Abstract
Recent genetic studies have identified common variation in susceptibility loci that stratify lifetime risks of breast cancer and may inform prevention and screening strategies. However, whether these loci have similar implications for women treated with tamoxifen or raloxifene (SERMs) is unknown. We conducted a matched case–control study of 592 cases who developed breast cancer and 1,171 unaffected women from 32,859 participants on SERM therapy enrolled on NSABP P-1 and P-2 breast cancer prevention trials. We formed a quantitative polygenic risk score (PRS) using genotypes of 75 breast cancer-associated single nucleotide polymorphisms and examined the PRS as a risk factor for breast cancer among women treated with SERMs. The PRS ranged from 3.98 to 7.74, with a one-unit change associated with a 42 % increase in breast cancer (OR = 1.42; P = 0.0002). The PRS had a stronger association with breast cancer among high-risk women with no first-degree family history (OR = 1.62) compared to those with a positive family history (OR = 1.32) (Pintx = 0.04). There was also suggestion that PRS was a stronger risk factor for ER-positive (OR = 1.59, P = 0.0002) than ER-negative (OR = 1.05, P = 0.84) breast cancer (Pintx = 0.10). Associations did not differ by tamoxifen or raloxifene treatment, age at trial entry, 5-year predicted Gail model risk or other clinical variables. The PRS is a strong risk factor for ER-positive breast cancer in moderate to high-risk individuals treated with either tamoxifen or raloxifene for cancer prevention. These data suggest that common genetic variation informs risk of breast cancer in women receiving SERMs.
Keywords: SERMs, Mammographic density, Polygenic risk score, Single nucleotide polymorphisms (SNPs)
Introduction
Primary prevention of breast cancer remains a major goal for reducing the burden associated with this disease. Two large breast cancer prevention trials of selective estrogen receptor modulators (SERMs) including the National Surgical Adjuvant Breast and Bowel Project (NSABP) P-1 placebo-controlled trial of tamoxifen [1] and double-blind NSABP P-2 trial comparing raloxifene to tamoxifen, showed that these agents reduce the risk of breast cancer among women with a 5-year predicted breast cancer risk of at least 1.66 by 50 % after five years of therapy [2]. Follow-up of the P-2 trial at a median exposure of 81 months suggested that long-term raloxifene use was 76 % as effective for preventing invasive disease, but had less toxicity than tamoxifen [3]. Thus, both tamoxifen and raloxifene are viable prevention strategies for women at high risk of breast cancer [4].
Almost 80 confirmed common genetic susceptibility loci for breast cancer have been identified to date [5–20]. Taken together, these validated loci are estimated to explain up to 14 % of familial breast cancer risk [5]. Two recent studies showed that a polygenic risk score (PRS) composed of 76–77 of these genetic loci can identify individuals at increased breast cancer risk in the general population [21, 22]. Specifically, those at highest risk by the PRS had a 1.8-fold increased risk for breast cancer relative to the second quartile of PRS, and those in the lowest quartile had a reduced risk (0.6 fold) of breast cancer [21]. The PRS association with breast cancer was stronger among those with ER-positive compared to ER-negative disease and effectively stratified breast cancer risk in women both with and without a family history of breast cancer [22].
It is not clear, however, whether these common genetic variants will also be risk factors for breast cancer among high-risk women treated with SERMs for breast cancer prevention, given the large risk reduction associated with SERMs. We present the first report to evaluate a comprehensive set of 75 established breast cancer susceptibility loci, in the context of a PRS, as a risk factor for breast cancer among high-risk women from NSABP P-1 and P-2 trials taking raloxifene and tamoxifen for breast cancer prevention. We also examined whether the influence of the PRS on breast cancer differs by type of SERM, extent of family history, ER-positive compared to ER-negative breast cancer, and other clinical characteristics.
Methods
Study populations
The study population consisted of a nested case–control sample within the NSABP P-1 and P-2 trials [23] including 596 breast cancer cases who developed breast cancer while on SERM therapy and 1,171 matched controls selected from the 32,859 participants enrolled in P-1 and P-2 breast cancer prevention trials. Controls were matched to cases on trial and treatment arm (P-1 tamoxifen, P-2 tamoxifen, P-2 raloxifene), age at trial entry, categories of 5-year predicted breast cancer risk based on the Gail model [24], history of lobular carcinoma in situ, history of atypical hyperplasia, and time on study (controls on study at least as long as the matched breast cancer case) (Table 1) [23]. Each study obtained informed consent and had ethics and institutional approvals.
