Abstract
Ribosomal DNA sequences of the second internal transcribed spacer (ITS-2) and 28S ribosomal DNA (618 bp) of Fasciolagigantica collected from cattle and buffaloes from four different geographical locations of India, were characterized for genotyping. ITS-2 sequence was analyzed in 28 worms that was typical of F. gigantica and differed at six positions, with one of these being a distinguishing deletion (T) at the 327th position in F. gigantica relative to F. hepatica. However, Fasciola specimens also showed intraspecies sequence polymorphism in the ITS-2, with two different ITS-2 sequences existing in the ribosomal DNA (rDNA) array within a single Fasciola worm. One of the sequences was identical to that of F. gigantica and the other showed extensive sequence polymorphism in the ITS-2. Using BspH1-restriction fragment length polymorphism, six variable ITS-2 sequences in F. gigantica were identified within these parasite specimens and were found distributed in these four geographical regions. 28S rDNA sequence of 24 flukes, collected from the above four geographical regions, showed a single nucleotide polymorphism at 284th nucleotide (G/A). Analyzing the sequence data of 28S rDNA of F. gigantica available from some African and Asian countries for this polymorphic 284th nucleotide position, it is proposed that there are two basic lineages of the F. gigantica for 28S rDNA existing in the fluke populations from five African and several Asian countries.
Keywords: Fasciola gigantica, 28S ribosomal DNA, ITS-2, Genetic characterization
Introduction
Fasciolosis, caused by the liver flukes Fasciola gigantica and F. hepatica, is a major problem for livestock farming due to huge losses caused by this parasitic infection as well as a public health concern due to its zoonotic potential. Fasciola hepatica, is the commonest and most widespread species in temperate regions while F. gigantica is found in tropical countries of Africa and Asia. The geographical co-existence and even, the presence of hybrid/introgressed forms that have both nuclear and mitochondrial sequences of these two species have been described (Agatsuma et al. 2000; Ali et al. 2008; Itagaki et al. 2005a, b, 2009; Peng et al. 2009). Studies have revealed the existence of a range of morphological forms and different ploidies such as diploid, triploid and mixoploid fasciolids (Itagaki et al. 2009; Le et al. 2008; Peng et al. 2009). Pure and mixed forms of Fasciola adults in human and ruminants, particularly in Asian countries like Iran (Ashrafi et al. 2006), China (Huang et al. 2004; Lin et al. 2007; Peng et al. 2009), Korea (Itagaki et al. 2005a), Japan (Itagaki et al. 2005b; Terasaki et al. 1998) and Vietnam (Itagaki et al. 2009; Le et al. 2008) have already been reported. A variety of molecular methods have been applied to the study of F. hepatica populations such as polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of multiple genes (ribosomal-Marcilla et al. 2002; mitochondrial-Walker et al. 2007), random amplified polymorphic DNA (RAPD) variability (Semyenova et al. 2003) and microsatellite markers (Hurtrez-Bousses et al. 2004). SNP assays on rDNA have been used by a number of investigators to resolve the differences in fasciolids at the interspecies level (Marcilla et al. 2002; Semyenova et al. 2005; Vara-Del Rio et al. 2007; Prasad et al. 2008) and for determining diversity within F. hepatica based on variations in mitochondrial DNA (Semyenova et al. 2006; Walker et al. 2007). The 5′ region of 28S rDNA has been used for PCR-RFLP analysis for distinguishing F. hepatica and F. gigantica (Marcilla et al. 2002).
Fasciolosis caused by F. gigantica is common in sheep, cattle and buffaloes in India (Sanyal 2004). While genetic variation of Fasciola isolates from animals and humans is well documented in other parts of the world, no systematic documentation on the molecular characterization of Fasciola spp. is available from India. Gunasekar et al. (2008) elucidated the molecular variability in the DNA fingerprinting pattern of F. gigantica isolates of cattle, buffalo and goats by RAPD-PCR and observed that cattle and buffalo isolates differed from the goat isolate. Another group (Prasad et al. 2008, 2009, 2011) studied ITS sequences of Fasciola species prevalent in the north-eastern region of India. This study was conducted in four different geographical regions of India that showed extensive polymorphism in the ITS-2 sequence of F. gigantica prevalent in these regions.
