Abstract
The polarized distribution of neuronal proteins to axons and dendrites relies upon microtubule-binding proteins such as CRMP, directed motors such as kinesin UNC-104/Kif1A, and diffusion barriers such as ankyrin. The causative relationships between these molecules are unknown. We show here that Caenorhabditis elegans CRMP (UNC-33) acts early in neuronal development, together with ankyrin (UNC-44), to organize microtubule asymmetry and axon-dendrite sorting. In unc-33 and unc-44 mutants, axonal proteins are present in dendrites and vice versa, suggesting bidirectional failures of axon-dendrite identity. UNC-33 protein is localized to axons by unc-44, and enriched in a region that resembles the axon initial segment. unc-33 and unc-44 establish the asymmetric dynamics of axonal and dendritic microtubules; in their absence, microtubules are disorganized, the axonal kinesin UNC-104 invades dendrites, and inappropriate UNC-104 activity randomizes axonal protein sorting. We suggest that UNC-44 and UNC-33 direct polarized sorting through their global effects on neuronal microtubule organization.
The asymmetric microtubule cytoskeleton is essential for axon-dendrite specification in development and for polarized protein sorting in mature neurons. The role of microtubules in axon specification, the developmental process that generates a single axon per neuron, has been characterized extensively in cultured hippocampal neurons and to a lesser extent in vivo 1, 2. Pharmacological manipulation of microtubule stability alters the number of axons per neuron and biases the selection of one process as the future axon 3, and conversely, molecules that affect axon specification regulate microtubules. The microtubule-binding CRMP protein, also called UNC-33, TOAD-64, Ulip, DRP, or TUC, is linked to early events in axonal development in vivo and in vitro 1. The founding CRMP protein, UNC-33, affects axon guidance and elongation in C. elegans 4. The best-characterized vertebrate CRMP, now known as CRMP2, promotes axonal specification in cultured mammalian neurons: overexpression of CRMP2 causes neurons to form multiple axons at the expense of dendrites, and dominant-negative fragments or siRNAs against CRMP2 cause the selective loss of axons 5-7. CRMP proteins interact with tubulin heterodimers and microtubules, and promote microtubule assembly in vitro 7. CRMP2 also binds the kinesin light chain (KLC) subunit of Kinesin-1, and acts as an adapter for the transport of tubulin dimers, the actin regulators Sra-1/WAVE, and TrkB into axonal growth cones 5, 8, 9.
In mature neurons, asymmetric microtubules regulate polarized protein sorting. For example, only axonal microtubules support the polarized transport of synaptic vesicles by the Kinesin-3/Kif1A/UNC-104 family 10. Both kinesins and their cargo contribute to polarized transport, although the complete nature of the sorting code has not been determined. Axonal microtubules are generally oriented with their plus ends away from the cell body 11, 12, whereas dendritic microtubules can have the same orientation, the opposite orientation, or mixed polarity, depending on the cell type and distance from the cell body 13, 14. Axonal and dendritic microtubules are also distinguished by their relative stability, association with microtubule-associated proteins, and post-translational modifications 3, 10. Axonal protein sorting is reinforced by the axon initial segment (AIS), a region near the cell body that acts as a membrane diffusion barrier and the action potential initiation zone 15, 16. In mammals, the giant actin-binding protein ankyrinG resides at the AIS and maintains its integrity 17, 18. AnkyrinG supports axon functions by preventing dendritic kinesins and their cargo from entering axons, but appears not to affect the properties of dendrites 18-20.
Although CRMPs, Kif1A, and ankyrinG all regulate axon development or function, the relationships between them have not been examined in vivo or in vitro. The biological effects of mammalian CRMPs, ankyrins, and kinesin3/Kif1A seem to be distinct, but are obscured by the fact that all belong to multigene families, with members that can have overlapping or antagonistic functions 10, 21-23. C. elegans mutants in unc-33, its sole CRMP homolog, and unc-44, its sole ankyrin homolog, share defects in locomotion, axon elongation, and axon guidance 4, 24-27. We show here that C. elegans unc-33 acts with unc-44 to direct polarized sorting of a wide array of neuronal proteins, in part by regulating the conserved kinesin-3/KIF1A protein UNC-104. Contrary to expectations, unc-33 and unc-44 mutants affect microtubule dynamics and protein sorting in dendrites as well as axons. Our results suggest that unc-33 and unc-44 establish the polarized microtubule cues that drive neuronal transport.
Results
Axonal proteins appear in dendrites in unc-33/CRMP mutants
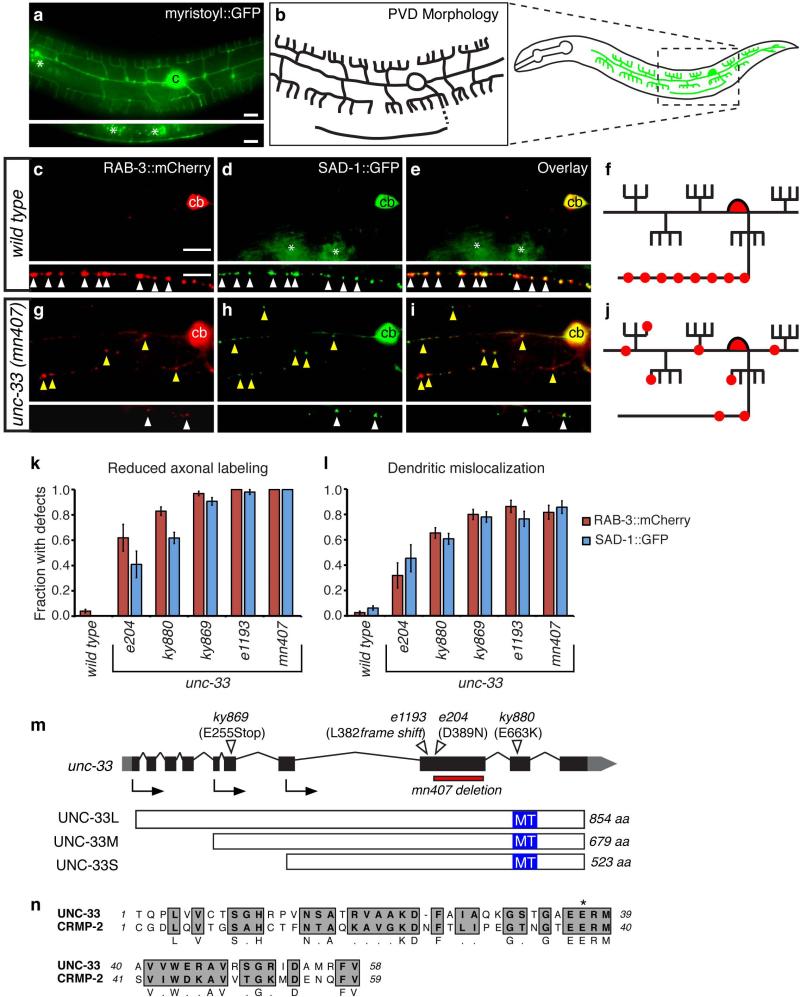
The C. elegans PVD sensory neurons have well-defined axons and dendrites that facilitate visualization of polarized protein localization (Fig. 1a,b) 28. Each PVD has an axon that grows ventrally and then anteriorly in the ventral nerve cord, and lateral dendrites that branch elaborately to circle the body 28. PVD presynaptic specializations are restricted to the axon in the ventral nerve cord 29. To generate axonal markers in PVD, a des-2 promoter fragment 30 was used to express two fluorescently tagged presynaptic molecules, RAB-3:: mCherry and SAD-1::GFP. RAB-3 is a Rab GTPase that labels a subset of synaptic vesicles 31, and SAD-1 is a presynaptically localized serine/threonine kinase that affects presynaptic differentiation and neuronal polarity 32. Both markers were localized to axonal PVD puncta in the ventral nerve cord, and were either faint or undetectable in PVD dendrites (Fig. 1c-f).
Figure 1. unc-33 mutants mislocalize presynaptic proteins to dendrites.
(a,b) PVD neuron morphology. (a) des-2::myristoyl::GFP marker in an L4 animal (b) Diagrams of PVD morphology. The unbranched ventral process is the axon.
