Abstract
BACKGROUND
Early identification of mutations may guide patients with metastatic colorectal cancer toward targeted therapies that may be life prolonging. The authors assessed tumor genotype correlations with clinical characteristics to determine whether mutational profiling can account for clinical similarities, differences, and outcomes.
METHODS
Under Institutional Review Board approval, 222 patients with metastatic colon adenocarcinoma (n = 158) and rectal adenocarcinoma (n = 64) who underwent clinical tumor genotyping were reviewed. Multiplexed tumor genotyping screened for >150 mutations across 15 commonly mutated cancer genes. The chi-square test was used to assess genotype frequency by tumor site and additional clinical characteristics. Cox multivariate analysis was used to assess the impact of genotype on overall survival.
RESULTS
Broad-based tumor genotyping revealed clinical and anatomic differences that could be linked to gene mutations. NRAS mutations were associated with rectal cancer versus colon cancer (12.5% vs 0.6%; P < .001) and with age ≥56 years (7% vs 0.9%; P = .02). Conversely, v-raf murine sarcoma viral oncogene homolog B (BRAF) mutations were associated with colon cancer (13% vs 3%; P = .024) and older age (15.8% vs 4.6%; P = .006). TP53 mutations were associated with rectal cancer (30% vs 18%; P = .048), younger age (14% vs 28.7%; P = .007), and men (26.4% vs 14%; P = .03). Lung metastases were associated with PIK3CA mutations (23% vs 8.7%; P = .004). Only mutations in BRAF were independently associated with decreased overall survival (hazard ratio, 2.4; 95% confidence interval, 1.09–5.27; P = .029).
CONCLUSIONS
The current study suggests that underlying molecular profiles can differ between colon and rectal cancers. Further investigation is warranted to assess whether the differences identified are important in determining the optimal treatment course for these patients.
Keywords: colorectal cancer, mutation, clinicopathologic, NRAS, BRAF
INTRODUCTION
Colorectal cancer (CRC) is the third most common form of cancer in the United States, with 143,460 estimated cases diagnosed in 2012 and 51,690 deaths.1 Colon and rectal cancers are often described as a single entity, but they differ in natural history and management. Local recurrence is significantly higher in rectal cancer,2 and rectal cancers often metastasize first to lung, whereas colon cancers metastasize first to liver.3 Treatment for locally advanced colon cancer often involves surgery followed by adjuvant chemotherapy; whereas, for rectal cancer, treatment involves preoperative chemoradiation followed by surgery and adjuvant chemotherapy. Whether these differences can be explained by the underlying biology, embryologic derivation, anatomy, and/or surgical technique is unclear.
It has been demonstrated that specific molecular alterations in tumors correlate significantly with patient and tumor characteristics. In lung adenocarcinoma, activating epidermal growth factor receptor (EGFR) mutations are identified most commonly in Asian female nonsmokers.4 In CRCs, v-raf murine sarcoma viral oncogene homolog B (BRAF) mutations have been associated with right-sided high-grade tumors in older patients.5 Tumor genotyping is rapidly being integrated into routine clinical care to gain a better understanding of the patient’s prognosis and to define the most appropriate treatment. Our institution has been performing clinical mutational profiling on patients with CRC since 2009. This testing represents a subset of patients with advanced metastatic disease for whom testing would assist in identifying targeted therapies for that individual. We retrospectively evaluated mutational signatures for correlation with clinical characteristics, anatomic site, and patient outcomes in the lower gastrointestinal tract.
MATERIALS AND METHODS
Patient Population
Under Institutional Review Board approval, we reviewed all patients (n = 222) with metastatic adenocarcinoma of the colon (n = 158) or rectum (n = 64) who received mutational profiling as part of their standard care between April 2009 and July 2011. Patients were chosen to undergo testing at the discretion of their treating physician. Patients with follow-up <6 months were excluded to provide adequate time for assessment of patient outcomes. Patients were initially diagnosed between January 1980 and February 2011 and were diagnosed with metastatic disease between January 1999 and February 2011. The distinction of colon versus rectal cancer was made by the location proximal or distal to the rectosigmoid junction, as determined by the patient’s treating physician.
