Abstract
Two C57BL/6 mice colonies maintained in two rooms of the same specific pathogen-free (SPF) facility were found to have different gut microbiota and a mucus phenotype that was specific for each colony. The thickness and growth of the colon mucus were similar in the two colonies. However, one colony had mucus that was impenetrable to bacteria or beads the size of bacteria—which is comparable to what we observed in free-living wild mice—whereas the other colony had an inner mucus layer penetrable to bacteria and beads. The different properties of the mucus depended on the microbiota, as they were transmissible by transfer of caecal microbiota to germ-free mice. Mice with an impenetrable mucus layer had increased amounts of Erysipelotrichi, whereas mice with a penetrable mucus layer had higher levels of Proteobacteria and TM7 bacteria in the distal colon mucus. Thus, our study shows that bacteria and their community structure affect mucus barrier properties in ways that can have implications for health and disease. It also highlights that genetically identical animals housed in the same facility can have rather distinct microbiotas and barrier structures.
Keywords: Bacteria, Colon, Intestine, MUC2, Mucus
See also: H Li et al (February2015)
Introduction
The distal small intestine and the large intestine are the reservoirs for an enormous and complex community of micro-organisms, the gut microbiota. The density of intestinal microbes forms a gradient along the intestine with few bacteria in the upper small intestine and up to 1012 bacteria per gram of faeces in distal colon 1. The microbiota is diverse and typically made up of in total 500–1,000 species with at least 160 species that are shared among individuals with two phyla, the Bacteroidetes and Firmicutes being dominant 2, 3. Less abundant phyla are Proteobacteria, Verrucomicrobia, Actinobacteria, Fusobacteria and Cyanobacteria. We have co-evolved with our microbiota and developed a finely tuned co-existence, with mutual benefits and ways to avoid harmful effects on the host. The microbiota, for example, aids in food digestion and development of the immune system 4. Together with the mucus layers, a stable microbiota prevents pathogenic bacteria from establishing host infections 5, 6.
Mucus covers the intestinal epithelium, but the principle of this mucus protection is solved differently along the intestinal tract 7. The mucus in the small intestine fills up the space between the villi and covers these, but is not attached to the epithelium and has a structure that can allow particles as large as bacteria to penetrate 8. The mucus protection in this site acts as a diffusion barrier with a high concentration of antibacterial products close to the epithelium and few bacteria reaching near the cell surface 9, 10. The small intestinal mucus not only excludes the microbiota, but is also important for immune system development 11, 12, 13. The higher bacterial load in colon and the slow transit time requires a different protective strategy. Here, mucus forms a physical barrier separating bacteria from the host, which is especially important and developed in the distal colon. The secreted mucus in distal colon forms a stratified inner layer of mucus that is able to keep the bacteria at a distance (> 50 μm in mice, > 200 μm in human) from the epithelium 14, 15. The mucus is structurally built around the MUC2 mucin that after secretion from the goblet cell expands dramatically and forms sheets of MUC2 networks that stack and form the stratified mucus layer 16. Failure of this protective barrier to separate bacteria from the epithelium is observed in a number of murine colitis models 15, 17. Muc2-deficient mice totally lack a protective inner mucus layer and have direct contact of bacteria with the epithelium and these mice develop severe colitis 14. Mice with mutations in the Muc2 gene also spontaneously develop inflammation 18. Deficiencies in decorating the mucus with O-glycans cause a faster degradation of the Muc2 mucin network resulting in increased bacterial contact with the cell surface and development of spontaneous colitis 19.
The mucus protection system observed in mouse is also present in human colon where an even thicker inner mucus layer separates bacteria from the tissue 15. Ulcerative colitis (UC) patients with active disease have bacteria in contact with the epithelium, whereas patients with UC in remission show a more mixed picture 15. The possibility that the microbiota stimulates the host to develop a mucus layer with improved protective properties has not been explored.
We have studied the colon mucus barrier in different wild-type C57BL/6 mouse colonies housed and bred separately in the same specific pathogen-free (SPF) mouse facility. The mucus phenotypes were compared with wild normally free-living mice caught in their natural habitat to elucidate the protective properties of natural occurring mucus. The bacterial composition in the two mouse colonies was analysed by 16S rRNA gene sequencing, which revealed specific bacterial compositions in the two colonies. Bacterial transmission of the mucus phenotypes was shown by colonizing germ-free animals with the flora from the two colonies.
Results
Bacterial composition in the two husbandries
In the same SPF animal facility, two different C57BL/6 colonies have been bred in different rooms. These two C57BL/6 colonies were named as Room 1 and Room 2, where our previous results on mucus properties have been based on the strain in Room 1, which was derived from Taconic 14. The two colonies were maintained on different diets with different composition, but we controlled for the impact of diet, at least within one generation of offspring, by feeding the mice in the two colonies the same chow (Food A, standard chow or Food B, autoclaved chow) from weaning. On Food A, the mice bred in Room 1 were significantly heavier compared to mice bred in Room 2 (P = 0.022, Supplementary Fig S1A). The stool of mice fed Food A was hard and formed distinct pellets, while the stool in animals fed Food B had a looser consistency and did not form hard pellets.
The microbiota composition of mice in the two rooms was analysed by 16S rRNA gene sequencing of bacteria in the lumen of ileum, distal colon (called lumen) and caecum as well as bound to the mucus on the tissue after washing the ileum and distal colon (called mucus). The sequencing data have been deposited to the European Nucleotide Archive (ENA) with accession number PRJEB7982. The mucus bacteria thus reflected the bacteria either attached to the outer border of the inner colon mucus layer, within the inner colon mucus in colon, or attached to the epithelial cells of the small intestine. Analysing the data using QIIME revealed that the microbiota differed between Room 1 and Room 2. Principal Coordinates Unweighted UniFrac analysis 20 showed separation of ileum and colon samples along the first component, whereas the two rooms clearly separated along PC2 on Food A (Fig1A and B). The difference remained also when mice were given Food B (Supplementary Fig S1). When bacterial diversity was studied (Supplementary Figs S2 and S3), the ileum showed the lowest diversity and the distal colon the highest. The microbiota throughout the gut thus clearly differed between the two colonies housed in adjacent rooms in the same animal facility.
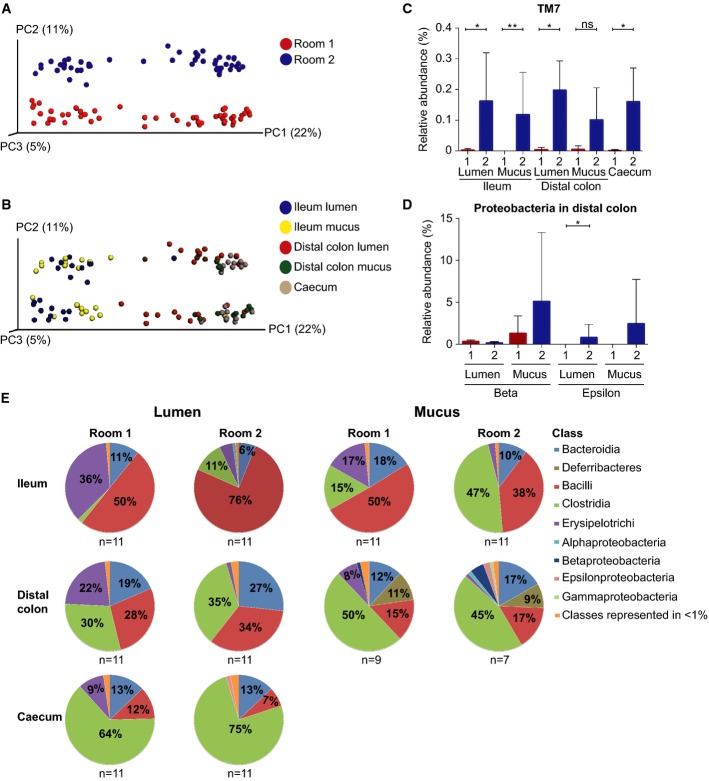
Figure 1.