(c-j) Representative images of RAB-3::mCherry and SAD-1::GFP in PVD neurons of wild type (c-f) and unc-33(mn407) (g-j) animals. For each set of fluorescence micrographs, the top panel is the maximum intensity projection of dendritic focal planes and the bottom panel is the maximum intensity projection of axonal focal planes. White and yellow arrowheads indicate axonal and dendritic puncta, respectively; ‘cb’ labels the PVD cell body, and asterisks mark gut autofluorescence. Anterior is at left and dorsal is up in all panels. Scale bars, 10 μm.
(k,l) Quantification of axonal localization defects (k) and dendritic mislocalization defects (l) of RAB-3::mCherry and SAD-1::GFP (n>30 animals/genotype). The fraction of animals with qualitative defects is shown. Error bars indicate standard error of proportion (s.e.p.). Alternative quantification of fluorescence intensity per animal is provided in Supplementary Fig. 1.
(m) unc-33 gene structure showing exons (black boxes), introns (lines), and untranslated regions (gray boxes), and lesions in unc-33 alleles. Arrows denote the positions of start codons for alternative unc-33 transcripts. Three UNC-33 protein isoforms (long, medium and short) are depicted with predicted microtubule-assembling domains in blue. (n) Predicted microtubule-assembling domain of UNC-33, with rat CRMP-2. Gray regions highlight conserved residues; asterisk denotes the conserved glutamate mutated to lysine in unc-33(ky880).
Several screens for altered localization of these presynaptic proteins yielded unc-33 alleles, identified based on map position, visible phenotypes, and failure to complement existing unc-33 alleles (see Methods). Two new unc-33 alleles and three existing unc-33 alleles all disrupted the distribution of RAB-3::mCherry and SAD-1::GFP in PVD, reducing fluorescence in axons and increasing it in dendrites to result in a nearly random distribution of presynaptic proteins (Fig. 1g-l; further quantification in Supplementary Fig. 1). The mutants followed the allelic series: wild type > e204 > ky880 > ky869 ≥ e1193 = mn407 (Fig. 1k,l; Supplementary Fig. 1). In strong alleles, all animals showed significant redistribution of axonal proteins into PVD dendrites. RAB-3::mCherry and SAD-1::GFP were usually colocalized to stable puncta both in axons and in dendrites, suggesting that presynaptic proteins retained their local relationships (Supplementary Fig 1g).
A broader survey of axonal markers demonstrated that unc-33 affected sorting of many axonal proteins in multiple neuron types. The tagged axonally-enriched proteins SNN-1/synapsin, APT-4/α-adaptin, and SYD-2/Liprin-α were all mislocalized to PVD dendrites in unc-33 mutants (Supplementary Fig. 1 and data not shown). Axonal markers were also mislocalized to dendrites in FLP mechanosensory neurons, AVE interneurons, and AWC and ASI chemosensory neurons (Supplementary Fig. 2 and data not shown). The full set of mislocalized proteins included synaptic vesicle proteins, plasma membrane proteins, and cytoplasmic proteins, which in each case accumulated in stable puncta in dendrites as well as axons.
unc-33 mutants have defects in axon guidance and elongation 24, 27, raising a concern that their protein localization defects could be an indirect result of misguided or shortened axons. PVD axon guidance to the ventral nerve cord was normal in most unc-33(mn407) (80%, n=52) and unc-33(ky880) (100%, n=37) animals, but most PVD axons terminated prematurely (Supplementary Fig. 3g), as did FLP axons in the head (Supplementary Fig. 3a). unc-34 (Ena/VASP) mutants had similar termination defects in PVD and FLP, but were nearly normal in RAB-3::mCherry and SAD-1::GFP localization (Supplementary Fig. 3a-i), arguing that premature axon termination cannot explain the mistargeting of axonal proteins in unc-33 mutants.
UNC-33L acts in development to establish axonal polarity
unc-33 encodes three protein isoforms with alternative N-termini and common C-terminal sequences: UNC-33L (long), UNC-33M (medium), and UNC-33S (short) 4, 27. The canonical allele unc-33(e204) is a missense mutation that affects all isoforms; two likely null alleles disrupting all isoforms are unc-33(e1193), a frameshift mutation, and unc-33(mn407), a 500 base pair deletion 27. Sequencing of the new alleles revealed that unc-33(ky869) generates a nonsense codon in the long and medium forms of unc-33 (Fig. 1m), and unc-33(ky880) generates a glutamate to lysine missense mutation in all isoforms, in the region corresponding to the microtubule (MT)-stabilizing domain of CRMP-2 (E663 in UNC-33L) 7 (Fig. 1n).
The strong mutant phenotype of unc-33(ky869), which should spare the short form of unc-33 (Fig. 1k-m), suggested that the long or medium isoform is required for activity. To test these isoforms individually, full-length untagged cDNAs encoding either UNC-33L or UNC-33M were expressed from pan-neuronal promoters and introduced into unc-33(mn407) null mutants. UNC-33L, but not UNC-33M, fully rescued unc-33 locomotion, egg-laying, and polarized localization of RAB-3::mCherry and SAD-1::GFP in PVD (Fig. 2a-b, Supplementary Fig. 1, and data not shown). The combined results from the unc-33(ky869) mutation and unc-33(mn407) rescue suggest that UNC-33L is sufficient and probably essential for unc-33 activity.
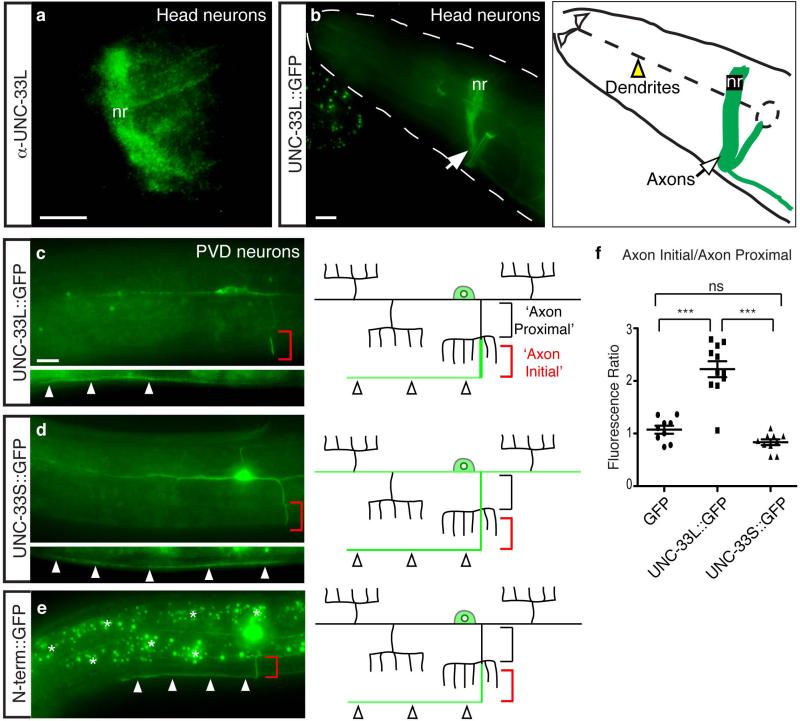
Figure 2. UNC-33L functions in PVD during the establishment of polarity.
(a,b) Isoform-specific and cell autonomous rescue of axonal protein localization in PVD neurons of unc-33 mutants. Quantification of axonal localization defects (a) and dendritic mislocalization defects (b) of RAB-3::mCherry and SAD-1::GFP were as in Fig. 1; n>25 animals per genotype.
(c-e) Development of PVD neurons, visualized with des-2::myrisotyl::GFP marker, with schematic diagrams at right. In c, two focal planes are shown for the late L2/early L3 stage animal to show the two neurites being elaborated. White arrowheads in (c) and (d) indicate the primary processes of PVD, ‘cb’ marks the PVD cell body, and asterisks mark gut autofluorescence. Anterior is at left and dorsal is up in all panels. Scale bar, 10 μm. (f) Quantification of axonal protein localization in PVD after heat shock-driven UNC-33L expression in unc-33(mn407) mutant animals (n>25 animals/condition), with a corresponding developmental time line. Rescue was defined as correct localization of RAB-3::mCherry in adult PVD neurons. All error bars indicate s.e.p.
unc-33 reporters and UNC-33 proteins are expressed in many or all neurons, but not in non-neuronal cells 4, 27. An UNC-33L cDNA under a pan-neuronal promoter fully rescued the unc-33 phenotype (Fig. 2a,b). More selective expression of UNC-33L only in PVD and FLP neurons resulted in near-complete rescue of their axonal sorting defects, consistent with cell-autonomous action of unc-33 (Fig. 2a,b; Supplementary Fig. 1, 2).