Table 1.
Characteristics of cases and matched controls within National Surgical Adjuvant Breast and Bowel Project P-1 and P-2 Trials
| Controls (N = 1,171) | Cases (N = 592) | |
|---|---|---|
| NSABP Trial | ||
| P-1 | 153 (13.1 %) | 79 (13.3 %) |
| P-2 | 1,018 (86.9 %) | 513 (86.7 %) |
| Type of breast event | ||
| Invasive breast cancer | 0 (0 %) | 453 (76.5 %) |
| DCIS | 0 (0 %) | 139 (23.5 %) |
| Estrogen receptor status (Invasive breast cancer only) | ||
| Unknown | 21 (4.6 %) | |
| Negative | 119 (26.3 %) | |
| Positive | 313 (69.1 %) | |
| Treatment | ||
| Tamoxifen | 628 (53.6 %) | 318 (53.7 %) |
| Raloxifene | 543 (46.4 %) | 274 (46.3 %) |
| Age (years) at entry | ||
| Mean (SD) | 59.9 (7.34) | 59.9 (7.27) |
| Median | 59.0 | 59.0 |
| <55 | 287 (24.5 %) | 146 (24.7 %) |
| 55–59 | 339 (28.9 %) | 170 (28.7 %) |
| 60–64 | 261 (22.3 %) | 137 (23.1 %) |
| 65+ | 284 (24.3 %) | 139 (23.5 %) |
| Five-year predicted breast cancer risk by the Gail model | ||
| Mean (SD) | 4.8 (2.41) | 4.9 (2.50) |
| Median | 4.2 | 4.5 |
| <=2.00 % | 67 (5.7 %) | 33 (5.6 %) |
| 2.01–3.00 % | 247 (21.1 %) | 121 (20.4 %) |
| 3.01–5.00 % | 365 (31.2 %) | 183 (30.9 %) |
| >5.00 % | 492 (42.0 %) | 255 (43.1 %) |
| History of LCIS at entry | ||
| No | 957 (81.7 %) | 480 (81.1 %) |
| Yes | 214 (18.3 %) | 112 (18.9 %) |
| History of atypical hyperplasia at entry | ||
| No | 897 (76.6 %) | 436 (73.6 %) |
| Yes | 274 (23.4 %) | 156 (26.4 %) |
| History of hysterectomy at entry | ||
| No | 602 (51.4 %) | 316 (53.4 %) |
| Yes | 569 (48.6 %) | 276 (46.6 %) |
| Number of first-degree relatives with breast cancer | ||
| 0 | 351 (30.0 %) | 198 (33.4 %) |
| 1 | 569 (48.6 %) | 268 (45.3 %) |
| ≥2 | 251 (21.4 %) | 126 (21.3 %) |
| Body mass index | ||
| Mean (SD) | 28.4 (5.86) | 28.6 (6.08) |
| Q1 | 24.2 | 24.2 |
| Median | 27.5 | 27.5 |
| Q3 | 31.9 | 31.9 |
| Range | 15.4–68.2 | 16.5–57.2 |
SD Standard deviation, DCIS Ductal carcinoma in situ, LCIS Lobular carcinoma in situ
Genotyping
The genotypes of 75 published breast cancer single nucleotide polymorphisms (SNPs) (Supplementary Table 1) were obtained from a genome-wide association study (GWAS) of cases and controls. Genotyping was performed by the RIKEN Center for Integrative Medical Science using the Illumina Human610-Quad BeadChip and genotypes are currently available through dbGAP (dbGaP Study Accession number is phs000305.v1.p1) [23, 25]. Four cases were ineligible due to low DNA quantity (n = 2) or quality (n = 2) for a total of 592 cases for GWAS analyses. Imputation was performed using Beagle and all samples from the version 2 of the 1000 Genomes data May 2011 [26] as a reference [27]. Of the 77 SNPs previously shown associated with breast cancer [5–20] (Supplementary Table 1), 75 were available and used to form the PRS. Of these, genotypes on 36 SNPs were imputed and had a quality score r2 > 0.4, with the majority (n = 33 of 36) above r2 > 0.8.