- Unweighted UniFrac-based PCoA plot of gut microbial communities from mice in Room 1 (red) and Room 2 (blue) at all intestinal locations.
- Unweighted UniFrac-based PCoA plot of mice gut microbial communities from both rooms at different gut segments; ileum lumen (blue), ileum mucus (yellow), distal colon lumen (red), distal colon mucus (green) and caecum (brown) (data as in A).
- The relative abundance (%) of the phylum TM7 in ileum lumen Room 1 (n = 11) and Room 2 (n = 11), ileum mucus Room 1 (n = 11) and Room 2 (n = 11), distal colon lumen Room 1 (n = 11) and Room 2 (n = 11), distal colon mucus Room 1 (n = 9) and Room 2 (n = 7), and caecum Room 1 (n = 11) and Room 2 (n = 11).
- The relative abundance (%) of the phylum Proteobacteria (beta and epsilon) in distal colon lumen and mucus in Room 1 (n = 11 in lumen and n = 9 in mucus) and Room 2 (n = 11 in lumen and n = 7 in mucus).
- Class level microbiota composition in mice from Room 1 and Room 2. The mean relative abundance (%) of the most abundant bacterial classes in ileum lumen Room 1 (n = 11) and Room 2 (n = 11), ileum mucus Room 1 (n = 11) and Room 2 (n = 11), distal colon lumen Room 1 (n = 11) and Room 2 (n = 11), distal colon mucus Room 1 (n = 9) and Room 2 (n = 7), and caecum Room 1 (n = 11) and Room 2 (n = 11). Classes represented as less than 1%: Other, DA052, Actinobacteria, Coriobacteriia, Cytophagia, Flavobacteriia, Sphingobacteriia, (Saprospirae), 4C0d-2, Chloroplast, Deferribacteres, ZB2, Alphaproteobacteria, Betaproteobacteria, Epsilonproteobacteria, Deltaproteobacteria, Gammaproteobacteria, TM7-3, Mollicutes, Verrucomicrobiae.
Data information: In (C) and (D), data are presented as mean ± SD. Wilcoxon's rank-sum test was conducted to compare over- or under-representation of the phylum. To correct for multiple testing, the P-values were converted to false discovery rate values (Q-values). *P < 0.001, Q < 0.001, **P < 0.001, Q < 0.01.
Source data are available online for this figure.
The different microbiota was reflected in significant differences at multiple taxonomical levels (Table1). However, the relative amounts of the dominant phyla (Firmicutes and Bacteroidetes) did not significantly differ between the two rooms at any location throughout the gut (Supplementary Fig S4, Table1). The major difference between the lumen- and mucus-associated microbiota was found in the distal colon where the phyla Deferribacteres had higher abundance in both rooms (Supplementary Fig S4, Table1).
Table 1.
The mean of the relative abundance of dominant phyla, classes and genera in ileum, distal colon, and caecum samples obtained from mice bred in Room 1 or Room 2
| Ileum lumen | Ileum mucus | Distal colon lumen | Distal colon mucus | Caecum | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Room 1 | Room 2 | Room 1 | Room 2 | Room 1 | Room 2 | Room 1 | Room 2 | Room 1 | Room 2 | |
| n = 11 | n = 11 | n = 11 | n = 11 | n = 11 | n = 11 | n = 9 | n = 7 | n = 11 | n = 11 | |
| mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | mean % (SD) | |
| Firmicutes | 88 (6) | 92 (6) | 81 (9) | 88 (9) | 80 (10) | 71 (8) | 73 (11) | 62 (24) | 85 (3) | 83 (3) |
| Bacilli | 50 (19) | 76 (15) | 50 (23) | 38 (24) | 28 (12) | 34 (19) | 15 (11) | 17 (14) | 12 (5) | 7 (4) |
| Lactobacillus | 50 (19) | 75 (15) | 50 (21) | 38 (24) | 27 (12) | 34 (19) | 15 (11) | 16 (9) | 12 (5) | 7 (4) |
| Clostridia | 2 (1) | 11 (11) | 15 (23)** | 47 (28)** | 30 (20) | 35 (22) | 50 (22) | 45 (28) | 64 (11)** | 75 (5)** |
| Unknown genus (family unknown) | < 1 | < 1 | 1 (2) | < 1 | 10 (9) | 12 (14) | 18 (10) | 16 (17) | 27 (13) | 29 (19) |
| Other | < 1 | < 1 | 2 (4) | < 1 | 10 (8) | 9 (6) | 18 (11) | 13 (14) | 21 (7) | 19 (16) |
| Ruminococcus | 0.1 (0.01)*** | 0.3 (0.2)*** | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 0.7 (0.3)*** | 1 (1)*** |
| Candidatus arthromitus (Segmented filamentous bacteria) | 0.04 (0.12)*** | 1 (2)*** | 11 (23) | 43 (29) | 0.01 (0.02)*** | 0.2 (0.3)*** | 0.03 (0.1)*** | 1 (1)*** | < 1 | < 1 |
| Anaerostipes | 0.06 (0.05)** | –** | 0.2 (0.2)** | –** | 2 (2)** | < 1** | 0.8 (0.8)*** | –*** | 0.8 (0.6)** | –** |
| Dehalobacterium | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 0.2 (0.1)** | 0.6 (0.2)** |
| Erysipelotrichi | 36 (16)* | 5 (6)* | 17 (13)** | 3 (3)** | 22 (14)* | 2 (2)* | 8 (7) | 1 (1) | 9 (7)* | 0.6 (0.7)* |
| Allobaculum | 35 (17)** | 5 (6)** | 15 (13) | 3 (3) | 22 (14)** | 2 (2)** | 7 (7) | 1 (1) | 9 (7)** | 0.4 (0.4)** |
| Other | 1 (1)** | –** | < 1 | < 1 | 0.5 (0.8)*** | 0.003 (0.01)*** | 0.2 (0.3)*** | –*** | 0.2 (0.2)** | –** |
| Bacteroidetes | 11 (6) | 6 (5) | 18 (9) | 10 (8) | 19 (10) | 27 (8) | 12 (6) | 17 (14) | 13 (4) | 13 (3) |
| Bacteroidia | 11 (5) | 6 (5) | 18 (9) | 10 (8) | 19 (10) | 27 (8) | 12 (6) | 17 (14) | 13 (4) | 13 (3) |
| Unknown genus (family S24-7) | 11 (6)** | 6 (5)** | 16 (10) | 10 (8) | 15 (9) | 14 (6) | 9 (5) | 10 (10) | 8 (2)** | 5 (1)** |
| Bacteroides | < 1 | < 1 | < 1 | < 1 | 0.4 (0.4)** | 4 (3)** | < 1 | 3 (2) | 0.4 (0.2)** | 2 (1)** |
| Parabacteroides | < 1 | < 1 | < 1 | < 1 | 0.3 (0.3)** | 4 (4)** | < 1 | 2 (2) | 0.3 (0.2)** | 2 (2)** |
| Prevotella | – | < 1 | < 1 | – | 0.01 (0.01)** | 0.3 (0.4)** | < 1 | < 1 | –** | 0.1 (0.1)** |
| Odoribacter | – | – | < 1 | – | < 1 | < 1 | < 1 | < 1 | 0.4 (0.3)** | 0.6 (0.5)** |
| Proteobacteria | 0.1 (0.1)* | 1 (1)* | < 1 | 1 (1) | < 1 | 1 (2) | 3 (3) | 10 (16) | 0.7 (0.4)*** | 2 (2)*** |
| Alphaproteobacteria | – | < 1 | < 1 | < 1 | – | < 1 | < 1 | 1 (2) | < 1 | < 1 |