We identified the developmental stage at which unc-33 acts by providing unc-33 function at different times using a heat shock promoter. PVD neurons are born during the L2 larval stage, extend an axon and two primary dendrites during L2 and early L3 stages, and elaborate dendritic branches during the late L3 and L4 stages (Fig. 2c-e). We observed axonal accumulation of RAB-3::mCherry and SAD-1::GFP beginning in late L3 (Supplementary Fig. 4). hsp::unc-33L rescued adult unc-33 defects after heat shock in L2 or L3 stages, but little rescue was observed after heat shock in L1, L4, or young adult stages (Fig. 2f). These results suggest that unc-33 acts in PVD neurons near the time when axon-dendrite polarity is established, and cannot restore polarized protein transport at later developmental times.
UNC-33L has been reported to be enriched in nerve ring axons 27. We generated a new polyclonal antibody against UNC-33L and confirmed its axonal localization in the nerve ring and ventral cord, and its absence from sensory dendrites (Fig. 3a). A similar pattern of axonal enrichment was observed with an internally-tagged UNC-33L::GFP protein that rescued unc-33 locomotion and protein localization defects (Fig. 3b). UNC-33L::GFP expressed under the des-2 promoter labeled PVD axons consistently, but was minimally expressed in primary PVD dendrites and was undetectable in dendrite branches (Fig. 3c). Within the PVD axon, UNC-33L::GFP was enriched in a segment of the axon near, but not adjacent to, the PVD cell body (Fig. 3c). When expressed separately, the unique N-terminal UNC-33L domain was enriched in axons, but not in this small axon segment, whereas the smaller UNC-33S protein was randomly distributed to axons and dendrites (Fig. 3d-f). Enrichment of UNC-33L::GFP in axonal segments near the cell body was also observed in FLP and AWC neurons (data not shown).
Figure 3. UNC-33L is enriched in PVD axons.
(a) UNC-33L immunoreactivity in nerve ring of wild-type animal.
(b) Biologically active UNC-33L::GFP protein expressed from a pan-neuronal promoter in wild type animal, showing localization in nerve ring axons and absence from sensory dendrites. nr, nerve ring.
(c-e) Representative images of UNC-33L::GFP, UNC-33S::GFP, and UNC-33L N-terminus::GFP proteins in wild type PVD neurons, with schematic diagrams at right. Red brackets indicate region of UNC-33L enrichment in axon, and arrowheads show expression in ventral nerve cord. Black brackets indicate proximal segment of PVD axon used for comparing fluorescence intensities in (f).
(f) Quantification of UNC-33L::GFP, UNC-33S::GFP, and GFP fluorescence, expressed as ratio of ‘axon initial’ domain to ‘axon proximal’ domain. Error bars indicate s.e.m. *** p < 0.001 according to the Bonferroni t-test, ns, not significant. Anterior is at left and dorsal is up in all panels. Scale bars, 10 μm.
Misplaced UNC-104/KIF1A randomizes axonal protein sorting
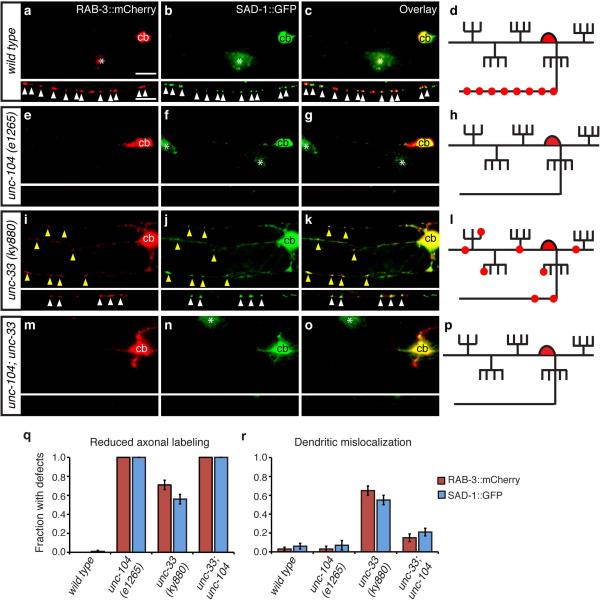
The appearance of axonal proteins in dendrites in unc-33 mutants could result either from a passive redistribution of proteins through diffusion, or from active defects in polarized protein traffic. To distinguish between these mechanisms, we combined unc-33 mutations with mutations in unc-104/KIF1A, a conserved Kinesin-3 family member that mediates the axonal transport of presynaptic vesicles and other presynaptic proteins 33, 34. In unc-104 mutants, axonal proteins are trapped in the cell body, but dendritic proteins are localized normally 10, 35. As expected, RAB-3::mCherry and SAD-1::GFP markers were restricted to the PVD cell bodies in unc-104 mutants (Fig. 4a-h), a pattern distinct from the randomized distribution in unc-33 (Fig. 4i-l). An unc-33 unc-104 double mutant resembled unc-104, with RAB-3::mCherry and SAD-1::GFP restriction to the PVD cell body (Fig. 4m-r). Therefore, both the appropriate axonal localization and the inappropriate dendrite localization of synaptic proteins in unc-33 mutants require UNC-104, suggesting that the axonal UNC-104 kinesin gains access to dendrites in unc-33 mutants.
Figure 4. unc-104/KIF1A kinesin mislocalizes presynaptic proteins to dendrites in unc-33 mutants.
(a-p) Representative images of RAB-3::mCherry and SAD-1::GFP in PVD neurons of wild type (a-d), unc-104(e1265) (e-h), unc-33(ky880) (i-l), and unc-104 unc-33 (m-p) animals, with corresponding diagrams. For each set of fluorescence micrographs, the top panel is the maximum intensity projection of dendritic focal planes and the bottom panel is the maximum intensity projection of axonal focal planes. White and yellow arrowheads indicate axonal and dendritic puncta, respectively; ‘cb’ marks the PVD cell body, and asterisks mark gut autofluorescence. Anterior is at left and dorsal is up in all panels. Scale bar, 10 μm.
(q,r) Quantification of axonal localization defects (q) and dendritic mislocalization defects (r) of RAB-3::mCherry and SAD-1::GFP, as in Fig. 1; n>30 animals/genotype. Error bars indicate s.e.p.
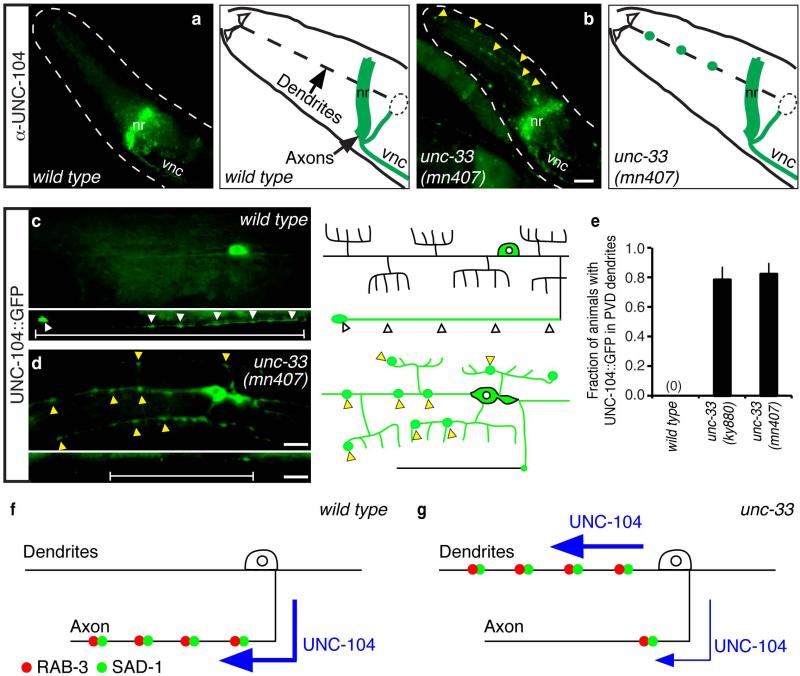
We examined UNC-104 localization directly using a monoclonal antibody that recognizes endogenous UNC-104 protein. In wild-type animals, UNC-104 immunoreactivity was detected in axon-rich regions such as the nerve ring, amphid commissure, and ventral nerve cord, but was not detected in anterior sensory dendrites or tail dendrites (Fig. 5a, Supplementary Fig. 5). These results suggest that endogenous UNC-104 protein is preferentially localized to axons.
