 10-probe panels drawn from the TCGA database (Supplementary Figure 1C).
cMethDNA assay
The cMethDNA assay is a refinement of the QM-MSP method used extensively by our laboratory and others (6, 7, 9, 10, 14). Here, a low fixed physiological level of recombinant gene-specific standard reference DNA is introduced into a constant volume of serum (50 copies of gene-specific standard in 300 μl serum) prior to purification of the DNA. This fixes a relatively high constant ratio of methylated DNA to reference DNA, quantified after multiplex and quantitative real-time PCR. Primer/probe sequences are in Supplementary Table 3.
Technical validation of the cMethDNA assay
Intra-Assay Testing
To directly compare the QM-MSP and cMethDNA assays, DNA from recurrent metastatic breast cancer patient sera was tested by both methods for the RASSF1A gene (Fig. 1B). The robustness of methylation values and frequency of detection of hypermethylated RASSF1A was higher with cMethDNA compared to the QM-MSP assay (p = 0.008, Wilcoxon signed-rank test). In a second experiment, 6 aliquots of serum from a single normal donor (300 μl serum per assay point) were spiked with a physiological range (0, 50, 200, 800 or 3200 copies) of fully methylated DNA from the breast cancer cell line, MDA-MB-453. DNA was purified, multiplexed and tested by both cMethDNA and QM-MSP methods for the 10-gene panel (excluding HIST1H3C, which is not methylated in this cell line) (Fig. 1C). Again, the cMethDNA assay reported significantly higher methylation than the QM-MSP method at all levels (50 copies, p = 0.008; 200 copies, p = 0.006; 800 copies, p = 0.006; 3200 copies, p = 0.004, one-sided t test, permuted) (Fig. 1C).
Reproducibility of replicates, comparison of extraction methods
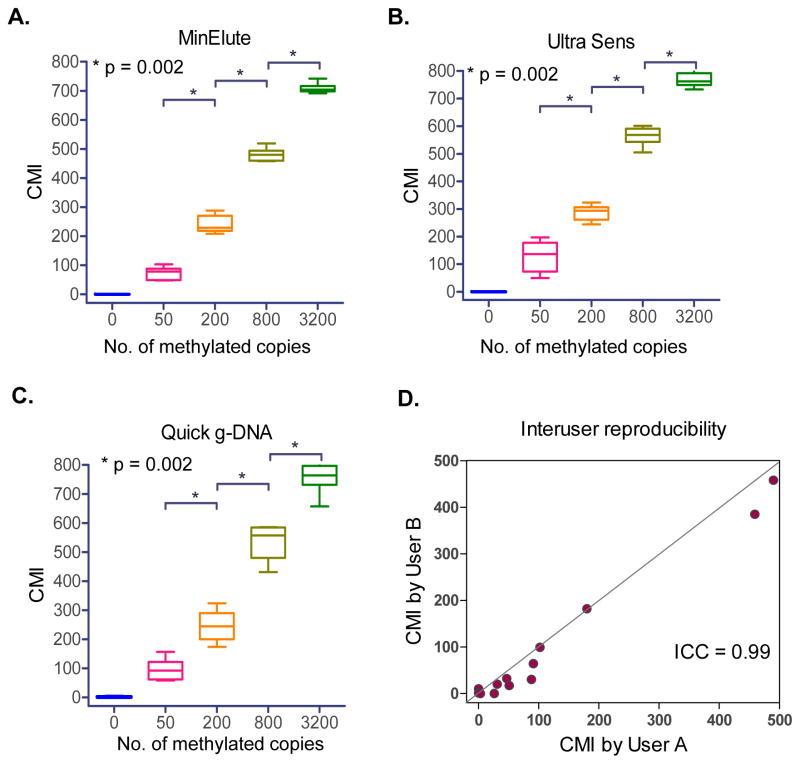
We performed a series of tests to establish linearity and sensitivity of the cMethDNA assay. Three different serum DNA extraction methods were evaluated (MinElute Virus Kit, Qiagen, UltraSens Virus Kit, Qiagen and Quick-gDNA Prep, Zymo Research). Methylated DNA was spiked as described above (Fig. 1C); six replicates per point per extraction method were tested by cMethDNA (Fig. 2A–C). The MinElute Virus Spin Kit method showed the highest reproducibility and the smallest inter-assay coefficients of variation (CV = 29%, 12%, 4.6%, 2.5%, for 50, 200, 800 or 3200 methylated DNA copies, respectively, Supplementary Table 4).