Statistical methods
The PRS was created using per allele odds ratios from the SNP associations with overall breast cancer (Supplementary Table 1) [5–20]. The PRS represented the combined effect of the 75 SNPs, regardless of departures from a multiplicative model, because there has been no evidence seen for SNP by SNP interactions [28]. Specifically, the log OR for each SNP was multiplied by the number of risk alleles and summed to generate a unique PRS for each person in the dataset [29]. For missing genotypes (0.05 %), the SNP was locally imputed within a 20 Mb region around the SNP, using Beagle v3.3.1 and 1000 Genomes, version 2 [26, 27]. The PRS approximated a normal distribution, and was included as a continuous measure (per one unit) in the conditional logistic regression risk model. For ease of presentation, the PRS score was also divided into quintiles based on the distribution among controls. Associations of PRS with breast cancer were examined with conditional logistic regression, accounting for the matched design.
Tests for differential associations of PRS by prevention agent (raloxifene vs. tamoxifen), family history (1 or more 1st degree relatives vs. 0 relatives), age at trial entry (<55 vs. ≥55), predicted 5-year risk based on the Gail model (<3.01 vs. ≥3.01 %), hysterectomy (no/yes), atypical hyperplasia (no/yes), and lobular carcinoma in situ (LCIS) (no/yes), with breast cancer were tested by creation of an interaction term between each covariate and the main effect of PRS. For ER-receptor status, age at onset (<55, 55–64, 65+) and type of breast cancer (ductal carcinoma in situ (DCIS) vs. invasive), we stratified cases (and their matched controls), fit conditional logistic regression models within each strata, and compared the odds ratios across strata by taking the difference in log OR, and dividing by the square root of the sum of the variances.
Results
Table 1 shows the characteristics of the cases and controls. There were 139 women with DCIS and 453 women with invasive breast cancer; 69 % of the invasive cases were ER-positive, 26 % were ER-negative, and 5 % had unknown ER status. A quarter of sample was less than age 55 at trial entry and over two-thirds had a five-year predicted risk score of greater than 3 % by the Gail model, indicative of a population at greater than average risk. Matching was successful on all variables (Table 1).