Figure 5. UNC-104 is mislocalized to dendrites in unc-33 mutants.
(a,b) Immunostaining of endogenous UNC-104 in wild-type (a) and unc-33(mn407) (b) animals, with corresponding schematic diagrams. Yellow arrowheads indicate UNC-104 immunoreactivity in sensory dendrite regions. nr, nerve ring; vnc, ventral nerve cord. (c,d) Localization of UNC-104::GFP in PVD neurons of wild-type (c) and unc-33 (mn407) (d) animals, with schematic diagrams. White arrowheads indicate UNC-104::GFP enrichment in PVD axons; yellow arrowheads indicate UNC-104::GFP in PVD dendrites.
(e) Quantification of animals with detectable UNC-104::GFP fluorescence in PVD dendrites (n>25 animals/genotype). Error bars indicate s.e.p.
(f,g) Inferred UNC-104/KIF1A activity in neurons of wild-type animals (f) and unc-33 mutants (g). Anterior is at left and dorsal is up in all panels. Scale bars, 10 μm.
By contrast, unc-33 mutants had substantial UNC-104 immunoreactivity in sensory dendrites and tail dendrites, as well as nerve ring and ventral cord axons (Fig. 5b, Supplementary Fig. 5). To confirm this observation at the single-neuron level, a functional UNC-104::GFP fusion protein 36 was expressed selectively in PVD and FLP neurons. In wild-type animals, UNC-104::GFP fluorescence was bright in axons and cell bodies, weak in primary dendrites, and absent from dendrite branches (Fig. 5c); in unc-33 mutants, UNC-104::GFP fluorescence was reduced in axons and increased in dendrites and dendrite branches (Fig. 5d,e). The combined genetic and cell biological results suggest that the mislocalization of axonal proteins in unc-33 mutants results from a failure to restrict UNC-104 kinesin to axons (Fig. 5f,g).
Cilia chemoreceptors are mislocalized in unc-33 mutants
To ask whether unc-33 affected sorting of dendritic proteins as well as axonal proteins, we examined the AWB chemosensory neuron, which has a dendrite that terminates in an anterior sensory cilium and an axon that grows into the nerve ring (Fig. 6a). ODR-10::GFP, a chemosensory G protein-coupled receptor, is enriched in AWB cilia and excluded from axons of wild-type animals37 (Fig. 6b). In unc-33 mutants, however, ODR-10::GFP was present in AWB axons as well as AWB cilia (Fig. 6b). The defect in ODR-10::GFP localization in unc-33 mutants was not as severe as the defect in axon protein localization, but it was consistently observed in multiple unc-33 alleles at both early and late larval stages (Fig. 6c).
Figure 6. A sensory chemoreceptor protein is mislocalized to axons in unc-33 mutants.
(a) Schematic diagram of AWB chemosensory neurons in the head. (b) Representative maximum projection fluorescence images showing ODR-10::GFP localization in AWB neurons of wild type, unc-33, unc-104, and unc-104; unc-33 double mutant animals. Yellow arrowheads indicate ODR-10::GFP in axons. Anterior is at left and dorsal is up in all images. Scale bar, 10 μm.
(c,d) Quantification of animals with ODR-10::GFP fluorescence in axons (n>30 animals per genotype). Error bars indicate s.e.p.
unc-104 axonal kinesin mutants do not normally affect ODR-10::GFP localization (Fig. 6b,d)35, and unc-33; unc-104 double mutants resembled unc-33 single mutants, with ODR-10::GFP in both axons and cilia (Fig. 6b.d). Thus ODR-10 mislocalization in unc-33 mutants was not caused by dysregulated UNC-104. The altered localization of ODR-10::GFP did, however, require odr-4, a gene that is normally required for ODR-10 exit from the endoplasmic reticulum and localization to cilia 37 (Supplementary Fig. 6). unc-101, an AP1 μ -adaptin subunit that restricts ODR-10::GFP to cilia 35, was also epistatic to unc-33 (Supplementary Fig. 6). These results suggest that mislocalized cilia proteins in unc-33 mutants are still regulated by odr-4 and unc-101.
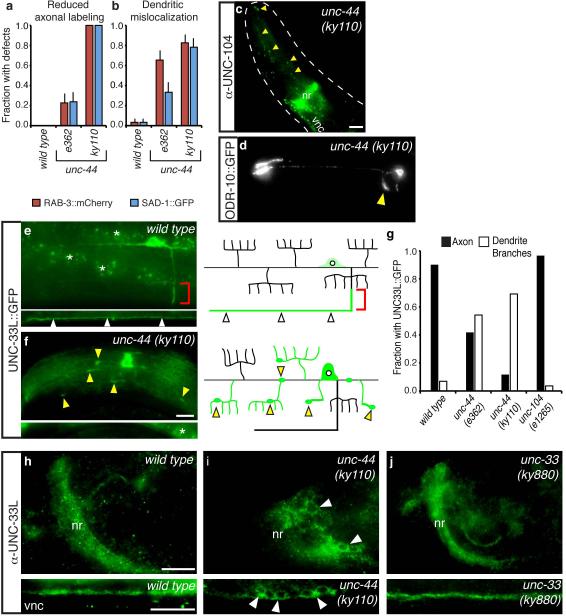
UNC-44/Ankyrin localizes UNC-33L to axons
The C. elegans unc-44 gene encodes its sole ankyrin homolog 26, and shares many functions with unc-33, including roles in axon growth and guidance 24, 27. We identified unc-44 mutants in several genetic screens for altered localization of axonal and dendritic proteins, suggesting that it might have functions related to those of unc-33. Indeed, examination of two unc-44 alleles with the markers described above revealed a close correspondence between unc-44 and unc-33. unc-44 mutants had reduced SAD-1::GFP and RAB-3::mCherry expression in PVD axons, and significant expression in dendrites (Fig. 7a,b). Similarly, UNC-104 kinesin was present in dendrites as well as axons of unc-44 mutants, and the cilia protein ODR-10::GFP was partly localized to axons in AWB neurons (Fig. 7c,d). unc-44 encodes at least ten proteins, of which two isoforms of >5000 amino acids are implicated in neuronal function 26; the mutants we examined are typical, but it is not known whether they are null alleles that affect all isoforms. Therefore, these experiments suggest common functions for unc-44 and unc-33, but may not detect all functions of unc-44.
Figure 7. unc-44/ankyrin mutations disrupt polarized protein sorting and UNC-33L localization.
(a,b) Axonal and dendritic mislocalization defects of RAB-3::mCherry and SAD-1::GFP in unc-44 PVD neurons, as in Fig. 1; n>25 animals/genotype.
(c) Immunostaining of endogenous UNC-104 in unc-44(ky110) animal; yellow arrowheads indicate UNC-104 immunoreactivity in sensory dendrite regions. nr, nerve ring; vnc, ventral nerve cord. Compare Fig. 5a,b.
(d) Representative maximum projection fluorescence image showing ODR-10::GFP localization in AWB neurons of unc-44(ky110) animal. Yellow arrowhead indicates ODR-10::GFP in axons. Compare Fig. 6b,c.
(e,f) UNC-33L::GFP protein in PVD neurons of wild-type (e) and unc-44(ky110) (f) animals. Red brackets indicate region of UNC-33L enrichment in proximal axon, white arrowheads indicate axonal UNC-33L::GFP, and yellow arrowheads indicate UNC-33L::GFP foci in dendrites and dendrite branches. Asterisks indicate gut autofluorescence.
(g) Quantification of animals with detectable UNC-33L::GFP in PVD axons and dendrite branches (n>25 animals/genotype). All error bars indicate s.e.p.
(h-j) Endogenous UNC-33L immunoreactivity in wild-type (h), unc-44(ky110) (i), and unc-33(ky880) (j) animals. Nerve ring (nr, top panels) and ventral nerve cord (vnc, bottom panels) are shown. Arrowheads indicate UNC-33L retained in neuronal cell bodies in unc-44 mutants. Anterior is at left and dorsal is up in all panels. Scale bars, 10 μm.
The suggestion that unc-44 and unc-33 have related functions was supported by an examination of UNC-33L protein. In unc-44 PVD neurons, UNC-33L::GFP was depleted from axons, and instead accumulated in neuronal cell bodies and dendrite branches (Fig. 7e-g). Immunostaining of endogenous UNC-33L confirmed that immunoreactivity was lost from the nerve ring and nerve cords, and enriched in cell bodies, throughout the nervous system of unc-44 mutants (Fig. 7h,i). Thus unc-44 recruits UNC-33L to axons, providing a molecular basis for the protein sorting defects of unc-44 mutants.