Materials and methods
Parasites
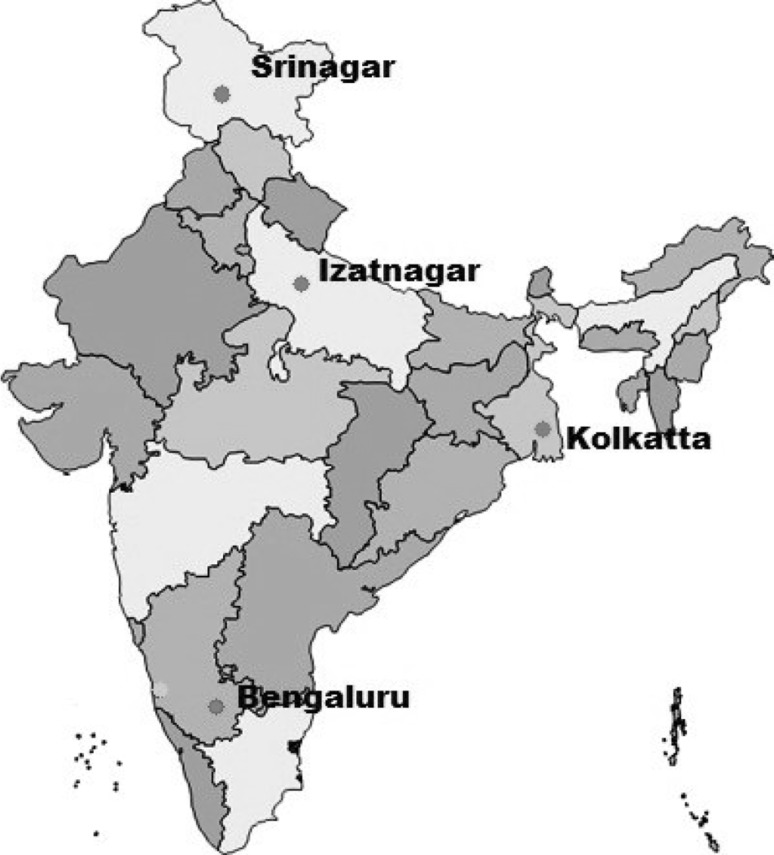
Twenty eight F.gigantica flukes collected from four different geographical regions of India were used in the ITS-2 characterization of the parasite (Fig. 1). These included 10 samples from Srinagar, retrieved exclusively from cattle (Bos taurus) and 18 samples from Bangalore, Izatnagar and Kolkata of cattle (n = 8) and buffalo (Bubalus bubalis) (n = 10) origin. For 28S ribosomal DNA characterization, 24 of the above 28 flukes were used in this study. After washing the flukes with normal saline, they were preserved in 70 % ethanol and stored at −20 °C until used for extraction of DNA.
Fig. 1.
Map of India showing the places where Fasciola specimens from cattle and buffaloes were collected and used in this study
Markers used for identification and genotypic analysis of the parasite
Genetic markers used in this study included sequences of the second internal transcribed spacer (ITS-2) and 28S rDNA (partial sequence), obtained by PCR. ITS-2 PCR products were digested with BspH1 restriction enzyme (New England BioLabs, UK) for generating RFLP pattern for species differentiation and identification of variable genotypes.
Extraction of genomic DNA, amplification, cloning and sequencing
Genomic DNA was extracted from individual adult worms using a commercial genomic DNA extraction kit (Promega Inc., USA) according to the manufacturer’s instructions. Genomic DNA was diluted to a working concentration of 100 ng/μl and 1 μl of this dilution was used as a template in a PCR of 25 μl to amplify the entire nuclear ITS-2 using primers (F:5′-GGTGGATCACTGGGCTCGTG-3′ and R:5′- TATGCTTAAATTCAGCGGGT-3′). The primers were designed on 5.8S and 28S rDNA sequences flanking the ITS-2 region to amplify a 550 bp fragment. A partial sequence of 28S rDNA gene, corresponding to 16-633 bp (618 bp) region, was amplified using forward and reverse primers (5′-ACGTGATTACCCGCTGAACT-3′ and 5′-CTGAGAAAGTGCACTGGCAAG-3′), designed on databank sequence (Accession No. AJ440785). Both the PCR amplified products were purified and cloned in a TA cloning vector (Invitrogen, USA) and recombinant plasmid DNA was subjected to custom sequencing on an automated sequencer using M13 forward and reverse primers.