Figure 2. cMethDNA Assay Validation.
A–C) Three serum purification methods (Qiagen MinElute, Qiagen Ultra Sens, and Zymo Research Quick g-DNA) were tested in conjunction with the cMethDNA method in normal serum aliquots spiked with 0–3200 methylated copies, prepared from one master stock. Replicates were purified and multiplexed separately. Box-Whisker plots show the median and full range of CMI (Y-axis) for replicates of each sample (X-axis). Statistical significance (Mann-Whitney test) is indicated by p-values. The % coefficient of variation (CV, a normalized measure of frequency distribution) is shown for each test in Supplementary Table 4. D) Inter-user reproducibility was evaluated for a set of thirteen patient serum samples processed independently by two investigators. User performance was evaluated for the 10-gene panel (Intra-class correlation coefficient = 0.99, 95% CI = 0.96–1.00).
Inter-User Reproducibility
The ability for two individuals to perform the cMethDNA assay and to arrive at similar results is important. For the 10-gene panel cumulative methylation levels obtained by two users were compared after independently extracting DNA and performing the cMethDNA assay on duplicate aliquots of patient cancer sera (N = 13). The inter-user reproducibility, tested for intra-class correlation coefficient showed strong agreement between users (ICC = 0.99; 95% CI = 0.96–1.00; Fig. 2D).
In summary, the cMethDNA approach results in an enhanced methylation signal, minimizes the effects of technical variability in purification of DNA, and since it uses spiked DNA as a standard, is not affected by potential day-to-day fluctuations in total serum DNA content that may be independent of changes in the tumor burden.
Detection of methylated DNA in serum of metastatic breast cancer patients
Assay specificity
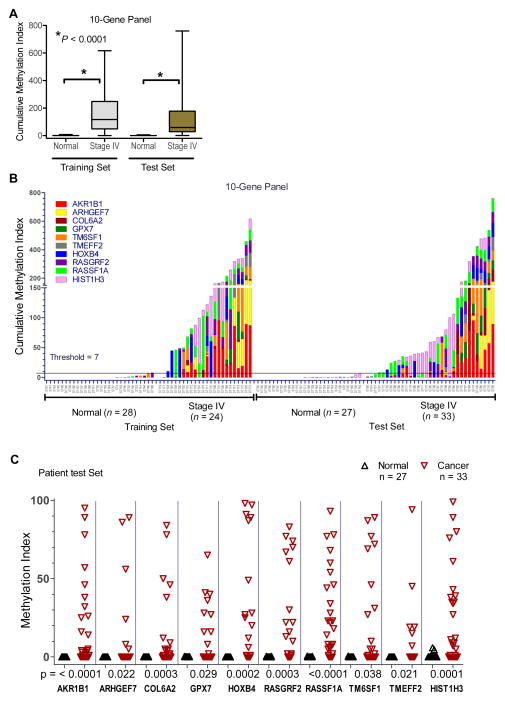
To verify the performance of the 10-gene marker panel in the cMethDNA assay for detection of circulating cell-free tumor DNA, we evaluated independent Training and Test sets of sera collected in prospective clinical trials of patients with recurrent stage IV breast cancer conducted at Johns Hopkins and through the TBCRC. The Training set consisted of 52 serum samples (28 normal and 24 cancer). Sera from cancer patients had significantly higher levels of methylated genes than women without cancer (median CMI = 117.3 and 0.04, respectively; p < 0.0001, Mann-Whitney test; Fig. 3A–B, Table 2A). Receiver Operating Characteristic (ROC) analyses identified a threshold of 6.9 units that maximized the sum of sensitivity and specificity in the Training set (Table 2B). The assay specificity was 96.4% (27/28, 95% CI = 81.7–99.9%) when sensitivity was 91.7% (22/24, 95% CI = 73.0–99.0%) for a likelihood ratio = 25.7 and an overall classification rate of 94% (49/52, 95% CI = 84–99%). The ROC analysis AUC = 0.95 (95% CI = 0.87–1.02; p < 0.0001). These findings were further verified in a Test set of patient samples (27 normal, 33 cancer) (Table 1, Supplementary Table 1A). Consistent with findings in the Training set, cumulative methylation levels were significantly higher in patients with metastatic breast cancer, compared to