UNC-33L localization to axons was normal in unc-33(ky880) mutants, which have a mutation in the predicted tubulin-binding domain (Fig. 7j). However, UNC-104 and synaptic proteins were mislocalized in unc-33(ky880), suggesting that localized UNC-33L is not sufficient for activity if it cannot bind tubulin. UNC-33L axonal localization was normal in unc-104 kinesin mutants (Fig. 7g). These results reveal a hierarchy in polarized protein traffic: UNC-33L localization requires unc-44, but not unc-104, whereas other axonal proteins are misrouted by mislocalized UNC-104.
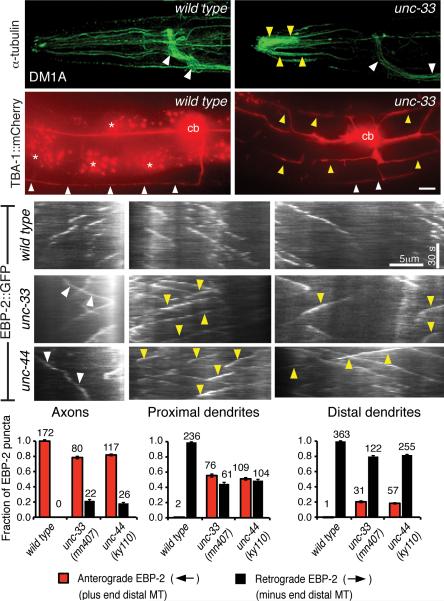
unc-33 and unc-44 organize the microtubule cytoskeleton
Their widespread defects in polarized protein sorting prompted an examination of the cytoskeleton in unc-33 and unc-44 mutants. C. elegans neurons are rich in microtubules, and a monoclonal antibody against α-tubulin strongly labeled the nervous system, including axons in the nerve ring and ventral nerve cord, dendrites, and cilia (Fig. 8a, Supplementary Fig. 7). This staining pattern was strikingly different in unc-33 and unc-44 mutants. First, tubulin immunoreactivity was enriched in the distal segments of dendrites, consistent with increased dendrite microtubules previously observed in unc-33 electron micrographs 24. Second, axonal tubulin staining in the nerve ring was strongly reduced (Fig. 8b, quantified in Supplementary Fig. 7). These results suggest that the overall organization of neuronal microtubules is altered in unc-33 and unc-44 mutants.
Figure 8. Microtubule defects in unc-33 and unc-44 mutants.
(a,b) Representative maximum intensity projections of confocal images of α-tubulin immunoreactivity in head neurons of wild type and unc-33(ky880) animals. White arrowheads indicate axon-rich nerve ring and ventral nerve cord, yellow arrowheads indicate distal sensory dendrites. unc-33(mn407) and unc-44(ky110) are shown in Supplementary Fig. 7, with quantification of immunostaining. (c,d) TBA-1::mCherry (α-tubulin) fluorescence in PVD neurons of representative wild type (c) and unc-33(mn407) (d) animals.
(e-g) Representative kymographs of moving EBP-2::GFP puncta in axons (e), proximal dendrites (f) and distal dendrites (g) of head neurons of wild type (top panel), unc-33(mn407) (middle) and unc-44(ky110) (bottom) animals. Time runs top to bottom; arrowheads mark mutant puncta moving against the normal direction for that process. Bar graphs show the fractions of puncta moving anterogradely and retrogradely with respect to the cell body. Error bars indicate s.e.p. Numbers above each column denote the number of puncta counted in the corresponding categories.
Staining with antibodies to different post-translationally modified forms of tubulin indicated that unc-33 and unc-44 abnormalities were restricted to certain microtubule populations. An anti-glutamylated tubulin antibody 38 labeled sensory cilia both in wild-type and in mutant animals, and staining of mechanosensory neurons with antibodies against acetylated α-tubulin 39 and detyrosinated α-tubulin was also normal, suggesting normal modification and localization of these specialized microtubule subsets (Supplementary Fig. 7c-h).
To examine tubulin distribution at the single cell level, an mCherry-tagged α-tubulin protein was expressed in PVD neurons. In wild type animals, α-tubulin::mCherry was consistently detected in axons and primary dendrites of PVD neurons, but not in dendrite branches, matching PVD microtubule distribution in electron micrographs (Fig. 8c)28. In unc-33 mutants, α-tubulin::mCherry fluorescence was significantly reduced in PVD axons, and brighter in dendrites and dendrite branches (Fig. 8d).
To monitor neuronal microtubule dynamics directly, we used the microtubule end-binding protein EBP-2 (EB1), whose preferential association with the plus ends of growing microtubules provides an indication of microtubule polarity and dynamics in vivo 40, 41. We expressed EBP-2::GFP in amphid sensory neurons, whose oriented axons and dendrites can be observed in a common focal plane, and examined moving EBP-2::GFP puncta by real-time imaging. In wild-type animals, all axonal EBP-2::GFP puncta moved away from neuronal cell bodies, whereas all dendritic puncta moved toward the cell bodies (Fig. 8g-i, Suppl. Movies 1-3). In unc-33 and unc-44 mutants, however, substantial bidirectional movement of both axonal and dendritic EBP-2::GFP was observed (Fig. 8g-i, Suppl. Movies 4-6). The effects on dendrites were strongest near the cell body, and weaker in distal regions near the cilia. Thus unc-33 and unc-44 contribute to the oriented growth of neuronal microtubules, affecting their polarity as well as their overall distribution.
Discussion
unc-33/CRMP and unc-44/ankyrin are essential for polarized neuronal sorting to both axons and dendrites in C. elegans. In unc-33 and unc-44 mutants, axonal transmembrane proteins (UNC-2), cytoplasmic proteins (SAD-1, SYD-2, SNN-1), and synaptic vesicle proteins (RAB-3) are randomized between axons and dendrites, and dendritic proteins are altered as well. By examining the localization of multiple proteins in wild-type and mutant backgrounds, we found that UNC-44 localizes UNC-33 to the axon, which increases the steady-state level of axonal microtubules and orients their asymmetric, plus-end distal growth, while preventing plus-end distal growth of dendritic microtubules. A requirement for UNC-33 at early developmental times suggests that this asymmetry establishes the lasting axon-dendrite polarity of the cell. Either UNC-33 itself or unc-33-dependent microtubule asymmetry directs the polarized recruitment of the plus-end directed axonal kinesin UNC-104/KIF1A, which recruits and delivers axonal cargo proteins. A loss of this polarized information in unc-33 and unc-44 mutants leads to a large-scale disruption of protein sorting, microtubule distribution, and kinesin activity in axons and dendrites.
Although unc-33, unc-44 and their vertebrate homologs have been implicated in axon specification and elongation, most functions of unc-33 and unc-44 were not predicted by previous studies 1, 2, 18, 20, 24, 27. CRMP2, the best-characterized CRMP protein, affects the number of axons per cell (the classical definition of “neuronal polarity” in vitro), but CRMP2 manipulations do not affect dendrites. We did not observe a reduction in axon number in unc-33 mutants, nor were axons converted into dendrites by morphological criteria. Instead, we found that axons and dendrites were morphologically recognizable, but profoundly defective in their cytoskeletal organization and protein localization. A likely explanation for this apparent discrepancy is the complexity of the mammalian CRMP family, which consists of several paralogous genes with multiple splice forms 21; as a result, complete disruption of mammalian CRMP function has not been attempted. It is also likely that neurons have different sources of polarity information in vivo than they have in vitro, such as external signals that modify intrinsic polarity signals 42, 43. We suggest that UNC-33 functions primarily to organize and polarize unique classes of axonal microtubules, and segregate them from dendrites. CRMP2 stimulates microtubule polymerization in vitro and promotes axonal transport of tubulin 5, 7, functions that could contribute to the axonal microtubule defects in unc-33 mutants.
Most properties of unc-44 mutants were also unexpected. Mammalian ankyrinG prevents the entry of dendritic proteins into axons 16, 18, 20, a function that it shares with unc-44. We found that unc-44 also excludes axonal proteins from dendrites. More significantly, the widespread microtubule defects in unc-44 mutants indicate that UNC-44 organizes the microtubule cytoskeleton in axons and dendrites, a function not previously ascribed to ankyrin. This function may arise from the ability of unc-44 to localize UNC-33L to axons, since there are extensive phenotypic similarities between unc-33 and unc-44 mutants.