Data analysis and phylogenetic tree construction
28S rDNA sequences of the F. gigantica flukes were aligned using MEGA 5.01 software (http://www.Megasoftware.net/mega5.01.html). Specific identification of the parasite was made by comparison with available sequences of the corresponding species in GenBank. The phylogenetic tree was obtained by maximum likelihood analysis using 500 bootstrap replicates (Douady et al. 2003). The ITS-2 sequences obtained from the different Indian isolates were aligned with the available ITS-2 sequences in GenBank and analyzed with Gene Tool and DNA Star software.
Results
Genotypic characterization based on the ITS-2 marker
Twenty eight specimens of Fasciola of cattle (n = 18) and buffalo (n = 10) origin, (collected from the above four regions) were sequenced for ITS-2 region. The sequence obtained for these different isolates was found to be identical to the ITS-2 sequence of F. gigantica and it differed from F. hepatica ITS-2 at six positions (207, 231, 270, 276, 327 and 334); where one distinguishing deletion of T nucleotide occurred at the 327th position. The substitution T → C at 207th and 231st, C → T at 270th and 276th and G → A at the 334th position clearly differentiated the species as F. gigantica. However, in addition to the F. gigantica ITS-2 sequence obtained in the above sequence analysis data, ITS-2 sequences with several nucleotide variations were also detected in the same flukes. These sequence variations were initially detected by BspH1 restriction digestion of the ITS-2 PCR products and subsequently confirmed by sequencing of these PCR products.
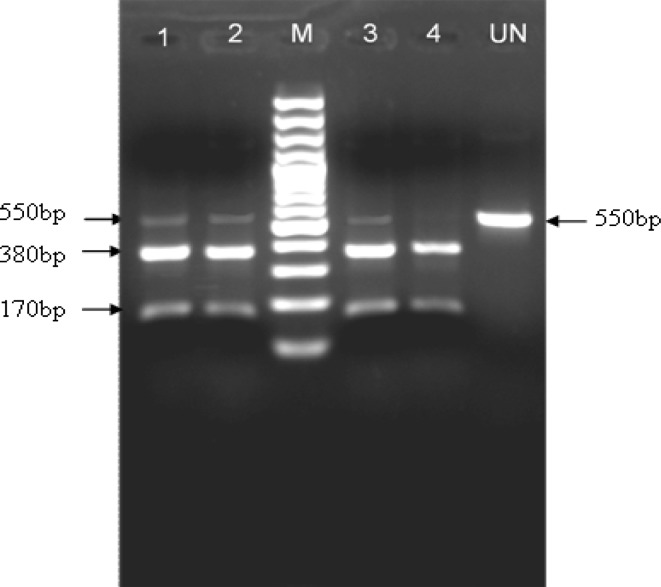
The ITS-2 PCR products amplified from the above Fasciola specimens on BspH1 restriction digestion revealed a restriction pattern of 380 and 170 bp fragments which is typical of F. gigantica. However, in the most of these fluke isolates, digestion of the ITS-2 PCR product with this enzyme produced a restriction profile different from that of F. gigantica and F. hepatica. BspH1 restriction enzyme digested the 550 bp ITS-2 product (flanked by the regions of 5.8S and 28S rDNA) into three fragments of 550, 380 and 170 bp (Fig. 2).The undigested fragment (550 bp but a very thin band, representing ~10 % of the PCR amplified DNA molecules) was excised from the gel and a total 13 of these undigested PCR products (550 bp) obtained from as many flukes were cloned and sequenced. Sequence analysis of these BspH1 un-digested ITS-2 PCR products (n = 13) showed polymorphisms at various nucleotides ranging from 1 to 25. Some of these ITS-2 products were showing one or two nucleotide changes from the typical ITS-2 sequence of F. gigantica while in others an extensive change of 12–25 nucleotides in each sequence was observed, corresponding neither to F. gigantica nor to F. hepatica. These variable ITS-2 sequences also showed length polymorphisms of 359, 362 and 363 bp, varying from the typical 361 bp length of F. gigantica ITS-2. Hence, BspH1 restriction digestion of ITS-2 PCR products could detect mixed sequences of ITS-2 being present in an individual worm, one corresponding to F. gigantica ITS-2 and the other showing variations at several nucleotides not corresponding to F. gigantica or F. hepatica. Interestingly, copy number of the ITS-2 sequence, typical of F. gigantica, was predominant in the ribosomal DNA array in each fluke, with polymorphic forms of the ITS-2 sequences being present in a miniscule copy number (depicted by the thickness of the BspH1 undigested 550 bp fragment). Based on the BspH1 restriction digestion, six variable ITS-2 sequences in F. gigantica were identified in the present study and were found distributed in the flukes from these four geographical regions (sequences deposited with GenBank with Accession Nos. JN541193, JN541194, JN541195, JN541196, JN541197 and JN541198).