Mutational Analysis
Clinical tumor genotyping was performed on all 222 patients using nucleic acids extracted from diagnostic formalin-fixed, paraffin-embedded tumor tissue using a modified Agencourt FormaPure method automated on a Beckman Coulter NXP workstation (Beckman Coulter, Pasadena, Calif). Mutational profiling simultaneously queried over 150 previously described hotspot mutations across 15 cancer genes, including v-akt murine thymoma viral oncogene homolog 1 (AKT1); adenomatosis polyposis coli (APC); BRAF; catenin (cadherin-associated protein) β1, 77 kDa (CTNNB1); EGFR; v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2); isocitrate dehydrogenase 1 (NAPD+), soluble (IDH1); v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT); Kirsten rat sarcoma viral oncogene homolog (KRAS); mitogen-activated protein kinase kinase 1 (MAP2K1); notch 1 (NOTCH1); neuroblastoma RAS viral oncogene homolog (NRAS); phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α (PIK3CA); phosphatase and tensin homolog (PTEN); and tumor protein 53 (TP53).6 This was performed using a custom-modified ABI PRISM SNaPshot Multiplex System (Applied Biosystems/Life Technologies Corporation, Carlsbad, Calif), as previously described.7 An earlier version of this assay, which was used on 74 of the specimens, did not test for IDH2 or MAP2K1 mutations. Specific mutations that were assessed using SNaP-shot are listed in Supporting Table 1 (see online supporting information). Testing of the tumor suppressor genes TP53, APC, and PTEN was limited to only the most common mutation sites (limiting coverage to 29%, 15%, and 15%, respectively, of all known somatic mutations).7
Statistical Analysis
The chi-square test was used to compare genotype frequency by tumor site, age at diagnosis (either greater than or equal to or less than the median age of 56 years), sex, family history of gastrointestinal cancer, history of polyps, stage at initial presentation, smoking status (former/current vs none), and site of first metastasis. A secondary exploratory analysis was performed to assess the impact of genotype mutation on overall survival (OS) and relapse-free survival (RFS) using Cox regression univariate and multivariate analyses and adjusting for age, sex, disease location (colon vs rectal), smoking history, and stage at presentation. The Kaplan-Meier method was used to estimate OS and RFS, and the results were compared using the log-rank test. OS was defined as the time from initial diagnosis to the date of death. Patients not known to have died were censored at date of last follow-up. RFS was calculated for those with stage I through III disease at diagnosis from the date of initial diagnosis to the date of first relapse. Because specific KRAS codons have been associated with inferior outcomes,8,9 each KRAS codon was independently assessed for OS and RFS using the log-rank test and was compared using patients with wild-type KRAS/BRAF (n = 118); patients who had concurrent BRAF mutations were excluded from analysis of the KRAS mutant patients, because BRAF mutations are known to be associated with poor outcome.5,10
RESULTS
Patient Characteristics
Patient characteristics are listed in Table 1. The median age at initial diagnosis was 56 years, and 58% of patients were men. Forty-three percent of patients had localized disease, and 56.3% had metastatic disease at diagnosis. Mutational profiling revealed that 9 patients (4%) had an NRAS mutation, 18 (8%) had an APC mutation, 23 (10%) had a BRAF mutation, 28 (13%) had a PIK3CA mutation, 47 (21%) had a TP53 mutation, and 81 (36%) had a KRAS mutation. No mutations were identified in AKT, CTNNB1, EGFR, FLT3, IDH1, IDH2, JAK2, KIT, or NOTCH1. Only 1 patient had a PTEN mutation; therefore, PTEN was not evaluated further in the correlative analysis. The 81 patients who had KRAS mutations included mutations in the following codons: glycine (G) to cysteine (C) substitution in codon 12 (G12C) (n = 8), G to aspartic acid (D) substitution in codon 12 (G12D) (n = 28), G to valine (V) substitution in codon 12 (G12V) (n = 14); G to D substitution in codon 13 (G13D) (n = 19), G to serine (S) substitution in codon 12 (G12S) (n = 5), G to alanine (A) substitution in codon 12 (G12A) (n =6), and G12V/G12C (n = 1). In total, 47 patients had more than 1 mutation, including 38 patients with 2 mutations and 9 patients with 3 mutations. Specifically, these combinations included 11 KRAS/TP53 mutations, 7 KRAS/APC mutations, 6 KRAS/PIK3CA mutations, 6 TP53/BRAF mutations, 3 KRAS/TP53/PIK3CA mutations, 3 KRAS/APC/PIK3CA mutations, 3 PIK3CA/BRAF mutations, 1 NRAS/TP53 mutation, 1 NRAS/TP53/PIK3CA mutation, 1 KRAS/PIK3CA/PTEN mutation, 1 PIK3CA/APC mutation, 1 APC/PIK3CA/BRAF mutation, 1 TP53/APC mutation, 1 TP53/PIK3CA mutation, and 1 APC/BRAF mutation.
TABLE 1.
Patient Characteristics
| Characteristic | No. of Patients (%) |
|---|---|
| Age: Median [range], y | 56 [29–84] |
| Sex | |
| Men | 129 (58) |
| Women | 93 (42) |
| Anatomic site | |
| Rectal | 64 (29) |
| Colon | 158 (71) |
| Initial stage at diagnosis | |
| I | 10 (4.5) |
| II | 22 (9.9) |
| III | 65 (29) |
| IV | 125 (56.3) |
| History of polyps | 30 (14) |
| Positive family history of GI cancer | 90 (41) |
| Smoking status | |
| None | 112 (50) |
| Former | 88 (40) |
| Current | 22 (10) |
| Specimen type for mutational analysis | |
| Pretreatment primary | 108 (49) |
| Post-treatment primary | 11 (5) |
| Metastatic site | 76 (34) |
| Primary recurrent disease | 7 (3) |
| Unspecified | 20 (9) |
| Overall frequency of mutations | |
| NRAS | 9 (4) |
| KRAS | 81 (36) |
| TP53 | 47 (21) |
| APC | 18 (8) |
| PIK3CA | 28 (13) |
| BRAF | 23 (10) |
| PTEN | 1 (0.5) |
Abbreviations: APC, adenomatosis polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homolog B; GI, gastrointestinal; KRAS, Kirsten rat sarcoma viral oncogene homolog; NRAS, neuroblastoma RAS viral oncogene homolog; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit a; PTEN, phosphatase and tensin homolog; TP53, tumor protein p53.