| Betaproteobacteria | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 1 (2) | 5 (8) | 0.2 (0.001)** | 0.1 (0.04)** |
| Deltaproteobacteria | 0.003 (0.01)* | 1 (1)* | 0.021 (0.028)* | 0.5 (0.5)* | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 |
| Desulfovibrio | –** | 1 (1)** | 0.003 (0.006)** | 0.5 (0.5)** | –** | 0.2 (0.2)** | 0.002 (0.005)*** | 0.3 (0.2)*** | 0.002 (0.005)** | 0.6 (0.3)** |
| Epsilonproteobacteria | – | < 1 | – | < 1 | –* | 0.02 (0.1)* | < 1 | 2 (5) | –** | 2 (2)** |
| Unknown genus (family Helicobacteraceae) | – | < 1 | < 1 | < 1 | –*** | 1 (1)*** | 0.002 (0.006) | 2 (5) | –*** | 2 (2)*** |
| Gammaproteobacteria | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 1 (1) | < 1 | < 1 |
| Actinobacteria | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 0.4 (0.2)** | 0.2 (0.1)** |
| TM7 | 0.002 (0.006)* | 0.2 (0.2)* | –** | 0.12 (0.14)** | 0.003 (0.01)* | 0.2 (0.1)* | < 1 | < 1 | 0.001 (0.003)* | 0.2 (0.1)* |
| Deferribacteres | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 11 (11) | 9 (10) | < 1 | < 1 |
| Deferribacteres | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 11 (11) | 9 (10) | < 1 | < 1 |
| Mucispirillum | < 1 | < 1 | < 1 | < 1 | < 1 | < 1 | 11 (11) | 9 (10) | < 1 | < 1 |
Statistical tests of over-or under-representation of bacterial lineages among sample groups (Room 1 versus Room 2) at each sample location were made at the phylum (bold), class (bold) and genus levels using Wilcoxon's rank-sum test. To correct for multiple testing, the P-values were converted to false discovery rate values (Q-values). (–) corresponds to no sequences present. For values < 1, the exact value is only given if significantly different.
P-value < 0.001, Q-value < 0.001,
P-value <0.001, Q-value < 0.01
P-value < 0.01, Q-value< 0.05.
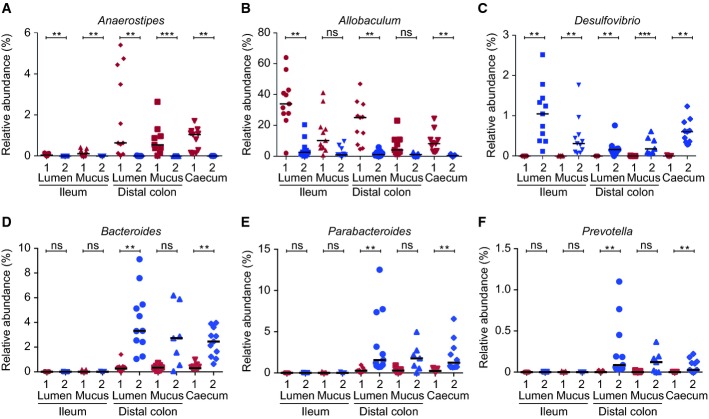
Comparing mice in the two rooms at the class and genus levels revealed pronounced differences (Food A, Figs1D–E and 2A–F, Table1). In Room 1, the genus Anaerostipes, within the Clostridia class, was found in significantly higher amounts at all locations throughout the gut (Fig2A, Table1). The Erysipelotrichi class was also found in higher abundances at all locations in Room 1 (Fig1E, Table1). The dominant genus within this class contributing to this difference was the genus Allobaculum, which was also found in higher relative abundances at all locations and significantly so in the lumen samples (Fig2B, Table1). Within the Bacteroidia class, an unknown genus (family S24-7) was found at significantly higher levels in Room 1 in both ileum lumen and caecum (Table1).
Figure 2.
- The relative abundance (%) of the genera Anaerostipes (A), Allobaculum (B), Desulfovibrio (C), Bacteroides (D), Parabacteroides (E) and Prevotella (F) in ileum lumen Room 1 (n = 11) and Room 2 (n = 11), ileum mucus Room 1 (n = 11) and Room 2 (n = 11), distal colon lumen Room 1 (n = 11) and Room 2 (n = 11), distal colon mucus Room 1 (n = 9) and Room 2 (n = 7), and caecum Room 1 (n = 11) and Room 2 (n = 11). Data are presented as mean ± SD. Wilcoxon's rank-sum test was conducted to compare over- or under-representation of the different genera. To correct for multiple testing, the P-values were converted to false discovery rate values (Q-values). *P < 0.001, Q < 0.001, **P < 0.001, Q < 0.01, ***P < 0.01, Q < 0.05.
Source data are available online for this figure.
In Room 2, the phylum TM7 was a small component but had a higher relative abundance at all locations (Fig1C, Table1). Proteobacteria was also generally more abundant in Room 2 (Supplementary Fig S4, Table1). Within this phylum, the classes Betaproteobacteria and Epsilonproteobacteria were found at higher levels in the mucus of mice in Room 2 and Epsilonproteobacteria were significantly more abundant in distal colon lumen and caecum (Fig1D, Table1). Within this class, an unknown genus of the Helicobacteraceae family was found at higher levels in Room 2 (Table1). Within the Deltaproteobacteria, one genus, Desulfovibrio, was found in significantly higher amounts at all locations of mice in Room 2 (Fig2C, Table1). The Clostridia class was significantly more abundant in ileum mucus and caecum samples of Room 2 (Fig1E, Table1). In ileum mucus, the dominant genus within the Clostridia class was Candidatus arthromitus (segmented filamentous bacteria, SFB), which was also found in higher levels in Room 2 (Table1). SFB also showed a small but significant difference between the rooms in the distal colon with higher levels in Room 2.
Also, within the Bacteroidia class, the genera Bacteroides, Parabacteroides, and Prevotella were found in significantly higher abundances in distal colon lumen and caecum samples in Room 2 (Fig2D–F, Table1).
The two diets used were tested by analysing animals in both locations also fed with Food B diet (autoclaved). Altering the diet from weaning gave some shift in the microbiota, but less than the difference between the two rooms. All mice on Food B showed higher relative abundances of the phylum Proteobacteria at all locations along the gut and in both rooms with significant differences in ileum lumen, ileum mucus and caecum in Room 1, and in distal colon lumen in both rooms (Supplementary Fig S5, Supplementary Table S1). The phylum TM7 was also found at higher levels at all gut locations and rooms when mice were fed Food B (Supplementary Table S1). The Erysipelotrichi class and the genus Allobaculum was found in higher relative abundances in ileum and distal colon, as well as in caecum when the mice were fed Food B in Room 1 (Supplementary Table S1).