The PRS based on 75 variants ranged from 3.98 to 7.74, with a median of 5.61. A one-unit change in PRS was associated with 42 % increase in breast cancer risk (OR = 1.42; 95 % CI 1.18–1.70). The PRS association with breast cancer risk was also evident when examining quintiles of the PRS (Table 2; Supplementary Fig. 1) (Ptrend = 0.0005). Relative to the middle quintile (5.52–5.78), women in the lowest quintile (3.98–5.17) were at a reduced risk of breast cancer (OR = 0.81; 95 % CI 0.59–1.12) while those in the highest PRS quintile (6.10–7.74) were at an increased risk (OR = 1.45; 95 % CI 1.06–1.98). This translates to a risk of 1.8 comparing highest to lowest quintiles.
Table 2.
Association of polygenic risk score (PRS) and breast cancer (n = 592 cases, 1,171 controls) within the National Surgical Adjuvant Breast and Bowel Project P-1 and P-2 trials
| Cases | Controls | Odds ratio (95 % CI) | P value* | |
|---|---|---|---|---|
| PRS quintiles | ||||
| Q1 (<5.17) | 95 (16.0 %) | 258 (22.0 %) | 0.81 (0.59–1.12) | |
| Q2 (5.18–5.51) | 116 (19.6 %) | 236 (20.2 %) | 1.07 (0.78–1.47) | |
| Q3 (5.52–5.78) | 114 (19.3 %) | 239 (20.4 %) | 1.00 (ref) | |
| Q4 (5.79–6.10) | 125 (21.1 %) | 227 (19.4 %) | 1.17 (0.85, 1.61) | |
| Q5 (>6.10) | 142 (24.0 %) | 211 (18.0 %) | 1.45 (1.06, 1.98) | 0.0005 |
P value for trend
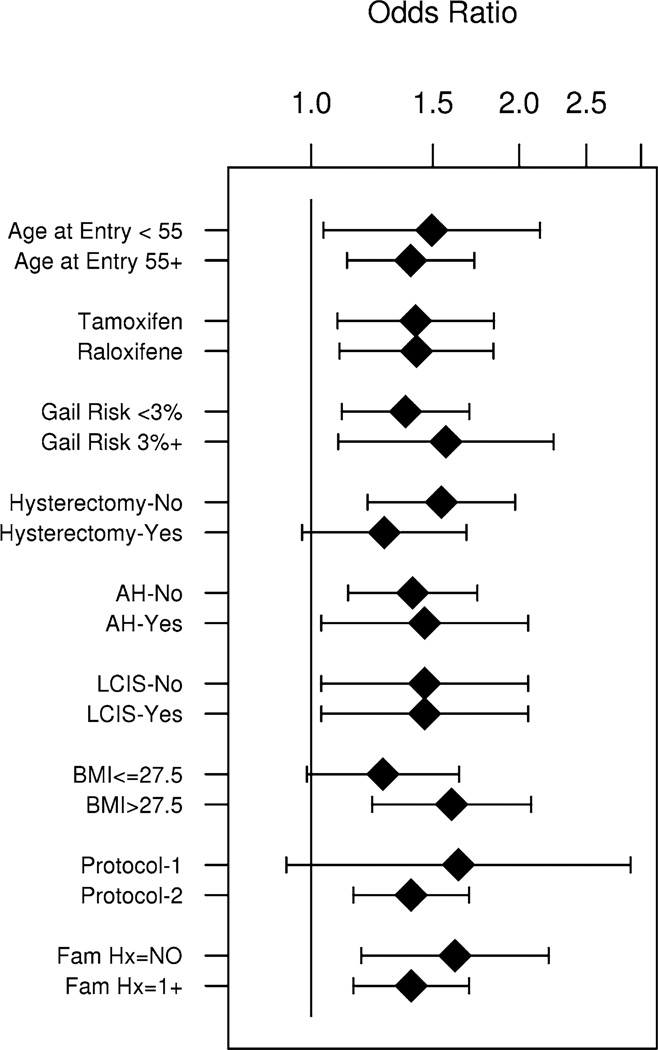
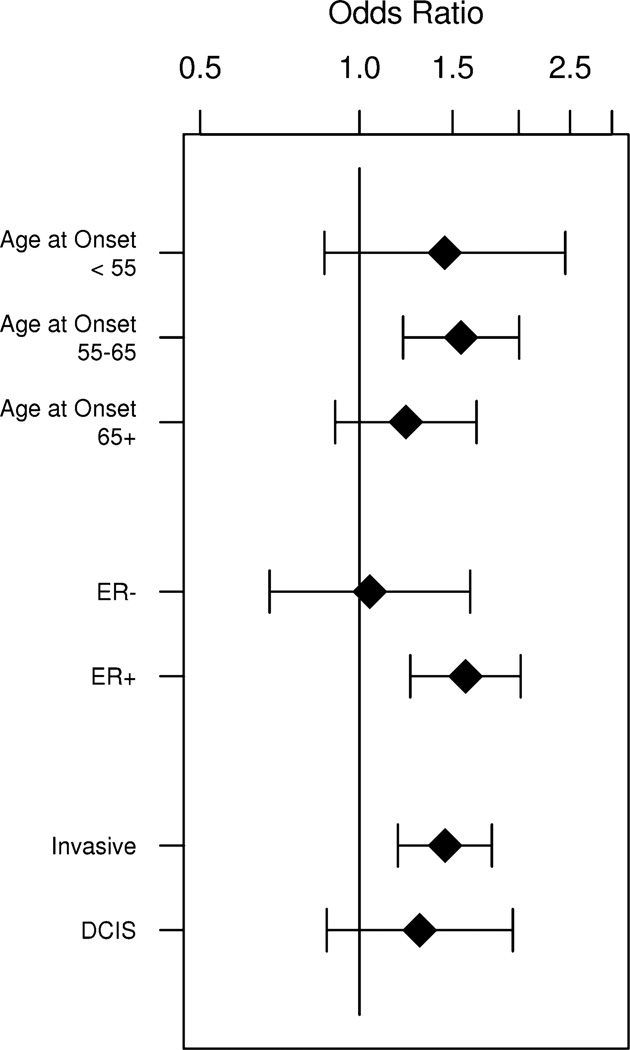
The association of PRS with breast cancer was similar across age at trial entry, treatment type, 5-year predicted risk, hysterectomy status, body mass index (BMI), presence of atypical hyperplasia, and LCIS (all P values for heterogeneity >0.15) (Fig. 1; Supplementary Table 2). However, there was evidence of a stronger association of PRS with breast cancer among women without a first-degree family history of breast cancer (OR = 1.62 per unit change in PRS, 95 % CI 1.18–2.21) compared to those with a positive family history (OR = 1.32, 95 % CI 1.06–1.66) (Pintx = 0.04) (Fig. 1). Further, the PRS appeared to be a stronger risk factor for ER-positive than ER-negative breast cancer, although the test for heterogeneity did not reach statistical significance (Pintx = 0.10) likely due to the limited number of ER-negative cases (n = 119). There was a 59 % increased risk with ER-positive breast cancer per unit change in PRS (OR = 1.59, 95 % CI 1.25–2.02, P = 0.0002), but only a 5 % increase in risk associated with ER-negative breast cancer (OR = 1.05, 95 % CI 0.68–1.62, P = 0.84) (Fig. 2; Supplementary Table 3). No differences were evident for age at onset or type of breast cancer (Fig. 2; Supplementary Table 3), although sample size was also limited for these comparisons.