Patients who had NRAS and BRAF mutations were further characterized. Mutations in NRAS were identified in codon 12 (n = 5), codon 61 (n = 3), and codon 13 (n = 1). These mutations included 5 from pretreatment primary tumors, 3 from sites of metastatic disease, and 1 from a post-treatment primary site. Eight patients with rectal cancer and 1 patient with colon cancer had NRAS mutations. The tumor in the patient with colon cancer was located in the descending colon, and he presented with metastatic disease at diagnosis. Among the 23 BRAF mutations, 12 were from pretreatment primary sites, 7 were from metastatic sites, 2 were from locally recurrent sites, and 2 were from unspecified sites. Twenty-one mutations were from patients with colon cancer. The 2 patients with rectal cancer who had BRAF mutations were both men and carried the common V600E mutation. One patient had stage III disease, and 1 had stage IV disease at diagnosis.
Mutational Profiles by Anatomic Site
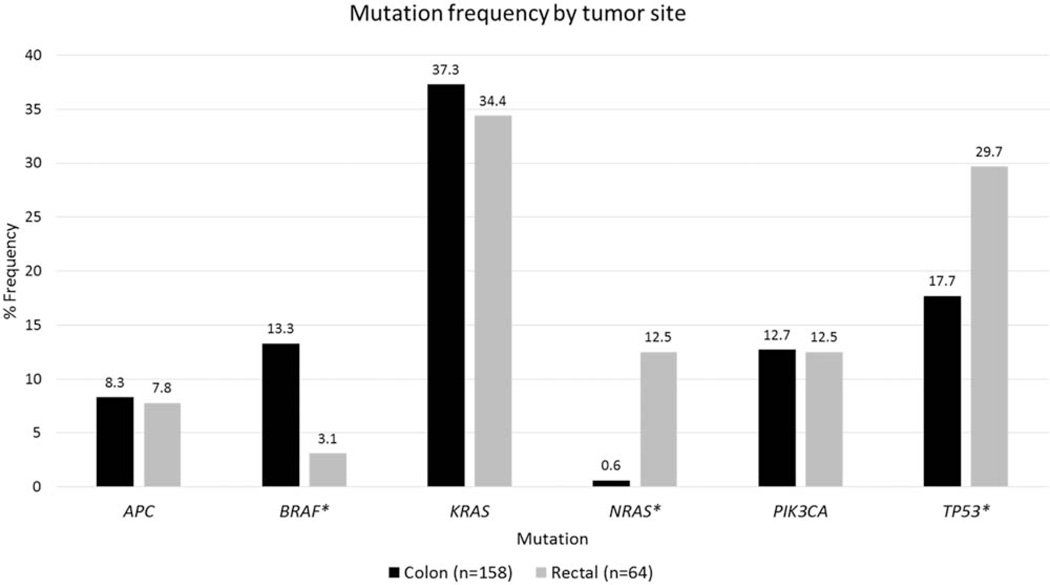
Figure 1 demonstrates the frequency of mutations identified by tumor site. There were significant differences between colon tumors and rectal tumors with regard to both NRAS and BRAF. Patients who had rectal cancer had a higher incidence of NRAS mutations compared with those who had colon cancer (12.5% vs 0.6%; P < .001). Conversely, BRAF mutations were more prevalent in patients who had colon cancer compared with those who had rectal cancer (13.3% vs 3.1%; P = .024). TP53 mutations were significantly more common in patients who had rectal cancer compared with those who had colon cancer (29.7% vs 17.7%; P = .048). There was no difference in the frequency of KRAS, APC, or PIK3CA mutations between patients with colon cancer versus rectal cancer.
Figure 1.
Mutation frequency is illusrated by tumor site. An asterisk denotes statistical significance (P < .05). The percentage frequency is listed above each column. APC indicates adenomatosis polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homolog B; KRAS, Kirsten rat sarcoma viral oncogene homolog; NRAS, neuroblastoma RAS viral oncogene homolog; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α; TP53, tumor protein p53.
Exploratory Analysis of Genotype and Clinical Characteristics
An exploratory analysis of additional clinical characteristics by genotype also was assessed (Table 2). NRAS mutations were associated with patients aged ≥56 years at diagnosis (7% vs 0.9%; P = .02) and with local recurrence as the first site of relapse (12.5% vs 3%; P = .03). BRAF mutations were associated with patients aged ≥56 years (15.8% vs 4.6%; P = .006) and with distant lymph node disease as the first site of relapse (26.2% vs 6.7%; P < .001), and they trended toward an association with women (15.1% vs 7%; P = .05) and an association with a personal history of polyps (19.4% vs 8.9%; P = .08). APC mutations were associated with nonsmokers (13.3% vs 2.8%; P = .004). PIK3CA mutations were associated with lung as the first site of relapse (23% vs 8.7%; P = .004) and trended toward an association in patients without polyps (14.1% vs 3.2%; P = .09). TP53 mutations were associated with men (26.4% vs 14%; P = .03) and patients aged <56 years (28.7% vs 14%; P = .007), and they trended toward an association in patients without a family history of gastrointestinal cancers (25% vs 15.6%; P = .09). Sites of first relapse in bone, liver, peritoneum, or other locations were not associated with any genotypes (data not shown), nor was stage at initial presentation. Genotypes and clinical correlations are summarized in Table 3.