It was thus concluded that the two C57BL/6 mice strains bred separately had stable, but different microbiota that were vertically transmitted. Food influenced the microbiota to a small extent. This raised the question of the effect of the different microbiota on the mucus thickness and properties.
Differences in the mucus quality in mice from the two husbandries
The thickness of the secreted mucus from mice in the two rooms was measured on small intestinal and distal colon explants mounted in a perfusion chamber after visualization of the mucus surface by charcoal 21. The small intestinal mucus is normally easily removed by aspiration, and the thickness was measured before (pre) and after (post) aspiration. No differences were observed between the rooms or diets (Fig3A). In colon, the mucus forms two layers; an outer non-attached and an inner mucus layer that is firmly attached to the epithelium 14. The thickness of the inner mucus layer as well as the mucus growth did not differ significantly between the two rooms on either food (Fig3B).
Figure 3.
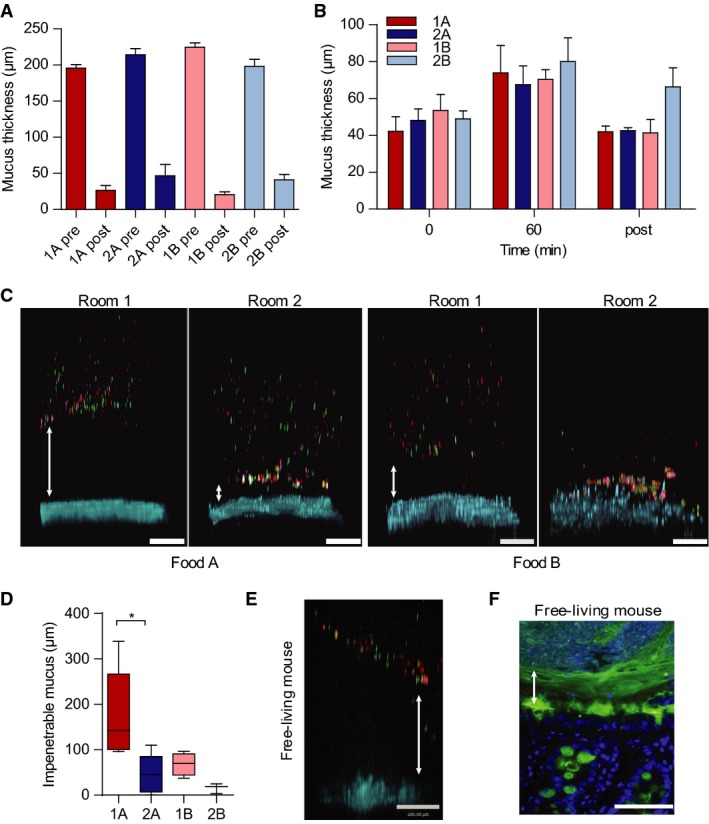
- Mucus thickness on explants from distal small intestine (pre) and mucus remaining after aspiration (post) in the different rooms (Room 1 and 2) with the different foods (Food A and Food B). Data are presented as mean ± SEM (n = 3).
- Mucus thickness measured during one hour on distal colon explants from mice in the two rooms (Room 1 and 2) with different foods (Food A and Food B). The mucus not attached to the epithelium was removed after 60 min and the attached mucus was measured (post). Data are presented as mean ± SEM (n = 3).
- Penetrability of beads (red 0.5 μm, purple 1 μm, green 2 μm) in mucus on explants from mice housed in Room 1 or Room 2 with either Food A or Food B. The epithelium is stained by Calcein violet (blue). Double arrows indicate impenetrable mucus. Scale bar is 100 μm.
- The impenetrable mucus as the mean distance of the 20 most penetrable beads shows a significant difference between the mice in the two rooms with Food A. Data are presented as boxplots with median and min–max whiskers. Mann–Whitney U-tests were performed to compare groups (Room 1 and 2) on each food (Food A: n = 5, Food B: n = 3–4, *P = 0.032).
- Mucus penetrability as in (C) on explants from free-living mice. Double arrow indicates impenetrable mucus. Scale bar is 100 μm.
- Immunostaining for Muc2 (green) of distal colon sections from free-living mice with Hoechst counterstain (blue). Double arrow indicates inner mucus layer without bacteria (visualized by DNA stain). Scale bar is 50 μm.
Source data are available online for this figure.
The size exclusion properties of the mucus can be assessed by allowing fluorescent beads, the size of bacteria, to sediment through the mucus formed on colonic explants 15. Using this system, the secreted mucus has previously been shown to separate the beads from the epithelium in both mouse and human biopsies. As shown before, mice from Room 1 secreted mucus that was impenetrable to the beads, whereas the mucus on explants from mice housed in Room 2 was more penetrable (Fig3C). Mice given food B showed increased penetrability in both groups, but a difference still remained between Room 1 and 2 mice (Fig3C). The penetrability of beads was estimated as the distance from the epithelium to the 20 closest located beads (Fig3D). This showed significantly more penetrable mucus with a thinner, impervious part of the inner mucus layer in Room 2.
Mice in Room 2 thus differed in mucus properties compared to mice in Room 1 that we had assumed to have a normal mucus layer. This raised doubts on what is a normal mucus layer. To address this, mice caught in their wild habitat were studied with the same methods as for mice from the two rooms. In free-living mice, we could typically observe a non-penetrable inner colon mucus layer that separated the beads from the epithelium even further than what was observed in mice from Room 1 (Fig3E). Tissue staining of distal colon tissue could also show bacteria well separated from the epithelium (Fig3F). Thus, free-living mice have mucus that is even better developed, thicker and more stratified with bacteria further separated from the epithelium than in Room 1.
Bacteria penetration in mucus and histology of mice in different husbandries
Muc2 mucin immunohistochemistry revealed that the mucus in fixed sections looked different in mice from Room 2 on Food A (Fig4A). When bacteria were localized in relation to the colon mucus using fluorescent in situ hybridization, bacteria penetrated the inner mucus layer at distinct locations. A separation between epithelium and bacteria was observed in other areas and most bacteria were still not in direct contact with the epithelial surface. A good separation of bacteria and epithelium by a stratified inner mucus layer was only present in mice from Room 1 (Fig4A). The inner mucus layer was after fixation observed to be thinner, and stratification was not evident in sections from animals fed Food B in both rooms, but the difference between the two rooms remained (Fig4B).
Figure 4.