women without cancer (p< 0.0001, Mann-Whitney test; Fig. 3A–B); Table 2A. Using the classification rule derived on the training data, the observed assay specificity was 100% (27/27, 95% CI = 87.2–100%) at a sensitivity was 90.9% (30/33; 95% CI = 75.7–98.1%) for an overall classification accuracy of 95% (57/60, 95% CI = 86–99%) and a likelihood ratio = 24.6 (Table 2B). As in the training set, the median frequency of methylation of most genes was high (median = 38%, range = 18–67%; Fig. 3C, Table 2C). RASSF1A had the highest incidence of methylation among all genes in both Training and Test sets (75% and 67%, respectively). Individually RASSF1A, HIST1H3C, RASGRF2, COL6A2, HOXB4 and AKR1B1 genes had the strongest performance in the cMethDNA assay (p = 0.0003 to p < 0.0001, Mann-Whitney Test of Training set) (Fig. 3C, Table 2C). A 6-gene panel of the six most robust biomarkers performed nearly as well as the 10-gene panel (Supplementary Figure 3). In normal samples, no significant age-dependent changes in cumulative methylation were observed (N = 55; One-way ANOVA, p = 0.8988 for quartiles; Table 1, Supplementary Table 1, Supplementary Fig. 4).
Figure 3. Evaluation of the 10-gene panel by cMethDNA assay.
Serum from patients with recurrent metastatic stage IV disease were assigned to Training and Test sets. A) Box plot shows that cancer sera display significantly higher median cumulative methylation than normal sera (p< 0.0001. Mann-Whitney). B) Cumulative cMethDNA assay values (CMI; Y-axis) were calculated for individual samples, each colored segment representing the methylation index for an individual gene. ROC analysis was performed on data collected from the Training set to define a normal laboratory methylation threshold (CMI = 7 units; Y-axis). C) Frequency of methylation for individual biomarkers in the 10-gene panel.
Scatter plot depicts gene methylation intensity (Y-axis, methylation index) for individual genes (X-axis) in the Test set for normal and cancer sera, indicated in the legend. The Mann-Whitney p-values are shown below each plot. Statistical analysis of Training and Test sets for the 10-gene panel and the methylation frequency of individual genes is shown in Table 2.
Table 2.
cMethDNA ability to detect circulating cell-free tumor DNA
| A. Cumulative Methylation Index (CMI;10 genes) in cancer and normal sera | Training Set | Test Set | ||
|---|---|---|---|---|
| Normal | Stage IV | Normal | Stage IV | |
|
| ||||
| N | 28 | 24 | 27 | 33 |
| Minimum | 0 | 0 | 0 | 1 |
| 25% Percentile | 0 | 49 | 0 | 28 |
| Median | 0 | 117 | 0 | 59 |
| 75% Percentile | 1 | 248 | 0 | 178 |
| Maximum | 7 | 616 | 6 | 760 |
| Mean | 1 | 165 | 1 | 144 |
| Lower 95% CI of mean | 0 | 99 | 0 | 76 |
| Upper 95% CI of mean | 2 | 230 | 1 | 213 |
| B. ROC Analysis | Training Set | Test Set |
|---|---|---|
|
| ||
| Area under the curve | 0.950 | 0.994 |
| 95% confidence interval | 0.874 – 1.027 | 0.984 – 1.005 |
| P-value | < 0.0001 | < 0.0001 |
| Sensitivity | 92% | 91% |
| Specificity | 96% | 100% |
| Likelihood ratio | 25.7 | 24.6 |
| Classification Accuracy | 94% | 95% |
| C. Methylation Frequency* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| AKR1B1 | ARHGEF7 | COL6A2 | GPX7 | HOXB4 | RASGRF2 | RASSF1A | TM6SF1 | TMEFF2 | HIST1H3 | |