TABLE 2.
Chi-Square Analysis of Mutations and Clinical Characteristics
| Genotype: Proportion of Patients (No.)a | ||||||
|---|---|---|---|---|---|---|
| Characteristic | NRAS | KRAS | TP53 | APC | PIK3CA | BRAF |
| Rectal cancer (64) vs colon cancer (158) | 12.5% (8) vs 0.6% (1) | 34.4% (22) vs 37.3% (59) | 29.7% (19) vs 17.7% (28) | 7.8% (5) vs 8.2% (13) | 12.5% (8) vs 12.7% (20) | 3.1% (2) vs 13.3% (21) |
| P | <.001 | .7 | .048 | .9 | .9 | .02 |
| Family history of GI cancer: Yes (90) vs no (132) | 3.3% (3) vs 4.5% (6) | 33.3% (30) vs 38.6% (51) | 15.6% (14) vs 25% (33) | 5.6% (5) vs 9.8% (13) | 15.6% (14) vs 10.6% (14) | 11.1% (10) vs 9.8% (13) |
| P | .7 | .4 | .09 | .3 | .3 | .8 |
| Men (129) vs women (93) | 4.7% (6) vs 3.2% (3) | 35.7% (46) vs 37.6% (35) | 26.4% (34) vs 14% (13) | 6.2% (8) vs 10.8% (10) | 14.7% (19) vs 9.7% (9) | 7% (9) vs 15.1% (14) |
| P | .6 | .8 | .03 | .2 | .3 | .05 |
| History of polyps: Yes (31) vs no (191) | 3.2% (1) vs 4.2% (8) | 41.9% (13) vs 35.6% (68) | 22.6% (7) vs 20.9% (40) | 9.7% (3) vs 7.9% (15) | 3.2% (1) vs 14.1% (27) | 19.4% (6) vs 8.9% (17) |
| P | .8 | .5 | .8 | .7 | .09 | .08 |
| Smoking history: Yes (109) vs no (113) | 5.5% (6) vs 2.7% (3) | 31.2% (34) vs 41.6% (47) | 22% (24) vs 20.4% (23) | 2.8% (3) vs 13.3% (15) | 10.1% (11) vs 15% (17) | 9.2% (10) vs 11.5% (13) |
| P | .3 | .1 | .8 | .004 | .3 | .6 |
| Stage at presentation: I (n = 10) vs II (n = 22) vs III (n = 64) vs IV (n = 125) | 10% (1) vs 4.5% (1) vs 3.1% (2) vs 3.2% (4) | 20% (2) vs 31.8% (7) vs 34.4% (22) vs 40% (50) | 10% (1) vs 27.3% (6) vs 17.2% (11) vs 23.2% (29) | 0 vs 9.1% (2) vs 10.9% (7) vs 7.2% (9) | 0 vs 22.7% (5) vs 12.5% (8) vs 12% (15) | 0 vs 9.1% (2) vs 15.6% (10) vs 8.8% (11) |
| P | .7 | .5 | .5 | .6 | .3 | .3 |
| Age at diagnosis: ≥56 y (114) vs <56 y (108) | 7% (8) vs 0.9% (1) | 34.2% (39) vs 38.9% (42) | 14% (16) vs 28.7% (31) | 5.3% (6) vs 11.1% (12) | 13.2% (15) vs 12% (13) | 15.8% (18) vs 4.6% (5) |
| P | .02 | .5 | .007 | .1 | .8 | .006 |
| Site of first recurrence Distant lymph node: Yes (42) vs no (180) | 0% (0) vs 5% (9) | 23.8% (10) vs 39.4% (71) | 26.2% (11) vs 20% (36) | 7.1% (3) vs 8.3% (15) | 14.3% (6) vs 12.2% (22) | 26.2% (11) vs 6.7% (12) |
| P | .14 | .06 | .04 | .8 | .7 | < .001 |
| Local recurrence (24 vs 198) | 12.5% (3) vs 3% (6) | 25% (6) vs 37.9% (75) | 16.7% (4) vs 21.7% (43) | 8.3% (2) vs 8.1% (16) | 12.5% (3) vs 12.6% (25) | 20.8% (5) vs 9.1% (18) |
| P | .03 | .2 | .6 | .9 | .9 | .08 |
| Lung (61 vs 161) | 4.9% (3) vs 3.7% (6) | 45.9% (28) vs 32.9% (53) | 21.3% (13) vs 21.1% (34) | 8.2% (5) vs 8.1% (13) | 23% (14) vs 8.7% (14) | 8.2% (5) vs 11.2% (18) |
| P | .7 | .07 | .9 | .9 | .004 | .5 |
Abbreviations: APC, adenomatosis polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homolog B; GI, gastrointestinal; KRAS, Kirsten rat sarcoma viral oncogene homolog; NRAS, neuroblastoma RAS viral oncogene homolog; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α; PTEN, phosphatase and tensin homolog; TP53, tumor protein p53.