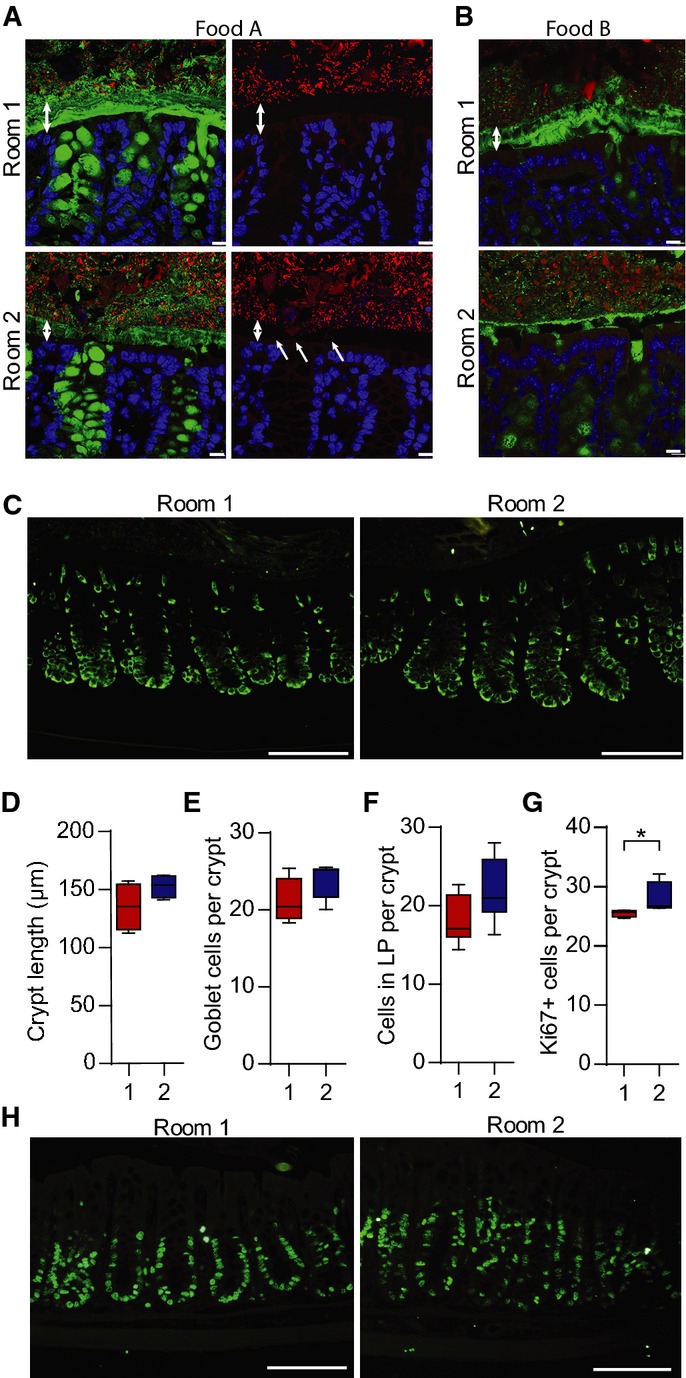
- Immunostaining of colon sections from mice in the two rooms with Food A (A) or Food B (B) using Anti-MUC2C3 (green) and FISH with a general bacterial 16S rRNA gene probe to detect bacteria (red) counterstained with Hoechst (blue). Double arrows mark the inner mucus layer that separates bacteria from the epithelium. Bacteria are found within the inner mucus layer (indicated by arrows) in mice housed in Room 2. Scale bar is 10 μm.
- Immunostaining of the Muc2 precursor before it gets O-glycosylated using the anti-apoMuc2 antiserum on mice from both rooms fed Food A. Scale bar is 100 μm.
- The crypt length was measured in distal colon on sections from mice in the two rooms on Food A (n = 4)
- The number of goblet cells per crypt was counted in distal colon of mice in the two rooms on Food A (n = 10).
- The number of cells in lamina propria between two crypts, as shown in Supplementary Fig S7, was counted in sections from mice in the two rooms on Food A (n = 10).
- The number of Ki67-positive cells per crypt was significantly different in distal colon of mice housed in Room 1 and Room 2 on Food A. Comparison of groups was performed using the Mann–Whitney U-test (n = 4, *P = 0.029).
- Ki67 immunostaining (green) on colonic sections from mice in Room 1 or Room 2 on Food A. Scale bars are 100μm.
Data information: In (D–G), data are presented as boxplots with median and min–max whiskers.
Source data are available online for this figure.
To analyse the reasons for the different mucus properties, proteomic analyses were performed showing no major differences between the rooms and a high correlation of the proteins identified in the two rooms (Supplementary Fig S6D and E). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD001479. Similar amounts were observed for major known mucus components such as Muc2, Fcgbp and Clca1 as well as for potential mucus processing proteins such as meprin β (Mep1b) and transglutaminase 3 (Tgase3) (Supplementary Fig S6A–C). Minor components involved in the mucosal immune defence systems such as defensin 20 (Defa20) and Immunoglobulin J-chain (IgJ) did not show significant differences (Supplementary Fig S6A and B). Inflammatory markers as S100a8, S100a9 and eosinophil cationic protein 3 were not detected and no difference in albumin or alpha 2-macroglobulin was found between the groups. The protein differences were small and not likely to explain the variable mucus properties.
The Muc2 mucin biosynthesis was assessed by staining the intracellular ER-localized non-glycosylated Muc2 precursor in mice from the two rooms on Food A. No differences were observed, suggesting that the level of Muc2 translation was similar in the two rooms (Fig4C). A tendency towards longer crypts in mice from Room 2 was also observed, but this did not reach significance (Fig4D). The number of goblet cells per crypt was counted, but no significant difference was reproducibly observed (Fig4E). As elongated colon crypts are often observed in mouse colitis models, the number of infiltrating immune cells in lamina propria was also counted. The number of these cells was higher in the Room 2 animals; however, it did not reach significance (Fig4G and Supplementary Fig S7A), but there was no major difference in the overall histology between the groups (Supplementary Fig S7B and C). To further address this difference, proliferative crypt cells were stained with the Ki67 antibody for dividing cells. The labelled cells were significantly increased in mice from Room 2 as compared to Room 1 (Fig4F and H). These findings did not demonstrate any typical colitis in any of the mouse colonies, but suggested that the gut microbiota may modulate colonic inflammation and cell proliferation in a relatively subtle way.
The two different mucus phenotypes were reproduced by colonization of germ-free mice
To test whether the altered microbiota was sufficient to explain the observed differences in the mucus phenotype in two mouse colonies, germ-free mice were colonized with caecal microbiota from mice in the two rooms. The separate groups of colonized mice were subsequently housed in either room. The colon mucus thickness of the colonized mice was similar to that of the mice conventionally raised in the same room (Fig5A). Mucus penetrability phenotypes were again different as a more penetrable inner mucus layer was observed in mice colonized with microbiota from Room 2, while mice colonized with microbiota from Room 1 showed a mucus layer impervious to beads (Fig5B and C). Immunohistochemistry revealed that the bacteria were separated from the epithelium by a well-stratified mucus layer in Room 1, but bacteria were in close proximity to the epithelium of mice colonized with microbiota from Room 2 (Fig5D). The composition of intestinal microbiota thus has a major influence on the properties of the inner colon mucus barrier.
Figure 5.