The live imaging of EB1 dynamics provides a first view of oriented microtubule growth in C. elegans neurons. A previous study using GFP-labeled kinesins as an indirect reporter of motor neuron microtubule polarity suggested that C. elegans axonal microtubules are oriented with distal plus ends, whereas dendritic microtubules are less polarized 44. Our results with EB1 in amphid neurons agree that axonal microtubules are oriented with distal plus ends, but suggest that dendritic microtubules are also oriented, with distal minus ends. This may reflect a difference between motor and sensory neurons: amphid dendrites have a distal basal body, not present in motor neurons, that could act as a microtubule organizing center. Indeed, in frog olfactory neurons, dendrites have oriented microtubules with distal minus ends associated with basal bodies 13.
The UNC-33L-enriched zone of C. elegans axons bears analogies with the vertebrate axon initial segment, which organizes the microtubule cytoskeleton and divides neurons into functional compartments. In most vertebrate neurons, the axon initial segment is a clearly demarcated diffusion boundary and the site of action potential initiation 15. In invertebrate neurons, the nature of the axon initial segment is less clear: a single process can split into axonal and dendritic regions far from the cell body, and spike zones can be distant from the soma 45, 46. C. elegans neurons do not have classical sodium-based action potentials or spike initiating zones, but they can have distinct axonal and dendritic regions. unc-44 represents a molecular connection to ankyrinG at the axon initial segment, and the region of UNC-33L enrichment represents a plausible subcellular site for the axon-dendrite boundary in several C. elegans neurons. Moreover, the vertebrate axon initial segment is enriched in unusual microtubule fascicles that are absent from other neuronal compartments 47, an observation that may relate to the microtubule-stabilizing functions of UNC-33 and CRMP2.
Although UNC-33L protein is largely excluded from dendrites, unc-33 mutant dendrites accumulate axonal proteins and excessive microtubules 24 and lose some dendritic proteins, in association with alterations in polarized microtubule growth. Thus unc-33 establishes a distinction between axons and dendrites that is essential for the molecular integrity of both compartments, either through self-organizing features of the cytoskeleton or through cascades of inappropriate protein sorting. Our results indicate that UNC-33L protects dendrites from axonal proteins by capturing the axonal kinesin UNC-104 and limiting its activity to axons. We speculate that UNC-33 regulates other kinesin motors as well, since it affects UNC-104-independent proteins such as ODR-10 and UNC-2, and CRMP2 acts as an adaptor for axonal Kinesin-1-dependent traffic 5, 9. A further examination of microtubule-dependent traffic in unc-33 mutants may support or extend this hypothesis.
Methods
Strains and Transgenes
Wild type nematodes were C. elegans variety Bristol, strain N2. Strains were cultured using standard techniques 25 at 21-23°C. Some strains were provided by the Caenorhabditis Genetics Center.
Germline transformation was carried out as described 48 using odr-1::DsRed (25 ng/μ1) or unc-122::DsRed (20 ng/μ1) as co-injection markers.
Localization of presynaptic proteins in PVD, FLP and AWC neurons
CX9797 kyIs445 [des-2::mCherry::RAB-3 (injected at 0.5 ng/μl), des-2::SAD-1::GFP (2 ng/μl), odr-1::DsRED (30 ng/μl)], CX10057 kyIs445; unc-33(e204), CX10072 kyIs445; unc-33(ky880), CX10396 kyIs445; unc-33(ky869), CX10599 kyIs445; unc-33(e1193), CX10409 kyIs445; unc-33(mn407), CX10053 kyIs445; unc-34(e315), CX9751 kyIs445; unc-104(e1265), CX11004 kyIs445; unc-104(e1265); unc-33(ky880), CX10242 kyIs445; unc-44(e362), CX9956 kyIs445; unc-44(ky110), CX10886 kyEx2811 [des-2::SYD-2::GFP (2 ng/μl), unc-122:DsRed (20 ng/μl)], CX9275 kyIs442 [odr-3::GFP::UNC-2, odr-3::mCherry::RAB-3, unc-122::GFP], CX10015 kyIs442; unc-33(ky869).
unc-33 rescue experiments
CX10603 kyIs445; unc-33(ky880); kyEx2658 [tag-168::UNC-33L (15 ng/μl), unc-122::DsRed (20 ng/μl)], CX10604 kyIs445; unc-33(ky880); kyEx2659 [tag-168::UNC- 33L (15 ng/μl), unc-122::DsRed (20 ng/μl)], CX10600 kyIs445; unc-33(ky880); kyEx2655 [mec-3::UNC-33L (25 ng/μl), unc-122::DsRed (20 ng/μl)], CX10560 kyIs445; unc-33(ky880); kyEx2619 [des-2::UNC-33L (25 ng/μl), unc-122::DsRed (20 ng/μl)], CX13244 kyIs445; unc-33(mn407); kyEx3861 [tag-168::UNC-33L (20 ng/μl), unc-122::DsRed (20 ng/μl)], CX13315 kyIs445; unc-33(mn407); kyEx3915 [tag-168::UNC-33M (20 ng/μl), unc-122::DsRed (20 ng/μl)], CX11626 kyIs445; unc-33(mn407); kyEx3131 [hsp16-41::UNC-33L (5 ng/μl), unc-122::DsRed (20 ng/μl)].
Analysis of PVD and FLP morphology
CX11480 kyEx3017 [des-2::myristoyl::GFP (7 ng/μl), unc-122::DsRed (20 ng/μl)], CX11329 kyEx3017; unc-33(ky880), CX11481 kyEx3017; unc-33(mn407).
Localization of UNC-33L::GFP, UNC-33S::GFP, and UNC-33L N-terminus::GFP
CX10992 unc-33(mn407); kyIs445; kyEx2875 [tag-168::UNC-33L::GFP (30 ng/ul), unc-122::DsRed (20 ng/ul)], CX11188 kyEx2968 [tag-168::UNC-33L::GFP (30 ng/ul), unc-122::DsRed (20 ng/ul)], CX11244 kyEx3012 [des-2::UNC-33L::GFP (2 ng/ul), des-2::mCherry (5 ng/ul), unc-122::DsRed (20 ng/ul)], CX13581 kyEx4110 [des-2::UNC-33S::GFP (0.5 ng/ul), unc-122::DsRed (20 ng/ul)], CX11461 unc-44(e362); kyEx3012, CX11777 unc-44(ky110); kyEx3012, CX11683 unc-104(e1265); kyEx3012, CX13652 kyEx4147 [des-2::UNC-33L N-terminus(326aa)::GFP (4 ng/μl), unc-122::DsRed (20 ng/ul)].
ODR-10::GFP localization in AWB neurons
CX3877 kyIs156 [str-1::ODR-10::GFP], CX10891 kyIs156; unc-33(ky880), CX10978 kyIs156; unc-33(ky869), CX10997 kyIs156; unc-33(mn407), CX3882 kyIs156; unc-104(e1265), CX11776 kyIs156; unc-104(e1265); unc-33(mn407), CX6093 kyIs156; odr-4(n2144), CX11868 kyIs156; odr-4(n2144); unc-33(mn407), CX4059 kyIs156; unc-101(m1), CX10994 kyIs156; unc-101(m1); unc-33(mn407).
UNC-104::GFP localization in PVD and FLP neurons
CX11551 kyEx3076 [des-2::UNC-104::GFP (1 ng/μl), des-2::mCherry (5 ng/μl), unc-122::DsRed (20 ng/μl)], CX11630 kyEx3076; unc-33(ky880), CX11733 kyEx3076; unc-33(mn407), CX11734 kyEx3076; unc-33(mn407).
Localization of mCherry::TBA-1 (α-tubulin) in PVD neurons
CX12489 kyEx3482 [des-2::mCherry::TBA-1 (2 ng/μl), des-2::myristoyl::GFP (5 ng/μl), unc-122::DsRed (20 ng/μl)], CX12487 unc-33(ky880); kyEx3480 [des-2::mCherry::TBA-1 (2 ng/μl), des-2::myristoyl::GFP (5 ng/μl), unc-122::DsRed (20 ng/μl)], CX12656 unc-33(mn407); kyEx3546 [des-2::mCherry::TBA-1 (2 ng/μl), des-2::myristoyl::GFP (5 ng/μl), unc-122::DsRed (20 ng/μl)].