Boldface values indicate statistically significant differences.
TABLE 3.
Summary of Genotype and Clinical Correlations
| Genotype | |||||
|---|---|---|---|---|---|
| Clinical Variable | NRAS | TP53 | BRAF | APC | PIK3CA |
| Site | Rectal | Rectal | Colon | NS | NS |
| Age | Older | Younger | Older | NS | NS |
| Sex | NS | Men | Women | NS | NS |
| Family history | NS | Negativea | NS | NS | NS |
| Polyps | NS | NS | Presenta | NS | Not presenta |
| Smoking status | NS | NS | NS | Nonsmokers | NS |
| Site of first recurrence | Local | NS | Distant lymph node | NS | Lung |
Abbreviations: APC, adenomatosis polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homolog B; NRAS, neuroblastoma RAS viral oncogene homolog; NS, no statistical significance; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α; TP53, tumor protein p53.
These findings only trended toward significance; all other factors had P values < .05.
Overall Survival and Relapse-Free Survival by Genotype
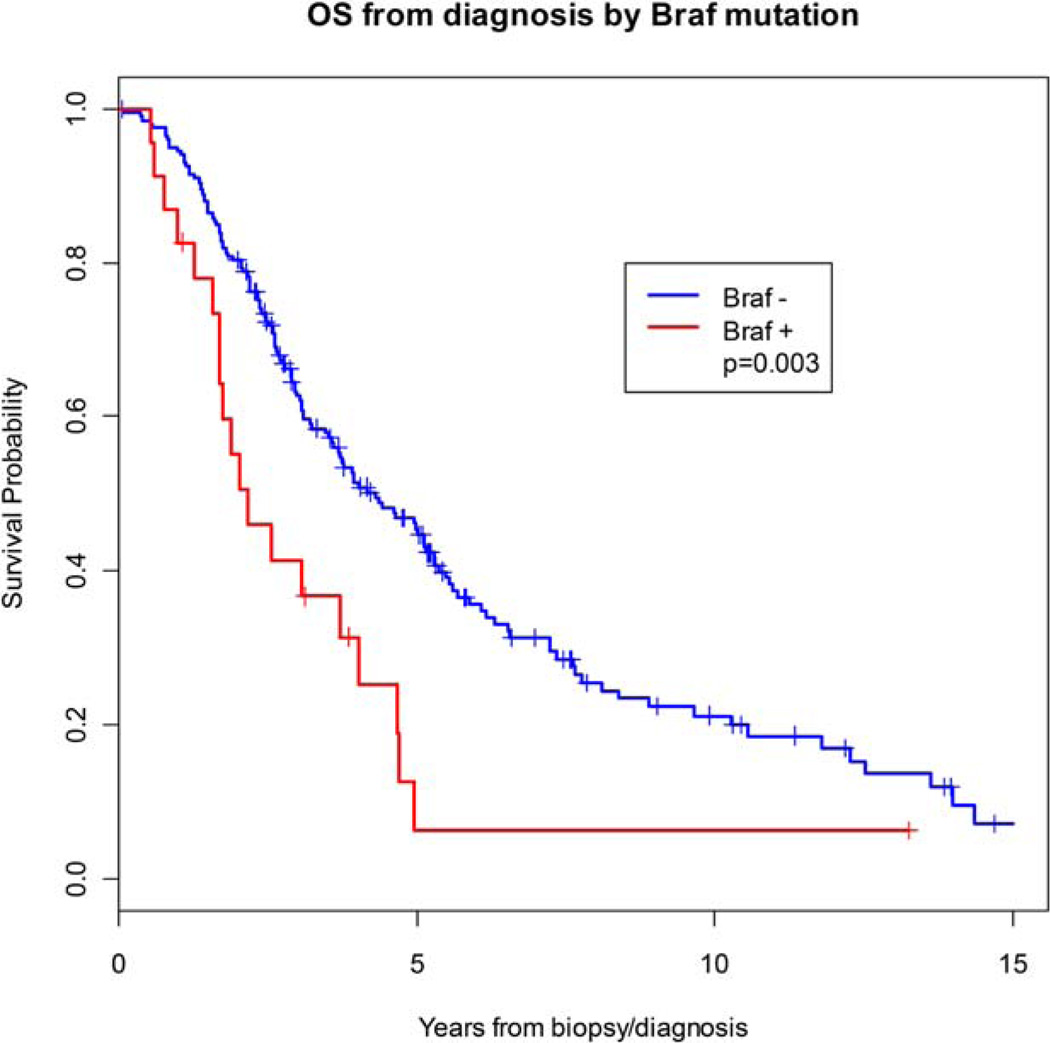
After a median follow-up of 91.2 months (range, 12.4–176.5 months), there were 154 deaths. Having a BRAF mutation was associated with a significantly inferior OS (hazard ratio, 3.13; 95% confidence interval, 1.56–6.29; P = .001). The estimated 5-year OS was 45.3% in BRAF-negative patients compared with 6.3% in BRAF-positive patients (P=.003) (Fig. 2). After adjusting for age, disease site, sex, smoking history, and stage at presentation, BRAF mutation remained a significant predictor of decreased OS (hazard ratio, 2.4; 95% confidence interval, 1.09–5.27; P = .029). No other genotypes were associated with OS. In the 97 patients who had localized disease at presentation, no mutations were associated with decreased RFS (Table 4).