Fig. 1.
Polygenic risk score (PRS) and breast cancer association (Odds ratios (OR) and 95 % confidence intervals) by clinical covariates
Fig. 2.
Polygenic risk score (PRS) and breast cancer association (Odds ratios (OR) and 95 % confidence intervals) by age at onset and tumor characteristics
Additional analyses examining the PRS and breast cancer association after adjustment for the two loci identified as breast cancer risk factors through a prior GWAS in this sample (rs10030044 at CTSO and rs8060157 at ZNF423) [23] showed no difference for the PRS and breast cancer association compared to the unadjusted results (data not shown).
Discussion
We present the first report to examine the influence of 75 common breast cancer susceptibility loci on breast cancer risk among women taking SERMs for primary prevention. Using genotyping data from women receiving tamoxifen and raloxifene in the NSABP P-1 and P-2 studies, we have shown that a PRS of the 75 loci is a risk factor for breast cancer in the presence of SERMs, with the risk of breast cancer ranging from OR = 0.59 to 1.98 for those with the lowest and highest PRS, respectively (compared to the average PRS). This finding suggests that the intrinsic risk of breast cancer associated with the common variants is maintained in the presence of the strong risk reducing effects of SERM treatment.
Although the PRS was a risk factor for women with and without a family history of breast cancer, we found that the association was stronger among women without a family history with 30 % increased risk per unit PRS in those with a family history, and 62 % increased risk in those without. This difference may reflect the fact that these SNPs explain a portion of familial breast cancer risk (estimated at 14 %) [5], thereby attenuating their influence on risk in this subgroup. One possible explanation is that a strong family history may reflect the presence of other more highly penetrant mutations that have a larger influence on breast cancer risk than these common genetic loci.
Our data suggested a stronger association of the PRS with ER-positive than with ER-negative breast cancer, although the differences did not reach statistical significance, likely due to the limited number of ER-negative breast cancers. The strong association with ER-positive breast cancer was expected, given that the majority of the 75 variants were originally identified in studies primarily comprised of ER-positive breast cancer [5–20]. In fact, only a small number of loci (LGR6, MDM4, 2p24.1, TERT, FTO, 19p13.31) have shown genome-wide significant (P < 5 × 10−8) associations with ER-negative breast cancer and risk estimates approximating 1.0 for ER-positive breast cancer (Supplementary Table 1) [7, 11, 30, 31]. Since SERMs are beneficial for prevention of ER-positive breast cancer, risk models incorporating a PRS that is strongly predictive of ER-positive cancer may allow better selection of women at high risk of ER-positive breast cancer who may benefit from SERM intervention. Furthermore, as the associations with breast cancer were similar in those taking either tamoxifen or raloxifene, it appears that the type of SERM may have little influence on the variant-associated risk. Thus, the PRS should also be evaluated as a risk factor for breast cancer in prevention trials or prospective patient populations treated with aromatase inhibitors, which are effective for prevention of ER-positive breast cancer [32, 33].
The absence of women on placebo or usual care in this study did not allow for examination of the interaction of the PRS and SERMs on risk. However, comparison with the few studies to date on the PRS and breast cancer association [21, 22] suggests that there is some attenuation of the association in the SERM-treated population. Although the PRS distributions are not directly comparable across these studies, a one-unit increase in PRS was associated with a 1.4-fold increased risk of breast cancer in this NSABP population, but associated with a 1.8-fold increase (95 % CI 1.6–2.1) in a general population [21]. Because of the known risk reduction associated with these SERMs, the SNPs (PRS) may not be as strong a risk factor in moderate to high-risk women on tamoxifen and raloxifene. In addition, the attenuation may be due in part to the large proportion of women with family history of breast cancer in the NSABP trials (70 %), for whom the PRS and breast cancer association was attenuated relative to those without a family history.
While the matched nature of the cases and controls precluded calculation of absolute risk and realistic area under the curve (AUC) estimates, the close matching in the two well-characterized clinical trials on a large number of clinical variables did allow evaluation of the PRS without potential confounding influences.