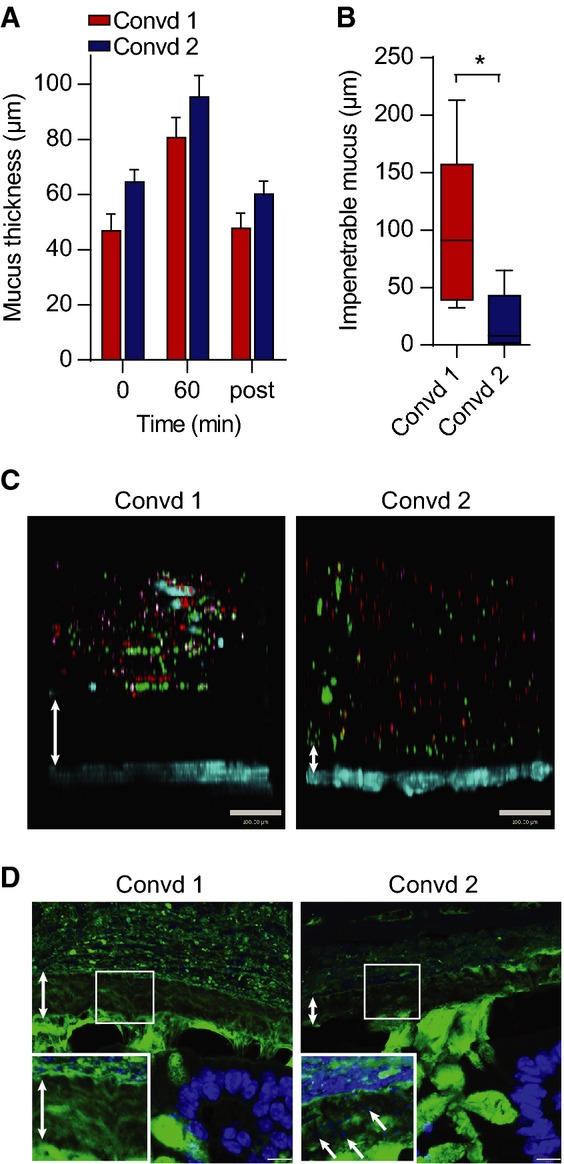
- Mucus thickness measured during 1 h on distal colon explants from mice colonized with flora from animals in Room 1 (Convd 1) or with flora from mice in Room 2 (Convd 2). Mucus not attached to the epithelium was removed after 60 min, and the attached mucus was measured (post). Data are presented as mean ± SEM (n = 4–5).
- Penetrability of beads in mucus on explants from mice colonized with flora from Room 1 (Convd 1) or Room 2 (Convd 2) presented as impenetrable mucus (distance to 20 most penetrating beads). There was a significant difference between the two groups analysed by the Mann–Whitney U-test (n = 5, *P = 0.032). Data are presented as a boxplot with median and min–max whiskers
- Pictures of Z-stacks of beads (red 0.5 μm, purple 1 μm, green 2 μm) penetrating the mucus on colonic explants from Convd 1 or Convd 2 mice. The epithelium is stained by Calcein violet (blue). Double arrows indicate impenetrable mucus. Scale bar is 100 μm.
- Immunostaining of colon sections of Convd 1 or Convd 2 mice using Anti-MUC2C3 (green) and Hoechst (blue) staining DNA and visualizing bacteria in mucus (inserts). Double arrows mark the inner mucus layer that separates bacteria from the epithelium. Arrows at an angle point to bacteria in the inner mucus. Scale bars are 10 μm.
Source data are available online for this figure.
Discussion
Genetically similar animals with slightly different stable and transmissible intestinal microbiota showed surprisingly large differences in the inner colon mucus layer. The mouse colony housed in Room 1 has been described previously 14 with a well-developed inner mucus layer that is impervious to bacteria. The other colony in Room 2 had much more permeable mucus, which demonstrates that mucus properties can vary in mice of the same strain that are maintained in identical hygiene conditions. To address which of the two mucus phenotypes that most closely resembled mammals in a real-life environment, we analysed caught free-living Mus musculus, that is the same species as experimental mice. Spanning a more diverse genetic as well as environmental background, we expected that these wild mice would show larger individual variability than the laboratory strains 22, 23, 24, 25. Wild mice do in addition have effects from parasites and viruses that are not present in laboratory animals. Nevertheless, these mice often had an even more developed mucus layer than observed in Room 1, which further supports the influence of the microbiota on mucus and that the Room 1 can be considered as a reasonable model. As the two mucus phenotypes in Room 1 and 2 were transmissible by colonizing germ-free mice with caecal contents, one can predict that the mucus phenotypes were almost solely dependent on the microbiota composition. The origin of the germ-free mice is more related to the origin of the animals in Room 2, but as they still were able to form a functional mucus barrier upon colonization with flora from animals in Room 1, genetic influences are less likely.
Sequencing the 16S rRNA gene from the two mouse colonies revealed significant bacterial differences. However, mechanisms for how bacteria affect the host epithelium and its production of mucus are far from understood. As there is a normal physical separation between the bacteria and epithelium, one possibility is that bacterial products, such as LPS and short chain fatty acids, diffuse through the mucus layer and affect the epithelium. As the vast majority of bacterial species identified were identical between the two colonies, the bacterial capacity to control mucus properties cannot be a general effect of all bacteria, but rather of specific microbes. Identifying the nature of these microbes and the products they secrete with a high capacity to stimulate mucus formation and improving mucus properties is of high priority.
The commensal bacteria live in the non-attached and expanded outer mucus of colon 26, where they can cleave off and utilize mucin glycans as energy source. The exoglycosidases are usually arranged in specialized genetic loci for utilizing specific types of substrates as, for example, starch (Sus) and xylose-containing polysaccharides (PUL) 27. As the mucin-decorating glycans are protecting the mucin polymeric network from degradation, it is important that the degradation process is not too fast. The importance of sufficiently complex mucin glycans was demonstrated in studies of mice with truncated glycans as found in O-glycan core 1 or core 3 deficient mice 19, 28. In especially the core 1 deficient mice, the inner mucus layer was more vulnerable and quickly dissolved allowing bacteria to reach the epithelium and by this trigger inflammation 19.
The animals in Room 1 with a more restricted bacterial contact with the epithelial cells due to a better mucus layer showed higher amounts of bacteria of the Erysipelotrichi class, mainly the genus Allobaculum. These bacteria have been suggested to have beneficial effects and this could also be the case for the mucus properties and formation 29. However, its increased abundance upon alteration to Food B in both rooms resulted in increased penetrability of the mucus. This would then maybe speak against a mucus promoting effect of Allobaculum. Our study corroborates previous studies showing an association between altered diets and an increase in Erysipelotrichi and Allobaculum, but various effects are shown for fat-enriched diets 29, 30, 31, 32. Together, our present results do not argue for any specific bacteria contributing significantly to the development of a better mucus layer.
Mouse colitis models typically show features like crypt elongation due to increased cell proliferation, infiltration of immune cells in lamina propria, thickening of muscularis propria and weight loss, all criteria used for scoring colitis 33, 34. Subtle changes in these parameters, as observed in Room 2, could indicate an immune stimulatory effect of the microbiota. This might be caused by the closer contact of bacteria with the epithelium in these mice due to mucus defects. This assumption is in line with the observations that colitis might be driven by extensive bacterial contact as shown both in mouse colitis models and in human UC patients 15. The gut microbiota is a prerequisite for inflammation in the gut and an imbalance in the microbiota in patients with inflammatory bowel disease (IBD) has been suggested 35.
Certain bacteria can, in contrast to promoting well-developed mucus, also destroy the mucus and its protective properties. Such capacities can be expected among pathogenic bacteria as these have developed specific mechanisms to circumvent the mucus protective system 36, 37, 38. We could not identify any bacteria that could be clearly linked to a more developed inner mucus layer and associated with the colony in Room 1. However, in Room 2, we found increased levels of (Beta-, Delta- and Epsilon-) Proteobacteria in distal colon mucus, the site for the observed mucus defects. Species of lower taxonomic levels within the Proteobacteria phylum have been correlated with colitis in a maternally transmitted fashion 39. The phylum Proteobacteria has been associated with human Crohn's disease 40. The class Deltaproteobacteria contains sulphur-reducing bacteria (SRB) that are also found at higher frequencies in patients with IBD as compared to healthy individuals 41. We found that higher levels of the genus Desulfovibrio, a SRB, were associated with a penetrable mucus phenotype in animals with a higher inflammatory tone. This is in line with the suggestion that SRB are linked to an inflammatory state of the gut 41, 42. We also found higher relative abundances of the genera Bacteroides and Prevotella in mice with less developed mucus, bacteria that have been found to be increased also in other colitis models or patients with IBD 39, 43.