Figure 2.
Overall survival is illustrated for patients with (+) and without (−) v-raf murine sarcoma viral oncogene homolog B (BRAF) mutations. The Kaplan-Meier method was used to estimate overall survival. Curves are compared using the log-rank test.
TABLE 4.
Estimated 5-Year Overall Survival and Relapse-Free Survival by Genotype
| Survival Rate (95% CI), % | ||||||
|---|---|---|---|---|---|---|
| 5-Year OS, n = 222 | 5-Year RFS, n = 97 | |||||
| Genotype | Mutation-Positive | Mutation-Negative | Log-Rank P | Mutation-Positive | Mutation-Negative | Log-Rank P |
| NRAS | 52 (16–79) | 41 (34–48) | .92 | 60 (13–88) | 4 (1–10) | .12 |
| KRAS | 37 (25–49) | 44 (35–52) | .47 | 3 (0.2–14) | 11 (5–19) | .10 |
| TP53 | 39 (24–54) | 42 (34–50) | .89 | 17 (4–37) | 5 (2–12) | .17 |
| APC | 55 (27–76) | 40 (33–48) | .40 | 11 (0.6–39) | 8 (4–15) | .83 |
| PIK3CA | 45 (25–63) | 41 (34–49) | .42 | 8 (0.5–29) | 7 (3–14) | .69 |
| BRAF | 6 (0.4–24) | 45 (38–53) | .003a | 8 (0.5–31) | 7 (3–14) | .56 |
Abbreviations: APC, adenomatosis polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homolog B; CI, confidence interval; KRAS, Kirsten rat sarcoma viral oncogene homolog; NRAS, neuroblastoma RAS viral oncogene homolog; OS, overall survival; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α; RFS, relapse-free survival; TP53, tumor protein p53.
Values in boldface indicate statistically significant differences.
Subgroup Analysis of KRAS Mutant Patients
When we compared the outcomes of patients who had specific KRAS mutations with the outcomes of patients who had KRAS/BRAF wild-type tumors, only the G13D mutation trended toward worse OS (P = .063).
DISCUSSION
This study profiles the mutational and clinical differences in patients with metastatic colon and rectal adenocarcinoma. Most importantly, of the 222 patients analyzed, a significant difference in the frequency of NRAS mutations was noted between patients with rectal cancer (12.5%) and those with colon cancer (0.6%). There were also significant differences in the rates of BRAF mutations and the subset of TP53 mutations screened for in colon cancers versus rectal cancers (13.3% vs 3.1% and 17.7% vs 29.7%, respectively). A clinical profile associated with certain mutations also was noted. NRAS mutations were identified in older patients with rectal cancer, whereas BRAF mutations were identified in older women patients with colon cancer. APC mutations were observed more commonly in nonsmokers. TP53 mutations were identified in younger men with rectal cancer.
The finding that NRAS mutations were identified almost exclusively in patients with rectal cancer is noteworthy. To our knowledge, this is the first study to identify NRAS as a predominantly rectal cancer mutation. In a study of 225 colorectal cancers, NRAS was detected in 5 patients (2.2%) in codons 12 and 61.11 It is noteworthy that no NRAS mutations were located in tumors of the proximal colon, whereas 4 mutations were located in tumors of the distal colon, and 1 mutation was located in a tumor of the rectum. In our study, 8 of 9 NRAS mutations were identified in rectal adenocarcinomas, whereas a single mutation was identified in a tumor of the descending colon. We also observed that NRAS mutations were more common in older patients. To our knowledge, this has not been described previously in CRC; however, it is noteworthy that this association has been observed in melanoma.12
Mutations in BRAF were identified most commonly in older women with colon cancer. These findings are concordant with prior studies.5,13 Although downstream pathway mitogen-activated protein kinase and extracellular signal-regulated kinase inhibitors are being actively investigated in early phase clinical trials, either alone or in combinations with other agents, it will be of interest to determine the impact of BRAF and NRAS mutations in predicting response.
TP53 mutations were identified more commonly in younger men with rectal cancer, consistent with reports that patients aged < 50 years are more likely to harbor TP53 mutations.14 This has been suggested as 1 of the earlier mutations once an adenoma has transitioned to cancer.15 Others have also observed that distal rather than proximal colon cancers have a higher incidence of TP53 mutations.16 The presence of an APC mutation was more common in nonsmokers than in smokers (13.3% vs 2.8%; P = .004). At least 2 prior studies reported no association with smoking and APC mutation in CRC, suggesting that this more likely is a sporadic mutation17,18 and is 1 of the earliest mutations in colon cancer progression.19
There are conflicting data regarding the similarities and differences between colon and rectal cancers at the molecular level. Different single nucleotide polymorphism profiles have been reported; and it has been demonstrated that protein expression of the cluster differentiation 44 standard isoform (CD44s), CD44 variant 6 (CD44v6), nuclear β-catenin, and CD68 can discriminate between left-sided and right-sided tumors.20–22 However, a gene expression study of microsatellite-stable colorectal tumors and a recent analysis of The Cancer Genome Atlas (TCGA) demonstrated only minor or no differences in colon cancers versus rectal cancers.23,24 In addition, this lack of difference in TCGA was only in the nonhypermutated specimens, in which copy number, methylation status, messenger RNA, and micro-RNA levels were assessed. Both colon and rectal cancers were analyzed together for somatic mutations, and the frequency of NRAS mutations among colon versus rectal adenocarcinomas was not described for that study. Furthermore, the high sensitivity of our SNaPshot assay relative to the depth of sequencing used in the next-generation studies may also contribute to these discrepancies.