In conclusion, this is the first study to examine a comprehensive PRS among a moderate to high-risk population receiving SERMs and to demonstrate a contribution of common genetic variation to the development of future breast cancers.
Supplementary Material
Acknowledgments
The authors thank the women who participated in the NSABP P-1 and P-2 clinical trials and provided DNA and consent for its use in genetic studies.
Funding This work was supported by grants from the National Cancer Institute (U10 CA12027, U10 CA69974, U10 CA37377, U10 CA69651, P50 CA116201, PGRN U19 GM61388), the Breast Cancer Research Foundation, the RIKEN Center for Integrative Medical Science, and the Biobank Japan Project which was funded by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Abbreviations
- AUC
Area under the curve
- BCAC
Breast Cancer Association Consortium
- BMI
Body mass index
- DCIS
Ductal carcinoma in situ
- GWAS
Genome-wide association study
- LCIS
Lobular carcinoma in situ
- NSABP
National Surgical Adjuvant Breast and Bowel Project
- PRS
Polygenic risk score
- SERMs
Selective estrogen receptor modulators
- SNP
Single nucleotide polymorphism
- ER
Estrogen receptor
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-014-3175-4) contains supplementary material, which is available to authorized users.
Contributor Information
Celine M. Vachon, Email: vachon.celine@mayo.edu, Division of Epidemiology, Department of Health Sciences Research, Mayo Clinic, 200 First Street SW, Rochester, MN, USA.
Daniel J. Schaid, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA
James N. Ingle, Division of Medical Oncology, Department of Oncology, Mayo Clinic, Rochester, MN, USA
D. Lawrence Wickerham, Section of Cancer Genetics and Prevention, Allegheny General Hospital, Pittsburgh, PA, USA; National Surgical Adjuvant Breast and Bowel Project (NSABP), Pittsburgh, PA, USA.
Michiaki Kubo, RIKEN Center for Integrative Medical Science, Yokohama, Japan.
Taisei Mushiroda, RIKEN Center for Integrative Medical Science, Yokohama, Japan.
Matthew P. Goetz, Division of Medical Oncology, Department of Oncology, Mayo Clinic, Rochester, MN, USA
Erin E. Carlson, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA
Soonmyung Paik, Section of Cancer Genetics and Prevention, Allegheny General Hospital, Pittsburgh, PA, USA.
Norman Wolmark, Section of Cancer Genetics and Prevention, Allegheny General Hospital, Pittsburgh, PA, USA; Department of Human Oncology, Allegheny General Hospital, Pittsburgh, PA, USA.
Yusuke Nakamura, Section of Hematology and Oncology, Department of Medicine, University of Chicago, Chicago, IL, USA.
Liewei Wang, Division of Clinical Pharmacology, Department of Molecular Pharmacology & Experimental Therapeutics, Mayo Clinic, Rochester, MN, USA.
Richard Weinshilboum, Division of Clinical Pharmacology, Department of Molecular Pharmacology & Experimental Therapeutics, Mayo Clinic, Rochester, MN, USA.
Fergus J. Couch, Division of Epidemiology, Department of Health Sciences Research, Mayo Clinic, 200 First Street SW, Rochester, MN, USA Division of Experimental Pathology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA.