Dynamic imaging of EBP-2::GFP in head neurons
CX14022 kyEx4341 [odr-8::EBP-2::GFP (12 ng/μl) unc-122::DsRed (20 ng/μl)], CX14028 unc-33(mn407); kyEx4341, CX14031 unc-44(ky110); kyEx4341.
Dual immunofluorescent labeling of α-tubulin and GFP
NG3146 gmIs18 [ceh-23::GFP (50 ng/μl)], CX12481 unc-33(mn407); gmIs18, CX12480 unc-44(ky110); gmIs18.
Standard molecular biology techniques were used to generate expression plasmids and cDNAs used for transgenes (described in detail in Supplementary Information).
Isolation and characterization of unc-33(ky880) and unc-33(ky869)
A strain expressing RAB-3::mCherry and SAD-1::GFP in PVDs (kyIs445) was mutagenized using ethylmethane sulfonate (EMS) according to standard procedures 25. F1s were cloned onto individual plates, and 40-60 F2 progeny from each F1 parent were screened under a compound fluorescence microscope. Mutants were chosen on the basis of dendritic mislocalization of RAB-3::mCherry and SAD-1::GFP, as detected by a Plan-Neofluar 40x objective on a Zeiss Axioplan2 microscope, resulting in the isolation of ky880.
Similarly, a strain expressing RAB-3::mCherry and UNC-2::GFP in AWCs (kyIs442) was mutagenized using EMS. Through an F1 clonal screen, ky869 was isolated by Y. Saheki as a mutant that showed reduced axonal localization of RAB-3::mCherry and UNC-2::GFP in AWC neurons. Further characterization uncovered a significant increase in dendritic localization of both markers in AWCs.
Mapping and rescue of ky880 and ky869
ky880 and ky869 were mapped to the middle of LGIV using single nucleotide polymorphisms in the CB4856 strain 49 and sequenced to identify unc-33 mutations. Both mutants failed to complement unc-33(e1193) and unc-33(mn407) null alleles for Unc and Egl behavior and for localization of RAB-3::mCherry and SAD-1::GFP in PVD and FLP neurons, but complemented the hypomorphic allele unc-33(e204) for all phenotypes. This intragenic complementation suggests that unc-33 has two interdependent functions, and is consistent with the reported oligomerization of UNC-33/CRMP proteins 27. A plasmid driving the expression of an UNC-33L cDNA under the regulation of the pan-neuronal tag-168 promoter rescued uncoordinated movement, egg-laying behavior, and RAB-3::mCherry and SAD-1::GFP localization in PVD neurons in unc-33(ky880) and unc-33(mn407) mutants.
Fluorescence microscopy and quantification
Animals were mounted on 2% agarose pads in 10 mM sodium azide. L4 larvae or young adult animals were used for analysis of fluorescently tagged markers in PVD, whereas L1 stage and L4 stage larvae were examined for the analysis of ODR-10::GFP in AWB neurons. Wild-type animals generally showed >15 bright RAB-3::mCherry and SAD-1::GFP puncta in PVD axons, no dendritic puncta, and faint or nonexistent fluorescence in the primary dendrites, with modest variations depending on expression level of the transgene. Animals were scored as having reduced axonal labeling if there were fewer than 12 bright puncta in PVD axons, and were scored as having ectopic dendritic labeling if bright RAB-3::mCherry or SAD-1::GFP puncta were present in PVD secondary dendrite branches. Several independent transgenic lines were examined for each transgene in each genetic background.
Z-stacks of fluorescent images were acquired on a Zeiss Axioplan2 imaging system or on a Zeiss LSM510 META laser scanning confocal imaging system. Wide-field and confocal z-stacks were processed using Metamorph or ImageJ (NIH) to obtain maximum intensity projections.
For the quantification of fluorescence intensities of RAB-3::mCherry and SAD-1::GFP in PVD axons and dendrites, fluorescent z-stacks were acquired under consistent detector settings using a Hamamatsu Photonics C2400 CCD camera and Metamorph software, under a 40x Plan-Neofluar on the Zeiss Axioplan2 imaging system. ImageJ was used to measure fluorescence intensities of maximum projections of axonal and dendritic focal planes. For each animal, background intensity was subtracted and fluorescent clusters with signals above an arbitrary threshold were scored for total fluorescent intensity. 10-17 worms per genotype were scored.
For the quantification of UNC-33::GFP in PVD axonal domains, fluorescent z-stacks were obtained as above, using the 63x Plan-Apochromat objective on the Zeiss Axioplan2 system, and fluorescence measurements were performed using Metamorph. Z-stack planes with ‘axonal proximal domain’ and ‘axonal initial domain’ in focus were selected for maximum intensitiy projections. Background fluorescence was substracted for each image, and a line tool was used to measure the fluorescence along the axonal domains. This average fluorescence for each axonal segment was used to calculate the ‘axon initial’/‘axon proximal’ ratio.
Heat-shock experiments
Animals were synchronized by allowing bleached eggs to hatch overnight in M9 buffer (22 mM KH2PO4, 22 mM Na2HPO4, 85 mM NaCl, 1 mM MgSO4) without food; the resulting larvae, synchronized at the L1 larval stage, were then fed and allowed to develop at 21-23°C. Animals were provided a 2 hour pulse of heat-shock at 33°C at varying time points, then recovered at 21-23°C. For each heat-shock time point, a corresponding group of animals was examined using DIC on the Zeiss Axioplan2 microscope to confirm the developmental stage at which animals were being heat-shocked. Adult animals were scored for the localization of RAB-3::mCherry and SAD-1::GFP in PVDs.
EBP-2::GFP dynamic imaging
Nematodes were mounted on 2% agarose pads, anesthetized with 6 mM levamisole, and imaged using a Zeiss Axio Imager Z1 microscope and the AxioCam MRm camera. A 63x/1.4 Plan-Apochromat objective was used for acquisition. EBP-2::GFP movies were acquired using an exposure time of 800ms, every 1s, for 60-120 s. Movies were processed and kymographs were analyzed using ImageJ.
Immunofluorescence
Antibodies used as well as their corresponding dilutions are as follows: DM1A anti-α-tubulin (mouse monoclonal, Sigma) 1:400, GT335 anti-glutamylated tubulin (mouse monoclonal, a gift from C. Janke and B. Edde, CNRS) 1:500, 6-11B-1 anti-acetylated-α-tubulin (mouse monoclonal, Sigma) 1:500, AB3201 anti-detyrosinated-α-tubulin (rabbit polyclonal, Millipore) 1:200, TUB-1A2 anti-tyrosinated-α-tubulin (mouse monoclonal, Sigma) 1:200, G10362 anti-GFP (rabbit monoclonal, Invitrogen) 1:200, 24H11 anti-UNC104 (mouse monoclonal, S. Koushika, unpub.) 1:250. Rabbit polyclonal antisera (Q3201 and Q3203) against an UNC-33L – specific fragment of UNC-33 (amino acid 29 to amino acid 128 of UNC-33L sequence) were generated using genomic antibody technology by Strategic Diagnostics, DE, and used at 1:250 dilution for immunostaining. Antisera from both rabbits yielded similar UNC-33L localization results. The secondary antibodies, Alexa Fluor 488-conjugated goat anti-mouse IgG, Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 594-conjugated goat anti-mouse IgG (Invitrogen) were used at 1:400 or 1:500 dilutions.
Whole mount immunostaining was performed according to the peroxide tube fixation protocol, except that animals were fixed in 2% paraformaldehyde as described previously 50. Briefly, nematodes were rocked in the fixation solution for 20 min at room temperature prior to freezing, and overnight at 4°C post freezing. Fixed animals were permeabilized in TTB buffer (100 mM Tris pH 7.4, 1% Triton X-100, 1 mM EDTA buffer) containing 1% β-mercaptoethanol for 2 hrs at 37 C, washed with BO3 buffer (10 mM H3BO3 pH 9.4, 0.1 % Triton X-100), reduced by 15 min incubation with 10 mM dithiothreitol in BO3 buffer at 37°C, washed with BO3 buffer, and sulfhydryl groups were oxidized by gentle agitation treatment with BO3 buffer containing 1% H2O2 for 1 hour at 25°C. Samples were washed and incubated for 15-30 min with ABB buffer (PBS containing 0.5 % Triton X-100, 1 mM EDTA, 0.05% sodium azide and 0.1% BSA), and stored at 4°C in ABA buffer (PBS containing 0.5 % Triton X-100, 1 mM EDTA, 0.05% sodium azide and 1% BSA) until further use. Animals were gently rocked and incubated overnight at 4°C in primary antibody solutions in ABA buffer, washed with ABB buffer; rocked in secondary antibody solutions in ABA buffer for 2 hours at room temperature, and washed with ABB buffer. The resulting animals were mounted on 2% agarose pads containing 50 mM Tris (pH 8.5) and 5 mM MgCl2 for visualization under the microscope.