|
| ||||||||||
| Training Set: | ||||||||||
| Cancer (n = 24) | 15 (63%) | 7 (29%) | 9 (37%) | 5 (21%) | 11 (46%) | 12 (50%) | 18 (75%) | 10 (42%) | 6 (8%) | 12 (50%) |
| Control (n = 28) | 0 | 0 | 1 (4%) | 0 | 0 | 2 (7%) | 1 (4%) | 2 (7%) | 0 | 0 |
|
| ||||||||||
| Test Set: | ||||||||||
| Cancer (n = 33) | 14 (42%) | 6 (18%) | 12 (36%) | 10 (30%) | 12 (36%) | 15 (45%) | 22 (67%) | 12 (36%) | 6 (18%) | 18 (55%) |
| Control (n = 27) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (7%) |
Positive methylation defined as CMI > 7.0 (set by ROC)
Positive methylation defined as > 1 unit of Methylation Index (MI)
Utility of cMethDNA to monitor response to treatment
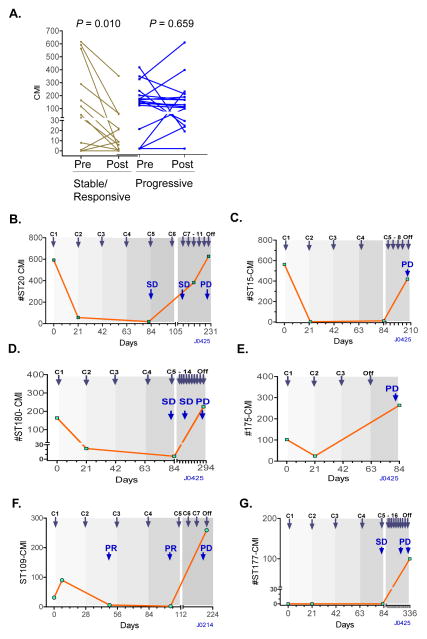
To determine if the cMethDNA assay might be used effectively to monitor response to chemotherapy, we evaluated sera collected from metastatic breast cancer patients who participated in two prospective clinical trials J0214, and J0425 (15). Serum samples (N = 58) were collected from 29 patients at baseline prior to initiation of treatment, and 18–49 days (median 21 days) after initiation of a new chemotherapy regimen (Supplementary Table 1B). The results showed a statistically significant decrease in median serum DNA methylation levels in patients having stable disease (SD) or with a therapeutic response (e.g. PR, partial response) according to RECIST criteria (p = 0.010)(Fig. 4A). This decrease in methylation was not observed in patients with progressive disease (PD; p = 0.659; Wilcoxon signed rank test) (Fig. 4A).
Figure 4. Monitoring response to treatment.
A) cMethDNA was performed on serum samples obtained from metastatic breast cancer patients at baseline (Pre) and after 18–49 days of therapy (Post). Data are plotted according to CMI in individual patients judged by RECIST criteria to have stable/responsive disease or progressive disease after 8–12 weeks. Pre- and post- difference in CMI was evaluated using Wilcoxon signed-rank test. B–G): Representative plots of CMI of patient (ST#) sera assessed by cMethDNA during the course of treatment. Patients received either 28 day cycles of docetaxel or 21 day cycles of capecitabine, indicated by shading. C: cycles of treatment; Imaging- and RECIST criteria- assessed progressive disease: PD; stable disease: SD; and partial response: PR.
Since a single time point post therapy may have limited potential to predict outcome as assessed by imaging several weeks to months later, we evaluated a subset of patients (13 of 29) with sera collected at three or more time points (total of 54 sera) during different cycles of the same therapy (Fig. 4B–G, Supplementary Fig 5, Supplementary Table 1B). Ten of thirteen patient sera showed methylation levels reflective of decreases in tumor burden during stable disease or partial response, and increasing levels of methylation during progressive disease as defined by RECIST criteria. These data show the potential of cMethDNA to reveal therapeutic response at an early time point during treatment and provide important information for clinicians that could aid decision making regarding further therapy.