Fig. 2.
BspH1 digestion of ITS-2 PCR product of four Indian isolates of F. gigantica; Srinagar (lane1), Bangalore (lane 2), Izatnagar (lane 3) and Kolkata (lane 4). Lane M 100 bp ladder DNA marker and UN undigested ITS-2 product
Genotypic characterization based on the 28S rDNA region
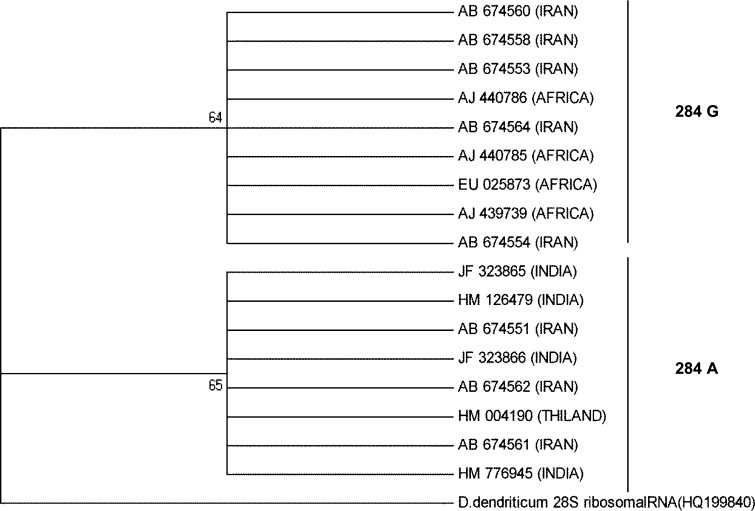
A total of 24 PCR products of 28S rDNA (covering 618 bp of 5′ end of the gene) generated from 6 flukes from each region on sequence analysis revealed one SNP at 284th nucleotide position. There was a transition of G→A at this nucleotide position in all the specimens sequenced. These sequences were aligned with 28S rDNA sequences of F. gigantica isolates available from some African and two Asian countries (Iran and Thailand). Phylogenetic analysis of aligned 28S ribosomal gene sequences (508 bp overlap at the 5′ end) showed two distinct clades (284A and 284G) on the basis of SNP at 284 nucleotide position (Fig. 3). One of the clade (284G) clustered all available African isolates and five Iranian isolates whereas another clade (284A) clustered all Indian, a Thailand and three Iranian isolates. All the Fasciola specimens, collected from four different geographical areas of India, belonged to the 284A clade. The sequence results of 508 bp overlap at the 5′ end of 28S rDNA indicated the existence of two main lineages for 28S ribosomal gene, proposed here as 284A and 284G, in the African and Asian countries (Table 1). The sequences of 28S rDNA of these four isolates were deposited in GenBank with Accession Nos. JF323865; JF323866; HM776945; HM126479.
Fig. 3.
Evolutionary relationship of 28S rDNA partial sequence of F. gigantica isolates of India (Srinagar, Bangalore, Kolkata and Izatnagar) with different isolates of Iran, Thailand and Africa (17 taxa) and one taxon of out group (28S ribosomal RNA sequence of Dicrocoelium spp.) established using maximum likelihood method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown above the branches
Table 1.