References
- 1.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90(18):1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 2.Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, Bevers TB, Fehrenbacher L, Pajon ER, Jr, Wade JL, 3rd, Robidoux A, Margolese RG, James J, Lippman SM, Runowicz CD, Ganz PA, Reis SE, McCaskill-Stevens W, Ford LG, Jordan VC, Wolmark N. Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP study of tamoxifen and raloxifene (STAR) P-2 trial. JAMA. 2006;295(23):2727–2741. doi: 10.1001/jama.295.23.joc60074. [DOI] [PubMed] [Google Scholar]
- 3.Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, Bevers TB, Fehrenbacher L, Pajon ER, Wade JL, 3rd, Robidoux A, Margolese RG, James J, Runowicz CD, Ganz PA, Reis SE, McCaskill-Stevens W, Ford LG, Jordan VC, Wolmark N. Update of the National Surgical Adjuvant Breast and Bowel Project Study of Tamoxifen and Raloxifene (STAR) P-2 Trial: Preventing breast cancer. Cancer Prev Res. 2010;3(6):696–706. doi: 10.1158/1940-6207.CAPR-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visvanathan K, Hurley P, Bantug E, Brown P, Col NF, Cuzick J, Davidson NE, Decensi A, Fabian C, Ford L, Garber J, Katapodi M, Kramer B, Morrow M, Parker B, Runowicz C, Vogel VG, 3rd, Wade JL, Lippman SM. Use of pharmacologic interventions for breast cancer risk reduction: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2013;31(23):2942–2962. doi: 10.1200/JCO.2013.49.3122. [DOI] [PubMed] [Google Scholar]
- 5.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Czene K, et al. Breast and Ovarian Cancer Susceptibility Collaboration. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, Chatterjee N, Garcia-Closas M, Gonzalez-Bosquet J, Prokunina-Olsson L, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver R, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, Miron P, Fasching PA, Brauch H, Chang-Claude J, Carpenter J, Godwin AK, Nevanlinna H, Giles GG, Cox A, Hopper JL, Bolla MK, Wang Q, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature Genet. 2013;45(4):392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, Lissowska J, Brinton L, Peplonska B, Southey MC, Hopper JL, McCredie MR, Giles GG, Fletcher O, Johnson N, dos Santos Silva I, Gibson L, Bojesen SE, Nordestgaard BG, Axelsson CK, Torres D, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 9.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, Aben KK, Strobbe LJ, Albers-Akkers MT, Swinkels DW, Henderson BE, Kolonel LN, Le Marchand L, Millastre E, Andres R, Godino J, Garcia-Prats MD, Polo E, Tres A, Mouy M, Saemundsdottir J, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 10.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, Eccles D, Evans DG, Fletcher O, Johnson N, dos Santos Silva I, Peto J, Stratton MR, Rahman N, Jacobs K, Prentice R, Anderson GL, Rajkovic A, Curb JD, Ziegler RG, Berg CD, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, Wang X, Ademuyiwa F, Ahmed S, Ambrosone CB, Baglietto L, Balleine R, Bandera EV, Beckmann MW, Berg CD, Bernstein L, Blomqvist C, Blot WJ, Brauch H, Buring JE, Carey LA, Carpenter JE, Chang-Claude J, Chanock SJ, Chasman DI, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43(12):1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45(4):371–384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M, Bergthorsson JT, Gudmundsson J, Aben KK, Strobbe LJ, Swinkels DW, van Engelenburg KC, Henderson BE, Kolonel LN, Le Marchand L, Millastre E, Andres R, Saez B, Lambea J, Godino J, Polo E, Tres A, Picelli S, Rantala J, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(6):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 14.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Luccarini C, Conroy D, Shah M, Munday H, Jordan C, Perkins B, West J, Redman K, Meyer KB, Haiman CA, Kolonel LK, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den Ouweland A, Houlston R, Ross G, Langford C, Pharoah PD, Stratton MR, Dunning AM, Rahman N, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, Wen W, Levy S, Deming SL, Haines JL, Gu K, Fair AM, Cai Q, Lu W, Shu XO. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, Zelenika D, Gut I, Heath S, Palles C, Coupland B, Broderick P, Schoemaker M, Jones M, Williamson J, Chilcott-Burns S, Tomczyk K, Simpson G, Jacobs KB, Chanock SJ, Hunter DJ, Tomlinson IP, Swerdlow A, Ashworth A, Ross G, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103(5):425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 18.French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S, Khan S, Maranian MJ, O’Reilly M, Hillman KM, Betts JA, Carroll T, Bailey PJ, Dicks E, Beesley J, Tyrer J, Maia AT, Beck A, Knoblauch NW, Chen C, Kraft P, Barnes D, Gonzalez-Neira A, Alonso MR, Herrero D, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92(4):489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, Dennis J, Wang