Patterns of methylation are retained between primary tumor and metastasis
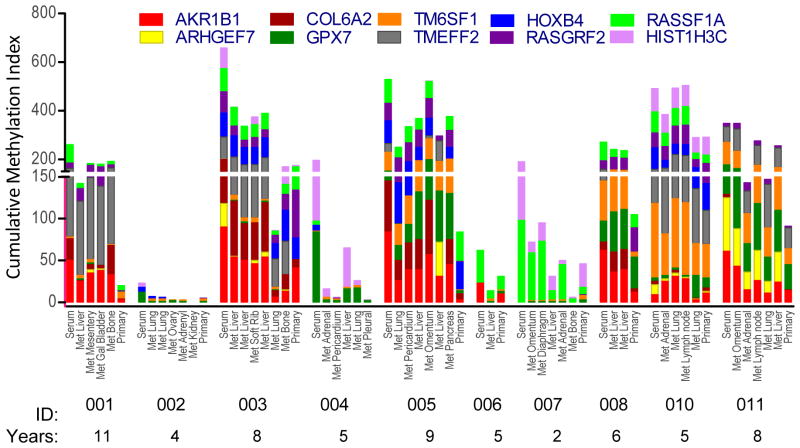
Previously, we observed similarity in the biomarker expression and methylation profiles of primary breast cancer and distant metastases collected from individuals within 4 hours of death (N = 10; 2–11 years after first diagnosis of breast cancer; age 33 to 79 years) (16). With the 10-gene marker panel, a striking concordance was observed between the methylated gene patterns in the samples from primary, metastases and serum of the same patient (Fig. 5). In this analysis serum was quantified with the cMethDNA assay, and tissues were quantified with the QM-MSP assay; Results are plotted on the same Y-axis, although cMethDNA is more robust. Comparative analyses indicated >50% of individual patient serum/tissue pairs or tissue/tissue pairs were concordant for ≥ 9 markers, and all had ≥ 6 matches (Supplementary Fig. 6). Since by chance alone, the median number of matches predicted is 7 (range = 3–9), based on the overall frequency of the genes evaluated, results indicate that methylation profiles were more patient-specific than random. Concordance between serum and primary and/or metastases was also observed in two other independent series of samples collected in clinical trials: 1) a subset of patients in the study Training set (10 of 24 patients; Table 1, Supplementary Table 1C, Supplementary Fig. 7A), and 2) from untreated patients diagnosed with de novo metastatic breast cancer (18 patients; Table 1, Supplementary Table 1C, in TBCRC 013, Supplementary Fig. 7B). As in the autopsy study, consistent profiles of hypermethylation were observed between the primary tumor, serum and distant metastasis samples, even though sera were tested with the cMethDNA method and tissues by the QM-MSP method.
Figure 5. Preservation of methylation patterns in primary tumor, serum, and distant metastases of individuals.
Specific gene methylation pattern (Y-axis) of the 10-gene panel was evaluated in primary tumor, multiple metastatic lesions and serum (X-axis) collected from 10 women at autopsy. The interval between surgery when primary tumor tissue was collected, and death when serum and metastases were collected, is indicated below the patient ID (2–11 years).
Collectively, these studies provide proof of principle that a core pattern of methylation is retained in the distant metastasis and serum of a given patient over time, perhaps decades.
Utility of the 10-gene panel to detect other tumor types
Careful analysis and selection of breast cancer markers determined in tissues allowed us to build the 10-gene panel, which was later confirmed in serum DNA from patients with metastatic breast cancer and validated in the independent TCGA breast cancer methylation tissue DNA array database. To determine if an in silico analysis of other cancer tissue databases will indicate the potential use of the 10-gene panel for detecting circulating methylated markers in other types of cancer, we examined several Infinium HumanMethylation27 array databases from TCGA, Supplementary Figure 8A–F). Based on ROC analyses, compared to breast (BRCA, AUC = 0.950), the panel performed with a high level of efficiency in three other tumor types, including lung (LUNG, AUC = 0.969), colon (COAD, AUC = 0.995), and rectum (READ, AUC = 0.997). However, the panel displayed poor performance with other tumor types, such as ovarian (OV, AUC = 0.668), kidney (KIRC, AUC = 0.725) and stomach (STAD, AUC = 0.792). Thus, the panel, similar to breast carcinomas, shows specificity for the highly prevalent lung and colorectal adenocarcinomas and may not be useful for others such as ovarian, kidney and stomach cancers.