Sites of first relapse by genotype also were analyzed. PIK3CA mutations were associated with lung, BRAF mutations were associated with distant lymph nodes, and NRAS mutations were associated with local recurrence as the first site of relapse. Tie et al reported a similar finding for PIK3CA mutations and lung metastases on univariate analysis, which trended toward significance on multivariate analysis.25 This raises the possibility that, although sites of first metastasis are more likely driven by anatomy, because the colon drains to the portal vein and the rectum drains to the vena cava through the inferior rectal vein,2 genotype may also play a role and should be further investigated.
We also assessed the association of genotype with clinical outcomes. BRAF was the only mutation associated with decreased OS. This has been demonstrated in multiple studies; and, in a recent meta-analysis of >11,000 patients with CRC, the presence of a BRAF mutation increased the risk of mortality greater than 2 times.5,10,26 When specific KRAS codons were analyzed, the G13D KRAS mutation trended toward significance for decreased OS compared with patients who had wild-type KRAS/BRAF. The G13D mutation has been associated with decreased OS in patients with CRC who received treatment with folinic acid (leucovorin), fluorouracil, and oxaliplatin (FOLFOX) plus panitumumab in 1 study8 but with improved survival compared with codon 12 mutations in a meta-analysis of patients who received cetuximab either alone or in combination with chemotherapy.27 Others have observed that the G12V mutation was associated with higher CRC-specific mortality,9 and the G12A mutation was associated with inferior OS in a pooled analysis.8 Our small numbers of individual codon mutations provided only limited insight.
There are additional points that merit further discussion. This was a retrospective, exploratory study and, thus, was strictly hypothesis-generating. Furthermore, because of the retrospective nature, there is likely selection bias among our patient cohort, and our results are only applicable to patients with metastatic disease, because these are the patients who are routinely offered mutation profiling at our institution. Only a portion of the known mutations in the tumor suppressor genes APC, TP53, NOTCH1, and PTEN were evaluated in our assay. Because of the limited coverage, the mutation rate of TP53 was only 21%, however it is well established that the mutation rate in CRC is closer to 40% to 50%.28 Therefore, other inactivating mutations in these genes probably were missed using this technique. In addition, the current study included patients who had genomic analysis performed on a mix of pretreatment and post-treatment primary tumors (as detailed in Table 1), sites of metastatic disease, and sites of local recurrences. It is possible that the mutational profile could change in the metastatic or post-treatment setting; however, it has been demonstrated that there is a high concordance of NRAS, KRAS, PIK3CA, BRAF, and TP53 mutations between primary and metastatic sites in CRC.25,29 Others have observed high concordance between pre-FOLFOX and post-FOLFOX treated CRCs when assessing KRAS, NRAS, BRAF, and PIK3CA.30 Data from those studies support the notion that sites of both primary and metastatic disease, as well as pretreatment and post-treatment tumors, may be valid sources for assessing mutational status. Finally, we did not have microsatellite instability information for the majority of patients with BRAF mutations, because we did not routinely test for this at our institution during the time these patients were evaluated; this can also provide important prognostic information.
Conclusions
In summary, this was an exploratory analysis of clinical correlations by genotype in patients with metastatic colon and rectal cancer. Notably, NRAS was identified predominantly in older patients with rectal cancer. This novel finding is important because, whereas the frequency in this study was only 12.5%, with over 40,000 rectal cancers per year in the United States, approximately 4800 patients may have NRAS mutations. Although we did not demonstrate inferior outcomes for patients with NRAS mutations, our numbers were small, and these patients warrant further investigation. Our study also confirmed the association of BRAF mutations with older patients who had colon cancer and with decreased survival. TP53 was correlated with young men who had rectal cancer, and APC was associated with non-smokers. Whereas some of these findings have been reported previously in single-institution studies, to our knowledge, no studies to date have provided a comprehensive assessment of these clinical correlates. These findings suggest there may be a genetic difference in colon and rectal adenocarcinomas. Furthermore, clinical profiles by genotype may aid clinicians in earlier identification of patients who harbor specific mutations and who may benefit from enrollment in clinical trials of targeted therapy in the metastatic setting. The long-term utility of this strategy is unknown and will depend on finding effective treatments for certain genotypes.
Supplementary Material
Acknowledgments
FUNDING SUPPORT
No specific funding was disclosed.