A more penetrable mucus layer was associated with a high prevalence of the bacterial phylum TM7. This is a phylum, with no cultivated representatives, commonly found in the oral flora at low levels 44, 45. This phylum has been linked to inflammatory conditions of the mouth such as periodontitis 46. The TM7 phylum is also found in human stool samples at low levels 47 and proposed to play a role in intestinal inflammation 48. The TM7 phylum has also been found in mice more susceptible to colitis caused by Helicobacter hepaticus 49. Both TM7 and Proteobacteria are found at increased levels in the altered flora after epithelial cell-specific ablation of MyD88 50. TM7 and the genus Prevotella are also increased in mice deficient in the inflammasome component Nlrp6 and suggested to be associated with inflammation 51. SFB is prominent in the ileum mucus as it can attach to the epithelium. These bacteria, capable of stimulating the immune system, are more common in Room 2 and could influence the mucus properties. However, it is considered to foster a normal intestinal homeostasis in healthy mice, something that is in contrast to the here observed less developed colon mucus 52. Together, there is a set of bacteria that might be possible to link to less developed colon mucus and bacteria closer to the epithelial cells. Whether any of the identified bacteria in Room 2 could be linked to the altered mucus properties observed in this room remains to be shown. The microbiota mucus sample strategy used may have reduced the observed differences in mucus microbiota as the amounts of attached mucus were not controlled for. Unfortunately, more elaborate mucus sampling techniques are difficult to perform. It is obvious from this study that the microbiota influences the properties of the mucus barrier and that this can be observed within the same animal house with animals of identical genetic background. This observation is very important to consider when analysing results from different laboratories and emphasize the importance of using littermate controls for most types of studies, also studies where this can have an indirect effect. The present results further emphasize the importance of careful interpretation upon comparison of phenotypes in different mouse colonies. The problems of comparing results generated with identical mice with different microbiota are evolving into a very central challenge to many fields of study and can jeopardize the basic concept of scientific research: the possibility to reproduce the results in different laboratories. Subtle changes towards inflammation in control animals may have an impact on results studying intestinal homeostasis. We have also observed different inner mucus phenotypes in animals from different vendors, further emphasizing the need for standardization and caution.
In conclusion, we demonstrate that different microbes have different effects on the properties of the colon mucus barrier. Even though the present study reveals subtle differences in the microbiota between the two mouse colonies, an increased inflammatory tone was observed in Room 2. Microbial changes with increased amounts of Proteobacteria and TM7 as observed in Room 2 argue for a set of bacteria that can be less favourable for an impenetrable inner colon mucus barrier. The results from the free-living mice strongly argue for the importance of a well-developed inner mucus layer that efficiently separates bacteria and host epithelium in a natural habitat.
Materials and Methods
A more detailed description can be found in Supplementary Experimental Procedures.
Animals
Wild-type female C57BL/6 mice were bred in two different environments (Room 1 and Room 2) in the same SPF unit fed either a standard chow diet (Food A: R34, Labfor, Lactamin, Stockholm, Sweden) or an autoclaved diet (Food B: 5021 Labdiet®, IPS, London, UK via Opend, Herfolge, Denmark). Germ-free mice were colonized with caecal flora from mice in the two rooms and treated as conventional animals onwards 53. Wild free-living house mice (Mus musculus) were caught alive in south-eastern Norway using Ugglan traps (Grahnab, Gnosjö, Sweden), approved by the Norwegian Environment Agency, and transported directly to the University of Gothenburg, approved by the Swedish Board of Agriculture. All animal experimental procedures were done in full compliance with Swedish animal welfare legislation and approved by the Swedish Laboratory Animal Ethical Committee in Gothenburg.
Sample collection for microbiota analysis
Using clean, sterile dissection tools, the ileum (Si8 only), distal colon and caecum were removed. The lumen content from Si8 and distal colon was removed by gently squeezing out the intestinal content in a collection tube. The intestinal segment was then gently flushed with 2 × 1 ml sterile PBS. The tissue was considered the mucus sample. The samples were immediately flash-freezed in liquid nitrogen and later stored at −80°C until analysis.
DNA extraction
DNA was extracted and homogenized with a FastPrep®-24 Instrument (MP Biomedicals) and then treated with lysozyme and precipitated. The dissolved DNA was treated with RNase A (Qiagen) and proteinase K in Buffer AL (Qiagen). The DNA was extracted using QIAamp DNA mini kit (Qiagen).
16S rRNA gene tag pyrosequencing and sequence analysis
Bacterial 16S rRNA gene sequences were amplified from each sample using the primers 27F (5′ AGAGTTTGATCCTGGCTCAG 3′) with Titanium Adaptor B and 338R (5′ TGCTGCCTCCCGTAGGAGT 3′) with Titanium Adaptor A and a sample-specific barcode sequence consisting of twelve nucleotides targeting the V1–V2 hypervariable region of the 16S rRNA gene using FastStart Taq DNA Polymerase (Roche). Triplicate PCRs were performed for each sample that were pooled and purified with AMPure beads (Becton Dickinson). The samples were amplified in PCR mixture-in-oil emulsions and sequenced from the 338r primer using Roche 454 FLX and Titanium chemistry (Roche) at the Science for Life Laboratories (Solna, Sweden). Post-processing of pyrosequencing data was done using QIIME software 1.7.0 package 54. The sequences were assigned to operational taxonomic units (OTUs) using UCLUST 55. The reads were aligned to the Greengenes Core reference alignment using PyNAST 56. The GG taxonomies were used to generate summaries of the taxonomic distributions of OTUs across different levels (phylum, order, family, genus and species A phylogenetic tree was built with FastTree 57 and used for estimates of α-diversity (Rarefaction curves, Chao1 58, Shannon diversity 59) and β-diversity (using unweighted UniFrac 20.
Mucus measurements
The thickness of the intestinal mucus was measured as described previously 21, 60. Mucus penetrability was measured as described previously 15, 21.
Tissue fixation and immunostaining
Pieces of ileum or colon with faecal material were fixed in Carnoy (methanol) and bacterial FISH and immunostainings were done with MUC2C3 antisera 14, apoMuc2 antisera 61 or Anti-Ki67 antibody (ab16667, Abcam) and DNA by Hoechst 34580 (Life technologies) as previously described 14. The sections were analysed for crypt length, number of lamina propria cells, number of goblet cells and number of Ki67-positive cells.
Proteomic analysis of mucus samples
The mucus samples removed after thickness measurements were solubilized in a guanidinium hydrochloride-based buffer and processed by the FASP method as described before. The samples were analysed by nano-reversed phase liquid chromatography (nRPLC) coupled to electrospray ionization–tandem mass spectrometry (ESI-MS/MS) in an LTQ-Orbitrap XL (Thermo Scientific) 62. Data from the MS/MS experiments were analysed with the MaxQuant 1.2.2.5 software 63. Relative protein amounts were quantified in ppm by intensity-based absolute quantification (iBAQ) 64.