DISCUSSION
We have derived an informative panel of novel gene markers for detecting tumor specific circulating, cell-free, methylated DNA in the sera of patients with metastatic breast cancer. Using this panel, we have demonstrated broad dynamic range, reproducibility, sensitivity, specificity, and accuracy of the cMethDNA assay for detecting methylated DNA in the vast majority of patients with newly diagnosed and recurrent Stage 4 breast cancer. We have also provided preliminary evidence for the potential of the cMethDNA assay to aid in monitoring disease during chemotherapy, and shown that a core methylation pattern typical of each primary tumor is retained in the metastatic lesions and serum over a long period of time (2–11 years). TCGA tumor tissue methylation arrays revealed that this panel of ten markers is methylated and likely to perform well to detect a variety of tumor types (i.e. lung, colon, rectal, and possibly uterine carcinomas and glioblastomas) and would have less utility for other tumors (i.e. ovarian, kidney, stomach). Shortly, planned studies will test the prognostic and predictive utility of this assay in an external sample set from larger prospective clinical trials.
The challenge to developing a reliable blood test for cancer has been finding the virtual “needle in the haystack” or desired methylated marker in the vast excess of unmethylated normal DNA. Our new cMethDNA assay overcomes this barrier by incorporating two innovations: First spiking of serum with physiological levels of recombinant standards matched to the gene of interest by size and homology at 5′ and 3′ ends that provide an internal reference for recovery and quantification of target DNA. Second, the inclusion of a multiplexed, pre-amplification step increases the dynamic range of detectability of the target gene and enables quantitation of a panel of markers in the amount of DNA normally used to evaluate one gene. The use of external standards has a further advantage in that it avoids the pitfalls inherent to endogenous reference DNAs, where total DNA may fluctuate up to 15-fold under healthy conditions independent of tumor burden, such as occurs in exercise overtraining (17, 18), trauma (19), surgery (20), sepsis (21), chronic inflammatory diseases (22–24), and pregnancy (25, 26).
Features critical to the high sensitivity and specificity of the cMethDNA assay are the identification of biomarkers through whole genome analysis of solid tumor and metastatic cancer sera, a careful filtration process, and the final selection of a panel of hypermethylated genes that were specifically and frequently methylated in breast cancer. Among the cancer-specific markers in our panel used for detecting cell-free DNA in serum by the cMethDNA assay and tissue DNA by QM-MSP, the HOXB4, RASGRF2, AKR1B1, TM6SF1, COL6A2, GPX7, HIST1H3C genes are being reported here for the first time in breast cancer. Interestingly, HOXB4 also hosts the regulatory region for miR-10a near its genome and is hypermethylated in 90% of hepatocellular carcinomas (27). In breast cancer, the cMethDNA assay had a classification accuracy of up to 95% that distinguished cancer versus normal sera with a sensitivity of over 90% and specificity of nearly 100%, which suggest a potential for greater clinical utility compared to CA27.29 (28), CEA (29), CTC (30) and other circulating tumor DNA tests (31, 32).
The application of cMethDNA as a non-invasive indicator of tumor burden and therapeutic response was tested using samples collected as secondary endpoints from three prospective clinical trials of women with metastatic breast cancer that was newly diagnosed or progressing after initial palliative chemotherapy. In a pilot study the cMethDNA assay was able to detect serum methylation levels that reflected tumor burden based on RECIST criteria. In the majority of cases changes in methylation tracked with disease progression. Investigations of sera during subclinical stages of breast cancer recurrence (adjuvant surveillance) are also needed. While imaging studies (in addition to clinical assessment) are the current standard to evaluate response after 8–12 weeks of initiating a new systemic, tests to monitor disease prognosis and predict therapy benefit earlier remain an unmet need. An ongoing trial, SWOG 0500, (NCT00382018) is evaluating the role of enumerating circulating tumor cells (CTC) in this setting, but may be hampered by the low sensitivity of that commercial assay. The cMethDNA assay appears to be promising in this setting, and we are embarking on a comparative analysis of the prognostic and predictive utility of cMethDNA, CA27.29, CEA and CTC, using prospectively collected serum to predict response to therapy in metastatic breast cancer. Investigations of sera obtained during the subclinical stages of breast cancer recurrence (adjuvant surveillance) are also needed.
In summary, the cMethDNA assay is a promising new liquid biopsy tool for detecting tumor specific cell free circulating DNA in a noninvasive manner. With further refinement, it could serve to monitor response to therapy, potentially prognosticate disease outcome, and serve as an early indicator of tumor recurrence.