For dual labeling of α-tubulin and soluble GFP in head neurons, the abovementioned protocol was modified as follows: animals were pre-chilled on ice 10 min prior to addition of fixative, and were incubated in fixative for 15 min at room temperature, followed by a flash freeze-thaw cycle, followed by incubation overnight at 4°C. Primary antibody and secondary antibody incubations were both carried out overnight at 4°C, followed by washes in ABB buffer at room temperature. Alexa Fluor 594 goat-anti-mouse IgG was used against DM1A anti-tubulin antibody, and Alexa Fluor 488 goat-anti-rabbit IgG was used against anti-GFP.
Fluorescence quantification of axonal versus dendritic immunofluorescence of α-tubulin and soluble GFP
Fluorescent z-stacks were acquired under consistent settings (30 ms and 50 ms exposures for α-tubulin and GFP respectively) with a Zeiss AxioCam MRm camera under the 40× Plan-Neofluar objective on a Zeiss Axio Imager Z1 scope. Stacks were processed using ImageJ to obtain maximum intensity projections of axonal planes (amphid commissure) and that of dendritic planes (distal dendrite region). Background fluorescence was substracted for each maximum projection image, after which axonal or distal dendrite regions were traced with a freehand tool in ImageJ. The same procedure was followed and similar regions of interest were selected for the α-tubulin and GFP images of each animal to obtain the corresponding total fluorescence intensities, and the ratio of the total fluorescence intensity in dendritic vs axonal regions was calculated.
Statistical analysis
Statistical analysis was performed using the t-test with Bonferroni correction or the Mann-Whitney U-test as appropriate.
Supplementary Material
Acknowledgements
We thank C. Ghenoiu, P. Nurse, Y. Saheki, S. Shaham, T. Starich, M. Tsunozaki, M. Heiman, A. Kelly and members of our labs for thoughtful advice and comments on the manuscript, C. Janke and B. Edde for antibodies, and the Caenorhabditis Genetics Center (CGC) for strains. C.I.B. and K.S. are Investigators of the Howard Hughes Medical Institute. This work was supported by the Howard Hughes Medical Institute.
Footnotes
Author Contributions
T.A.M. designed, conducted, and interpreted most experiments and wrote the paper; M.K., G.J.W., K.S., L.W., and J.E.S. conducted and interpreted individual experiments with unc-33 and unc-44 mutants; S.P.K. generated the UNC-104 antibody and helped design transport experiments; C.I.B. designed and interpreted experiments and wrote the paper.
References
- 1.Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 2.Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Herman RK, Shaw JE. Analysis of the Caenorhabditis elegans axonal guidance and outgrowth gene unc-33. Genetics. 1992;132:675–689. doi: 10.1093/genetics/132.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura T, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- 6.Inagaki N, et al. CRMP-2 induces axons in cultured hippocampal neurons. Nat Neurosci. 2001;4:781–782. doi: 10.1038/90476. [DOI] [PubMed] [Google Scholar]
- 7.Fukata Y, et al. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- 8.Arimura N, et al. Anterograde transport of TrkB in axons is mediated by direct interaction with Slp1 and Rab27. Dev Cell. 2009;16:675–686. doi: 10.1016/j.devcel.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Kawano Y, et al. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 11.Burton PR, Paige JL. Polarity of axoplasmic microtubules in the olfactory nerve of the frog. Proc Natl Acad Sci U S A. 1981;78:3269–3273. doi: 10.1073/pnas.78.5.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidemann SR, Landers JM, Hamborg MA. Polarity orientation of axonal microtubules. J Cell Biol. 1981;91:661–665. doi: 10.1083/jcb.91.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton PR. Ultrastructure of the olfactory neuron of the bullfrog: the dendrite and its microtubules. J Comp Neurol. 1985;242:147–160. doi: 10.1002/cne.902420202. [DOI] [PubMed] [Google Scholar]
- 14.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubb MS, Burrone J. Building and maintaining the axon initial segment. Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winckler B, Forscher P, Mellman I. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 1999;397:698–701. doi: 10.1038/17806. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–746. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedstrom KL, Ogawa Y, Rasband MN. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J Cell Biol. 2008;183:635–640. doi: 10.1083/jcb.200806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song AH, et al. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Sobotzik JM, et al. AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc Natl Acad Sci U S A. 2009;106:17564–17569. doi: 10.1073/pnas.0909267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn CC, et al. TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J Neurosci. 2003;23:2815–2823. doi: 10.1523/JNEUROSCI.23-07-02815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa Y, et al. Spectrins and ankyrinB constitute a specialized paranodal cytoskeleton. J Neurosci. 2006;26:5230–5239. doi: 10.1523/JNEUROSCI.0425-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brot S, et al. CRMP5 interacts with tubulin to inhibit neurite outgrowth, thereby modulating the function of CRMP2. J Neurosci. 2010;30:10639–10654. doi: 10.1523/JNEUROSCI.0059-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 25.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otsuka AJ, et al. An ankyrin-related gene (unc-44) is necessary for proper axonal guidance in Caenorhabditis elegans. J Cell Biol. 1995;129:1081–1092. doi: 10.1083/jcb.129.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuboi D, Hikita T, Qadota H, Amano M, Kaibuchi K. Regulatory machinery of UNC-33 Ce-CRMP localization in neurites during neuronal development in Caenorhabditis elegans. J Neurochem. 2005;95:1629–1641. doi: 10.1111/j.1471-4159.2005.03490.x. [DOI] [PubMed] [Google Scholar]
- 28.Albeg A, et al. C. elegans multi-dendritic sensory neurons: morphology and function. Mol Cell Neurosci. 2011;46:308–317. doi: 10.1016/j.mcn.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 30.Treinin M, Gillo B, Liebman L, Chalfie M. Two functionally dependent acetylcholine subunits are encoded in a single Caenorhabditis elegans operon. Proc Natl Acad Sci U S A. 1998;95:15492–15495. doi: 10.1073/pnas.95.26.15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonet ML, et al. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crump JG, Zhen M, Jin Y, Bargmann CI. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron. 2001;29:115–129. doi: 10.1016/s0896-6273(01)00184-2. [DOI] [PubMed] [Google Scholar]
- 33.Yonekawa Y, et al. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 35.Dwyer ND, Adler CE, Crump JG, L'Etoile ND, Bargmann CI. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 2001;31:277–287. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhou HM, Brust-Mascher I, Scholey JM. Direct visualization of the movement of the monomeric axonal transport motor UNC-104 along neuronal processes in living Caenorhabditis elegans. J Neurosci. 2001;21:3749–3755. doi: 10.1523/JNEUROSCI.21-11-03749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dwyer ND, Troemel ER, Sengupta P, Bargmann CI. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell. 1998;93:455–466. doi: 10.1016/s0092-8674(00)81173-3. [DOI] [PubMed] [Google Scholar]
- 38.Wolff A, et al. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol. 1992;59:425–432. [PubMed] [Google Scholar]
- 39.Siddiqui SS, Aamodt E, Rastinejad F, Culotti J. Anti-tubulin monoclonal antibodies that bind to specific neurons in Caenorhabditis elegans. J Neurosci. 1989;9:2963–2972. doi: 10.1523/JNEUROSCI.09-08-02963.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 41.Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-beta signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adler CE, Fetter RD, Bargmann CI. UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat Neurosci. 2006;9:511–518. doi: 10.1038/nn1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ou CY, et al. Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell. 2010;141:846–858. doi: 10.1016/j.cell.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolls MM, et al. Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyrand P, Weimann JM, Marder E. Multiple axonal spike initiation zones in a motor neuron: serotonin activation. J Neurosci. 1992;12:2803–2812. doi: 10.1523/JNEUROSCI.12-07-02803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palay SL, Sotelo C, Peters A, Orkand PM. The axon hillock and the initial segment. J Cell Biol. 1968;38:193–201. doi: 10.1083/jcb.38.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 49.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 50.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.