GenBank sequences referred to the defined lineages
| Gene | Databank accession number | Country origin | Proposed lineage |
|---|---|---|---|
| 28S rDNA | JF323865 | India | 284 A |
| JF323866 | India | ||
| HM776945 | India | ||
| HM126479 | India | ||
| HM004190 | Thailand | ||
| AB674551 | Iran | ||
| AB674561 | Iran | ||
| AB674562 | Iran | ||
| AB674553 | Iran | 284G | |
| AB674554 | Iran | ||
| AB674558 | Iran | ||
| AB674560 | Iran | ||
| AB674564 | Iran | ||
| EU025873 | Kenya | ||
| AY222245 | Senegal | ||
| AJ440785a | Burkina Faso | ||
| AJ440786 | Damanhour, Egypt | ||
| AJ439739 | Cape Verde, Santiago |
aSequence used as standard
Discussion
ITS-2 and 28S rDNA sequences were used for genetic characterization and identification of the Fasciola species prevalent in four geographical regions of India. Sequence analysis of the ITS-2 of Fasciola isolates of cattle and buffalo showed that mostly two types of ITS-2 sequences were present in an individual fluke in these four studied regions. Flukes showed ITS-2 sequence that was typical of F. gigantica. However, these flukes also revealed existence of ITS-2 sequences in the rDNA array showing extensive polymorphisms, not corresponding to either F. gigantica or F. hepatica. A total of six such variable ITS-2 sequences were found and some of which were showing a change of 12-25 nucleotides from F. hepatica and F. gigantica ITS-2. This limited study for ITS-2 variability conducted on a total of 13 specimens would indicate that there can be more number of polymorphisms prevailing in these ITS-2 sequences, once a larger fluke population is screened. Present study indicated that both pure F. gigantica and Fasciola with mixed ITS-2 genotype prevailed in the domestic ruminants in India. However, none of the flukes collected from these animals possessed ITS-2 sequence of F. hepatica.
Sequence alignment of the 28S rDNA from the available Fasciola isolates of Africa and Asia (Iran, Thailand and India) showed a distinct variation (A/G) at 284th nucleotide, grouping these Fasciola isolates into two distict clades of 284A and 284G. Based on this polymorphism at 284th position of the 28S rDNA, it is proposed that there are two basic lineages of F. gigantica for 28S rDNA that exists in the fluke populations from five African and several Asian countries. Teofanova et al. (2011) proposed existence of 2 lineages for 28S rDNA in F. hepatica in Eastern Europe. These were based on the polymorphism at 105th nucleotide in 28S rDNA in F. hepatica. However, in F. gigantica these lineages are proposed on the existence of variable 284th nucleotide of the 28S rDNA.
Identification of the Fasciola species by morphometry is difficult, as sometimes we encounter morphological forms of the parasite not typical of F. gigantica or F. hepatica. Molecular probes of ribosomal and mitochondrial genomes do resolve the ambiguities arising in the fluke identification by morphometry. Studies in the north-east region of India based on ITS-2 marker have indicated the existence of F. gigantica in the north-eastern part of the country but there are no reports on the polymorphic forms of ITS-2 prevalent in these flukes (Prasad et al. 2009, 2011). Previous studies have indicated existence of only F. gigantica in the plains of India and F. hepatica in the temperate climates of the Himalayan region of the country (Sharma et al. 1989). In an earlier study on classifying Fasciola species to F. hepatica or F. gigantica in India, Varma 1953 created a new species Fasciola indica for the flukes with morphology neither typical of F. gigantica nor F. hepatica. But, there has not been any further confirmation on the existence of F. indica in India. However, existence of this species in India can be scientifically validated, once the Fasciola species prevalent in India are identified based on the use of genetic markers and not on mere morphological parameters.
To conclude, present results indicate that besides pure F. gigantica, F. gigantica with mixed ITS-2 genotype circulates in these parts of the country. Based on the SNP at 284th nucleotide position in the 28S rDNA of F. gigantica, two basic lineages of the F. gigantica for 28S rDNA are proposed to exist in the fluke populations from five African and several Asian countries.
Acknowledgments
The authors are thankful to the Director, Indian Veterinary Research Institute, Izatnagar for providing financial support for this research work. We are also thankful to Mr. Ashok Kumar Mishra for his technical assistance in this work.