CONFLICT OF INTEREST DISCLOSURES
Dr. Borger, Dr. Dias-Santagata, Dr. Ellisen, and Dr. Iafrate are paid consultants for Bio-Reference Laboratories, the licensee of SNaPshot, the technology used in this study. Dr. Faris reports receiving personal fees from N-of-One.
We acknowledge Dr. Lipika Goyal for her assistance in reading the article.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Presented in part at the Annual Meeting of the American Society of Clinical Oncology; May 31 to June 3–7, 2011; Chicago, IL, June, 2011.
REFERENCES
- 1.American Cancer Society. Cancer Facts & Figures 2012. Atlanta, GA: American Cancer Society; 2012. [Google Scholar]
- 2.Hong TS, Clark JW, Haigis KM. Cancers of the colon and rectum: identical or fraternal twins? Cancer Discov. 2012;2:117–121. doi: 10.1158/2159-8290.CD-11-0315. [DOI] [PubMed] [Google Scholar]
- 3.Kornmann M, Staib L, Wiegel T, et al. Long-term results of 2 adjuvant trials reveal differences in chemosensitivity and the pattern of metastases between colon cancer and rectal cancer. Clin Colorectal Cancer. 2013;12:54–61. doi: 10.1016/j.clcc.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Linardou H, Dahabreh IJ, Bafaloukos D, Kosmidis P, Murray S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 2009;6:352–366. doi: 10.1038/nrclinonc.2009.62. [DOI] [PubMed] [Google Scholar]
- 5.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 6.Dias-Santagata D, Lam Q, Vernovsky K, et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: diagnostic and therapeutic implications [serial online] PLoS One. 2011;6:e17948. doi: 10.1371/journal.pone.0017948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeters M, Douillard JY, Van Cutsem E, et al. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31:759–765. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 9.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18:4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 11.Irahara N, Baba Y, Nosho K, et al. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19:157–163. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacker E, Nagore E, Cerroni L, et al. NRAS and BRAF mutations in cutaneous melanoma and the association with MC1R genotype: findings from Spanish and Austrian populations. J Invest Dermatol. 2013;133:1027–1033. doi: 10.1038/jid.2012.385. [DOI] [PubMed] [Google Scholar]
- 13.Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg M, Danielsen SA, Ahlquist T, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset [serial online] PLoS One. 2010;5:e13978. doi: 10.1371/journal.pone.0013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 16.Russo A, Bazan V, Iacopetta B, et al. The TP53 Colorectal Cancer International Collaborative Study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23:7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 17.Luchtenborg M, Weijenberg MP, Kampman E, et al. Cigarette smoking and colorectal cancer: APC mutations, hMLH1 expression, and GSTM1 and GSTT1 polymorphisms. Am J Epidemiol. 2005;161:806–815. doi: 10.1093/aje/kwi114. [DOI] [PubMed] [Google Scholar]
- 18.Diergaarde B, Vrieling A, van Kraats AA, van Muijen GN, Kok FJ, Kampman E. Cigarette smoking and genetic alterations in sporadic colon carcinomas. Carcinogenesis. 2003;24:565–571. doi: 10.1093/carcin/24.3.565. [DOI] [PubMed] [Google Scholar]
- 19.Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18:1967–1979. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- 20.Haug U, Poole EM, Xiao L, et al. Glutathione peroxidase tagSNPs: associations with rectal cancer but not with colon cancer. Genes Chromosomes Cancer. 2012;51:598–605. doi: 10.1002/gcc.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mates IN, Jinga V, Csiki IE, et al. Single nucleotide polymorphisms in colorectal cancer: associations with tumor site and TNM stage. J Gastrointest Liver Dis. 2012;21:45–52. [PubMed] [Google Scholar]
- 22.Minoo P, Zlobec I, Peterson M, Terracciano L, Lugli A. Characterization of rectal, proximal and distal colon cancers based on clinicopathological, molecular and protein profiles. Int J Oncol. 2010;37:707–718. doi: 10.3892/ijo_00000720. [DOI] [PubMed] [Google Scholar]
- 23.Sanz-Pamplona R, Cordero D, Berenguer A, et al. Gene expression differences between colon and rectum tumors. Clin Cancer Res. 2011;17:7303–7312. doi: 10.1158/1078-0432.CCR-11-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tie J, Lipton L, Desai J, et al. KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin Cancer Res. 2011;17:1122–1130. doi: 10.1158/1078-0432.CCR-10-1720. [DOI] [PubMed] [Google Scholar]
- 26.Safaee Ardekani G, Jafarnejad SM, Tan L, Saeedi A, Li G. The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis [serial online] PLoS One. 2012;7:e47054. doi: 10.1371/journal.pone.0047054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao C, Huang YF, Yang ZY, Zheng DY, Chen JZ, Tang JL. KRAS p.G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: a systematic review and meta-analysis. Cancer. 2013;119:714–721. doi: 10.1002/cncr.27804. [DOI] [PubMed] [Google Scholar]
- 28.Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat. 2003;21:271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- 29.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamoto Y, Tsuchihara K, Yoshino T, et al. KRAS mutations in primary tumours and post-FOLFOX metastatic lesions in cases of colorectal cancer. Br J Cancer. 2012;107:340–344. doi: 10.1038/bjc.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.