Supplementary Material
Acknowledgments
We thank Dustin VanDenBerg (JH) and Russell Towers (MSKCC) for technical assistance, and members of the JH blood repository. We thank the investigators and patients who participated in the Johns Hopkins trials J0214 (PI, A. C. Wolff) and J0425 (PI: A. C. Wolff), and in the trial TBCRC 005 (PI, A. C. Wolff) and 013 (PI, T. King) and David Euhus, Alan Rein and Bert Vogelstein for critically reviewing the manuscript.
Funding: This work was supported by grants from AVON Foundation for Research (SS and ACW), the Rubenstein family (SS), John A. Sellon Charitable Trust (SS), the Department of Defense Center of Excellence on “Targeting Metastatic Breast Cancer” grant W81XWH-04-1-0595 (SS), the Avon/NCI PFP 3P40 CA006973-41S (ACW), the Susan G. Komen for the Cure Grant BCTR0504444 (ACW), the Breast Cancer Research Foundation (ACW), and SKCCC Core grant P30 CA006973 (SS). We also thank the AVON Foundation for Research, Susan G. Komen for the Cure, and the Breast Cancer Research Foundation for their support of the TBCRC.
Footnotes
Author contributions: MJF, WWT, ZLB, SS conceived of and developed the cMethDNA assay; MJF, WWT and ZLB performed the assays; MJF, SS, ACW, CBU, KV, ZZ, CW, and LC designed the study; ACW, SJ, LAC, JNI provided serum samples from clinical trials at JH and TBCRC 005; PA provided samples from the RAP; JPL, MdB, TAK, JB, KM provided samples from TBCRC 013; MJ and SS wrote the paper; ZZ and LC performed the biostatistical analyses.
Competing interests: MJF, SS, WWT and ZLB are inventors of the cMethDNA assay, a US patent, P12014-02 claiming the benefit of U.S. Provisional Patent Application No.61/650,083, and an international patent, PCT/US13/42198 have been filed through JHU. QM-MSP for RASSF1A is protected by US patent 8,062,849.
References
- 1.Van De Voorde L, Speeckaert R, Van Gestel D, Bracke M, De Neve W, Delanghe J, et al. DNA methylation-based biomarkers in serum of patients with breast cancer. Mutat Res. 2012;751:304–25. doi: 10.1016/j.mrrev.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Muller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, et al. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–5. [PubMed] [Google Scholar]
- 3.Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, et al. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141–5. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- 4.Fiegl H, Jones A, Hauser-Kronberger C, Hutarew G, Reitsamer R, Jones RL, et al. Methylated NEUROD1 promoter is a marker for chemosensitivity in breast cancer. Clin Cancer Res. 2008;14:3494–502. doi: 10.1158/1078-0432.CCR-07-4557. [DOI] [PubMed] [Google Scholar]
- 5.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–32. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Euhus DM, Bu D, Ashfaq R, Xie XJ, Bian A, Leitch AM, et al. Atypia and DNA methylation in nipple duct lavage in relation to predicted breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1812–21. doi: 10.1158/1055-9965.EPI-06-1034. [DOI] [PubMed] [Google Scholar]
- 7.Locke I, Kote-Jarai Z, Fackler MJ, Bancroft E, Osin P, Nerurkar A, et al. Gene promoter hypermethylation in ductal lavage fluid from healthy BRCA gene mutation carriers and mutation-negative controls. Breast Cancer Res. 2007;9:R20. doi: 10.1186/bcr1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, et al. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011;458:73–84. doi: 10.1007/s00428-010-1013-6. [DOI] [PubMed] [Google Scholar]
- 9.Suijkerbuijk KP, van der Wall E, Vooijs M, van Diest PJ. Molecular analysis of nipple fluid for breast cancer screening. Pathobiology. 2008;75:149–52. doi: 10.1159/000123853. [DOI] [PubMed] [Google Scholar]
- 10.Fackler MJ, Rivers A, Teo WW, Mangat A, Taylor E, Zhang Z, et al. Hypermethylated genes as biomarkers of cancer in women with pathologic nipple discharge. Clin Cancer Res. 2009;15:3802–11. doi: 10.1158/1078-0432.CCR-08-1981. [DOI] [PubMed] [Google Scholar]