References
- Agatsuma T, Arakawa Y, Iwagami M, Honzako Y, Cahyaningsih U, Kang SY, Hong SJ. Molecular evidence of natural hybridization between Fasciola hepatica and F. gigantica. Parasitol Int. 2000;49:231–238. doi: 10.1016/S1383-5769(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ali H, Ai L, Song HQ, Ali S, Lin RQ, Seyni B, Issa G, Zhu XQ. Genetic characterization of Fasciola samples from different host species and geographical localities revealed the existence of F. hepatica and F. gigantica in Niger. Parasitol Res. 2008;102:1021–1024. doi: 10.1007/s00436-007-0870-7. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Valero MA, Panova M, Periago MV, Massoud J, Mas-Coma S. Phenotypic analysis of adults of Fasciola hepatica, Fasciola gigantica and intermediate forms from the endemic region of Gilan, Iran. Parasitol Int. 2006;55:249–260. doi: 10.1016/j.parint.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Douady CJ, Delsuc F, Boucher Y, Doolittle WF, Douzer EJ. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol Biol Evol. 2003;20:248–254. doi: 10.1093/molbev/msg042. [DOI] [PubMed] [Google Scholar]
- Gunasekar KR, Tewari AK, Sreekumar C, Gupta SC, Rao JR. Elucidation of genetic variability among different isolates of Fasciola gigantica (giant liver fluke) using random-amplified polymorphic DNA-polymerase chain reaction. Parasitol Res. 2008;103:1075–1081. doi: 10.1007/s00436-008-1095-0. [DOI] [PubMed] [Google Scholar]
- Huang WY, He B, Wang CR, Zhu XQ. Characterization of Fasciola species from Mainland China by ITS-2 ribosomal DNA sequence. Vet Parasitol. 2004;120:75–83. doi: 10.1016/j.vetpar.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Hurtrez-Bousses S, Durand P, Jabbour-Zahab R, Guegan JF, Meunier C, Bargues MD, Mas-Coma S, Renaud F. Isolation and characterization of microsatellite markers in the liver fluke (Fasciola hepatica) Mol Ecol Notes. 2004;4:689–690. doi: 10.1111/j.1471-8286.2004.00786.x. [DOI] [Google Scholar]
- Itagaki T, Kikawa M, Terasaki K, Shibahara T, Fukuda K. Molecular characterization of parthenogenic Fasciola sp. in Korea on the basis of DNA sequences of ribosomal ITS1 and mitochondrial NDI gene. J Med Vet Sci. 2005;67:1115–1118. doi: 10.1292/jvms.67.1115. [DOI] [PubMed] [Google Scholar]
- Itagaki T, Kikawa M, Sakaguchi K, Shimo J, Terasaki K, Shibahara T, Fukuda K. Genetic characterization of parthenogenic Fasciola sp. in Japan on the basis of the sequences of ribosomal and mitochondrial DNA. Parasitology. 2005;131:679–685. doi: 10.1017/S0031182005008292. [DOI] [PubMed] [Google Scholar]
- Itagaki T, Sakaguchi K, Terasaki K, Sasaki O, Yoshihara S, Van Dung T. Occurrence of spermic diploid and aspermic triploid forms of Fasciola in Vietnam and their molecular characterization based on nuclear and mitochondrial DNA. Parasitol Int. 2009;58:81–85. doi: 10.1016/j.parint.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Le TH, De NV, Agatsuma T, Nguyen TGT, Nguyen QD, McManus DP, Blair D. Human fascioliasis and the presence of hybrid/introgressed forms of Fasciola in Vietnam. Int J Parasitol. 2008;38:725–730. doi: 10.1016/j.ijpara.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Lin RQ, Dong SJ, Nie K, Wang CR, Song HQ, Li AX, Huang WY, Zhu XQ. Sequence analysis of the first internal transcribed spacer of rDNA supports the existence of the intermediate Fasciola between F. hepatica and F.gigantica in mainland China. Parasitol Res. 2007;101:813–817. doi: 10.1007/s00436-007-0512-0. [DOI] [PubMed] [Google Scholar]
- Marcilla A, Bargues MD, Mas-Coma S. A PCR-RFLP assay for the distinction between Fasciola hepatica and Fasciola gigantica. Mol Cel Probes. 2002;16:327–333. doi: 10.1006/mcpr.2002.0429. [DOI] [PubMed] [Google Scholar]