Q, Humphreys MK, Luccarini C, Baynes C, Conroy D, Maranian M, Ahmed S, Driver K, Johnson N, Orr N, dos Santos Silva I, Waisfisz Q, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Hall P, Czene K, Irwanto A, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44(3):312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, Healey S, Morrison J, Kartsonaki C, Lesnick T, Ghoussaini M, Barrowdale D, Peock S, Cook M, Oliver C, Frost D, Eccles D, Evans DG, Eeles R, Izatt L, Chu C, Douglas F, Paterson J, Stoppa-Lyonnet D, Houdayer C, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42(10):885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vachon CM, Pankratz VS, Scott CG, Haeberle L, Ziv E, Jensen MR, Brandt KR, Whaley DH, Olson JE, Heusinger K, Hack CC, Jud SM, Beckmann MW, Schulz-Wendtland R, Tice JA, Norman AD, Cunningham JM, Purrington KS, Easton DF, Sellers TA, Kerlikowske K, Fasching PA, Couch FJ. The contributions of breast density and common genetic variation to breast cancer risk. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/dju397. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Easton DF. Germline polymorphisms and susceptibility to breast cancer; 35th Annual CTCR-AACR San Antonio Breast Cancer Symposium; Dec 4–8, 2012.2012. [Google Scholar]
- 23.Ingle JN, Liu M, Wickerham DL, Schaid DJ, Wang L, Mushiroda T, Kubo M, Costantino JP, Vogel VG, Paik S, Goetz MP, Ames MM, Jenkins GD, Batzler A, Carlson EE, Flockhart DA, Wolmark N, Nakamura Y, Weinshilboum RM. Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov. 2013;3(7):812–825. doi: 10.1158/2159-8290.CD-13-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costantino JP, Gail MH, Pee D, Anderson S, Redmond CK, Benichou J, Wieand HS. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J Natl Cancer Inst. 1999;91(18):1541–1548. doi: 10.1093/jnci/91.18.1541. [DOI] [PubMed] [Google Scholar]
- 25.db GaP Genotypes and Phenotypes. A Genome-Wide Association Study in Participants Experiencing Breast Cancer Events in High-Risk Postmenopausal Women Receiving Selective Estrogen Receptor Modulators on NSABP Trials P-1 and P-2. A Collaboration Between the NIH Pharmacogenetics Research Network and the RIKEN Yokohama Institute Center for Genomic Medicine. 2013 http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000305.v1.p1. [Google Scholar]
- 26.McVean G. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. Genome-wide imputation ran February 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne RL, Herranz J, Michailidou K, Dennis J, Tyrer JP, Zamora MP, Arias-Perez JI, González-Neira A, Pita G, Alonso MR, Wang Q, Bolla MK, Czene K, Eriksson M, Humphreys K, Darabi H, Li J, Anton-Culver H, Neuhausen SL, Ziogas A, Clarke CA, Hopper JL, Dite GS, Apicella C, Southey MC, et al. A large-scale assessment of two-way SNP interactions in breast cancer susceptibility using 46,450 cases and 42,461 controls from the breast cancer association consortium. Hum Mol Genet. 2014;23(7):1934–1946. doi: 10.1093/hmg/ddt581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darabi H, Czene K, Zhao W, Liu J, Hall P, Humphreys K. Breast cancer risk prediction and individualised screening based on common genetic variation and breast density measurement. Breast Cancer Res. 2012;14(1):R25. doi: 10.1186/bcr3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, Lindblom A, Nevanlinna H, Greco D, Aittomaki K, Blomqvist C, Chang-Claude J, Vrieling A, Flesch-Janys D, Sinn HP, Wang- Gohrke S, Nickels S, Brauch H, Ko YD, Fischer HP, Schmutzler RK, Meindl A, Bartram CR, Schott S, Engel C, Godwin AK, et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72(7):1795–1803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, Soucy P, Fredericksen Z, Barrowdale D, Dennis J, Gaudet MM, Dicks E, Kosel M, Healey S, Sinilnikova OM, Bacot F, Vincent D, Hogervorst FB, Peock S, Stoppa-Lyonnet D, Jakubowska A, Radice P, Schmutzler RK, Domchek SM, Piedmonte M, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9(3):e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goss PE, Ingle JN, Ales-Martinez JE, Cheung AM, Chlebowski RT, Wactawski-Wende J, McTiernan A, Robbins J, Johnson KC, Martin LW, Winquist E, Sarto GE, Garber JE, Fabian CJ, Pujol P, Maunsell E, Farmer P, Gelmon KA, Tu D, Richardson H. Exemestane for breast-cancer prevention in postmenopausal women. N Engl J Med. 2011;364(25):2381–2391. doi: 10.1056/NEJMoa1103507. [DOI] [PubMed] [Google Scholar]
- 33.Cuzick J, Sestak I, Forbes JF, Dowsett M, Knox J, Cawthorn S, Saunders C, Roche N, Mansel RE, von Minckwitz G, Bonanni B, Palva T, Howell A. Anastrozole for prevention of breast cancer in high-risk postmenopausal women (IBIS-II): an international, double-blind, randomised placebo-controlled trial. Lancet. 2014;383(9922):1041–1048. doi: 10.1016/S0140-6736(13)62292-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.