- 11.Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, et al. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71:6195–207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly RM, Rudek MA, Garrett-Mayer E, Jeter SC, Donehower MG, Wright LA, et al. Docetaxel metabolism is not altered by imatinib: findings from an early phase study in metastatic breast cancer. Breast Cancer Res Treat. 2011;127:153–62. doi: 10.1007/s10549-011-1413-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fackler MJ, McVeigh M, Evron E, Garrett E, Mehrotra J, Polyak K, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107:970–5. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Fackler MJ, Lee JH, Choi C, Park MH, Yoon JH, et al. Basal-like breast cancer displays distinct patterns of promoter methylation. Cancer Biol Ther. 2010;9:1017–24. doi: 10.4161/cbt.9.12.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudek MA, Connolly RM, Hoskins JM, Garrett-Mayer E, Jeter SC, Armstrong DK, et al. Fixed-dose capecitabine is feasible: results from a pharmacokinetic and pharmacogenetic study in metastatic breast cancer. Breast Cancer Res Treat. 2013;139:135–43. doi: 10.1007/s10549-013-2516-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JM, Fackler MJ, Halushka MK, Molavi DW, Taylor ME, Teo WW, et al. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–46. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatouros IG, Destouni A, Margonis K, Jamurtas AZ, Vrettou C, Kouretas D, et al. Cell-free plasma DNA as a novel marker of aseptic inflammation severity related to exercise overtraining. Clin Chem. 2006;52:1820–4. doi: 10.1373/clinchem.2006.070417. [DOI] [PubMed] [Google Scholar]
- 18.Breitbach S, Tug S, Simon P. Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med. 2012;42:565–86. doi: 10.2165/11631380-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 19.Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000;46:319–23. [PubMed] [Google Scholar]
- 20.Szpechcinski A, Chorostowska-Wynimko J, Kupis W, Maszkowska-Kopij K, Dancewicz M, Kowalewski J, et al. Quantitative analysis of free-circulating DNA in plasma of patients with resectable NSCLC. Expert Opin Biol Ther. 2012;12 (Suppl 1):S3–9. doi: 10.1517/14712598.2012.668519. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care. 2006;10:R60. doi: 10.1186/cc4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liggett T, Melnikov A, Tilwalli S, Yi Q, Chen H, Replogle C, et al. Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J Neurol Sci. 2010;290:16–21. doi: 10.1016/j.jns.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raptis L, Menard HA. Quantitation and characterization of plasma DNA in normals and patients with systemic lupus erythematosus. J Clin Invest. 1980;66:1391–9. doi: 10.1172/JCI109992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beyer C, Pisetsky DS. The role of microparticles in the pathogenesis of rheumatic diseases. Nat Rev Rheumatol. 2010;6:21–9. doi: 10.1038/nrrheum.2009.229. [DOI] [PubMed] [Google Scholar]
- 25.Muller HM, Ivarsson L, Schrocksnadel H, Fiegl H, Widschwendter A, Goebel G, et al. DNA methylation changes in sera of women in early pregnancy are similar to those in advanced breast cancer patients. Clin Chem. 2004;50:1065–8. doi: 10.1373/clinchem.2003.030387. [DOI] [PubMed] [Google Scholar]
- 26.Tsui NB, Lo YM. Recent advances in the analysis of fetal nucleic acids in maternal plasma. Curr Opin Hematol. 2012;19:462–8. doi: 10.1097/MOH.0b013e328358e17a. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Wang S, Zhang YJ, Kappil MA, Chen Wu H, Kibriya MG, et al. Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics. 2012;7:1230–7. doi: 10.4161/epi.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan DW, Beveridge RA, Muss H, Fritsche HA, Hortobagyi G, Theriault R, et al. Use of Truquant BR radioimmunoassay for early detection of breast cancer recurrence in patients with stage II and stage III disease. J Clin Oncol. 1997;15:2322–8. doi: 10.1200/JCO.1997.15.6.2322. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen AC, Sorensen PD, Jacobsen EH, Madsen JS, Brandslund I. Sensitivity of CA 15-3, CEA and serum HER2 in the early detection of recurrence of breast cancer. Clin Chem Lab Med. 2013;51:1511–9. doi: 10.1515/cclm-2012-0488. [DOI] [PubMed] [Google Scholar]
- 30.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 31.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 32.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.