Statistical analysis
For all mucus, histology and immunostaining measurements data were analysed using a two-tailed Mann–Whitney U-test. For the microbiota analysis, significant differences were conducted using the Wilcoxon rank-sum test, and P-values were converted to false discovery rate values (Q-values) to correct for multiple testing in the R software (http://www.r-project.org/).
Data availability
The data behind the graphs are presented in a source data file at http://embor.embopress.org/. Microbiota 16S rDNA sequencing data have been deposited to the ENA sequence read archive under accession number PRJEB7982 (http://www.ebi.ac.uk/ena/data/view). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository 65 with the dataset identifier PXD001479.
Acknowledgments
We acknowledge the CCI unit at the University of Gothenburg for assistance with confocal microscopy and Frida Svensson for technical assistance. We acknowledge the PRIDE team for the deposition of the data to the ProteomeXchange Consortium. This work was supported by the Swedish Research Council (no. 7461, 21027, 22220-01-5) The Swedish Cancer Foundation, The Knut and Alice Wallenberg Foundation, IngaBritt and Arne Lundberg Foundation, Sahlgren's University Hospital (LUA-ALF), Wilhelm and Martina Lundgren's Foundation, Assar Gabrielsson's foundation, Clas Groschinskys foundation, Torsten och Ragnar Söderbergs Stiftelser, The Sahlgrenska Academy, National Institute of Allergy and Infectious Diseases (U01AI095473, U01AI095776-03:9006862), the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, and The Swedish Foundation for Strategic Research—The Mucus-Bacteria-Colitis Center (MBC) of the Innate Immunity Program.
Author contributions
MEVJ and GCH conceived the original idea; HEJ, AR-P, AS, AE, FS, FB, GCH and MEVJ designed the experiments; HEJ, AR-P, AS, AE, FS and MEVJ performed the experiments; HEJ, AR-P, AS, AE, FB, GCH and MEVJ analysed the data; PB and MB caught and assisted in the analysis of free-living mice; HEJ, AR-P, GCH. and MEVJ wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Source Data for Supplementary Figures
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3A, B, D
Source Data for Figure 4D, E, F, G
Source Data for Figure 5A, B
References
- Martins dos Santos V, Muller M, de Vos WM. Systems biology of the gut: the interplay of food, microbiota and host at the mucosal interface. Curr Opin Biotechnol. 2010;21:539–550. doi: 10.1016/j.copbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Human microbiome project consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2013;63:1069–1080. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroentero Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Hansson GC. Microbiology. Keeping bacteria at a distance. Science. 2011;334:182–183. doi: 10.1126/science.1213909. [DOI] [PubMed] [Google Scholar]
- Knoop KA, Miller MJ, Newberry RD. Trans-epithelial antigen delivery in the small intestine: different paths, different outcomes. Curr Opin Gastroenterol. 2013;29:112–118. doi: 10.1097/MOG.0b013e32835cf1cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103 + dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambort D, Johansson ME, Gustafsson JK, Nilsson HE, Ermund A, Johansson BR, Koeck PJ, Hebert H, Hansson GC. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci USA. 2012;109:5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, Sjovall H, Hansson GC. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE. 2010;5:e12238. doi: 10.1371/journal.pone.0012238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wei B, Wen T, Johansson ME, Liu X, Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson JK, Ermund A, Johansson ME, Schutte A, Hansson GC, Sjovall H. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol. 2012;302:G430–G438. doi: 10.1152/ajpgi.00405.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnenbrink M, Wang J, Hardouin EA, Kunzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22:1904–1916. doi: 10.1111/mec.12206. [DOI] [PubMed] [Google Scholar]
- Abolins SR, Pocock MJ, Hafalla JC, Riley EM, Viney ME. Measures of immune function of wild mice, Mus musculus. Mol Ecol. 2011;20:881–892. doi: 10.1111/j.1365-294X.2010.04910.x. [DOI] [PubMed] [Google Scholar]
- Bemark M, Boysen P, Lycke NY. Induction of gut IgA production through T cell-dependent and T cell-independent pathways. Ann N Y Acad Sci. 2012;1247:97–116. doi: 10.1111/j.1749-6632.2011.06378.x. [DOI] [PubMed] [Google Scholar]
- Boysen P, Eide DM, Storset AK. Natural killer cells in free-living Mus musculus have a primed phenotype. Mol Ecol. 2011;20:5103–5110. doi: 10.1111/j.1365-294X.2011.05269.x. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Holmen Larsson JM, Hansson GC. Microbes and Health Sackler Colloquium: the two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2010;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Gaia N, Johansson M, Stahlman M, Backhed F, Delzenne NM, Schrenzel J, Francois P, Cani PD. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- Martinez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der SM, de Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Takaishi H, Matsuki T, Nakazawa A, Takada T, Kado S, Asahara T, Kamada N, Sakuraba A, Yajima T, Higuchi H, et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int J Med Microbiol. 2008;298:463–472. doi: 10.1016/j.ijmm.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Post S, Subramani DB, Backstrom M, Johansson ME, Vester-Christensen MB, Mandel U, Bennett EP, Clausen H, Dahlen G, Sroka A, et al. Site-specific O-glycosylation on the MUC2 mucin protein inhibits cleavage by the Porphyromonas gingivalis secreted cysteine protease (RgpB) J Biol Chem. 2013;288:14636–14646. doi: 10.1074/jbc.M113.459479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarepour M, Bhullar K, Montero M, Ma C, Huang T, Velcich A, Xia L, Vallance BA. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2013;81:3672–3683. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Verma AK, Ahuja V, Paul J. Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. J Clin Microbiol. 2010;48:4279–4282. doi: 10.1128/JCM.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor SO, Ridgway K, Scovell L, Kemsley EK, Lund EK, Jamieson C, Johnson IT, Narbad A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 2003;69:1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA, Holland T, Cotton D, Hauser L, Keller M. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher T, Rehman A, Lepage P, Hellmig S, Folsch UR, Schreiber S, Ott SJ. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57:1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS ONE. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, Fenton LA, Bruno ME, Kaetzel CS. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5:501–512. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, Jin C, Wunderlich C, Wunderlich T, Eisenbarth SC, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci USA. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Host interactions with Segmented Filamentous Bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin Immunol. 2013;25:342–351. doi: 10.1016/j.smim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Nonparametric-estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- Hayek LC, Buzas MA. Surveying Natural Populations. New York: Columbia University Press; 1997. [Google Scholar]
- Gustafsson JK, Ermund A, Ambort D, Johansson ME, Nilsson HE, Thorell K, Hebert H, Sjovall H, Hansson GC. Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J Exp Med. 2012;209:1263–1272. doi: 10.1084/jem.20120562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson GC, Baeckstrom D, Carlstedt I, Klinga-Levan K. Molecular cloning of a cDNA coding for a region of an apoprotein from the ‘insoluble’ mucin complex of rat small intestine. Biochem Biophys Res Commun. 1994;198:181–190. doi: 10.1006/bbrc.1994.1026. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pineiro AM, Bergstrom JH, Ermund A, Gustafsson JK, Schutte A, Johansson ME, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastrointest Liver Physiol. 2013;305:G348–G356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Source Data for Supplementary Figures
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3A, B, D
Source Data for Figure 4D, E, F, G
Source Data for Figure 5A, B
Data Availability Statement
The data behind the graphs are presented in a source data file at http://embor.embopress.org/. Microbiota 16S rDNA sequencing data have been deposited to the ENA sequence read archive under accession number PRJEB7982 (http://www.ebi.ac.uk/ena/data/view). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository 65 with the dataset identifier PXD001479.