- Peng M, Ichinomiya M, Ohtori M, Ichikawa M, Shibahara T, Itagaki T. Molecular characterization of Fasciola hepatica, Fasciola gigantica, and aspermic Fasciola sp. in China based on nuclear and mitochondrial DNA. Parasitol Res. 2009;105:809–815. doi: 10.1007/s00436-009-1459-0. [DOI] [PubMed] [Google Scholar]
- Prasad PK, Tandon V, Biswal DK, Goswami LM, Chatterjee A. Molecular identification of the Indian liver fluke, Fasciola (Trematoda: Fasciolidae) based on the ribosomal internal transcribed spacer regions. Parasitol Res. 2008;103:1247–1255. doi: 10.1007/s00436-008-1121-2. [DOI] [PubMed] [Google Scholar]
- Prasad PK, Tandon V, Biswal DK, Goswami LM, Chatterjee A. Use of sequence motifs as barcodes and secondary structures of internal transcribed spacer 2 (ITS2, rDNA) for identification of the Indian liver fluke, Fasciola (Trematoda: Fasciolidae) Bioinformation. 2009;3:314–320. doi: 10.6026/97320630003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad PK, Goswami LM, Tandon V, Chatterjee A. PCR-based molecular characterization and in silico analysis of food-borne trematode parasites Paragonimus westermani, Fasciolopsis buski and Fasciola gigantica from Northeast India using ITS2 rDNA. Bioinformation. 2011;6:64–68. doi: 10.6026/97320630006064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal PK. Control of tropical fasciolosis in cattle and buffaloes in India at the backdrop of its integrated management. J Vet Parasitol. 2004;15:13–16. [Google Scholar]
- Semyenova SK, Morozova EV, Chrisanfova GG, Asatrian AM, Movsessian SO, Ryskov AP. RAPD variability and genetic diversity in two populations of liver fluke, Fasciola hepatica. Acta Parasitol. 2003;48:125–130. [Google Scholar]
- Semyenova SK, Morozova EV, Vasilyev VA, Gorokhov VV, Moskvin S, Movsessyan SO, Ryskov AP. Polymorphism of internal transcribed spacer 2 (ITS-2) sequences and genetic relationships between Fasciola hepatica and F. gigantica. Acta Parasitol. 2005;50:240–243. [Google Scholar]
- Semyenova SK, Morozova EV, Chrisanfova GG, Gorokhov VV, Arkhipov IA, Moskvin AS, Movsessyan SO, Ryskov AP. Genetic differentiation in Eastern European and Western Asian populations of the liver fluke, Fasciolahepatica, as revealed by mitochondrial nad1 and cox1 genes. J Parasitol. 2006;92:525–530. doi: 10.1645/GE-673R.1. [DOI] [PubMed] [Google Scholar]
- Sharma RL, Dhar DN, Raina OK. Studies on the prevalence and laboratory transmission of fascioliasis in animals in the Kashmir Valley. Brit Vet J. 1989;145:57–61. doi: 10.1016/0007-1935(89)90010-9. [DOI] [PubMed] [Google Scholar]
- Teofanova D, Kantzoura V, Walker S, Radoslavov G, Hristov P, Theodoropoulos G, Bankov I, Trudgett A. Genetic diversity of liver flukes (Fasciola hepatica) from Eastern Europe. Infect Genet Evol. 2011;11:109–115. doi: 10.1016/j.meegid.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Terasaki K, Moriyama-Gonda N, Noda Y. Abnormal spermatogenesis in the common liver fluke (Fasciola sp.) from Japan and Korea. J Vet Med Sci. 1998;60:1305–1309. doi: 10.1292/jvms.60.1305. [DOI] [PubMed] [Google Scholar]
- Vara-Del Rıo MP, Villa H, Martinez-Valladares M, Rojo-Vazquez FA. Genetic heterogeneity of Fasciola hepatica isolates in the northwest of Spain. Parasitol Res. 2007;101:1003–1006. doi: 10.1007/s00436-007-0574-z. [DOI] [PubMed] [Google Scholar]
- Varma AK. On Fasciola indica n.sp. with some observations on Fasciola hepatica and Fasciola gigantica. J Helminth. 1953;27:185–198. doi: 10.1017/S0022149X00026456. [DOI] [Google Scholar]
- Walker SM, Prodohl PA, Fletcher HL, Hanna REB, Kantzoura V, Hoey EM, Trudgett A. Evidence for multiple mitochondrial lineages of Fasciola hepatica (liver fluke) within infrapopulations from cattle and sheep. Parasitol Res. 2007;101:117–125. doi: 10.1007/s00436-006-0440-4. [DOI] [PubMed] [Google Scholar]