Abstract
Only a few distantly related mammals and birds have the trait of complex vocal learning, which is the ability to imitate novel sounds. This ability is critical for speech acquisition and production in humans, and is attributed to specialized forebrain vocal control circuits that have several unique connections relative to adjacent brain circuits. As a result, it has been hypothesized that there could exist convergent changes in genes involved in neural connectivity of vocal learning circuits. In support of this hypothesis, expanding on our related study (Pfenning et al. [2014] Science 346: 1256846), here we show that the forebrain part of this circuit that makes a relatively rare direct connection to brainstem vocal motor neurons in independent lineages of vocal learning birds (songbird, parrot, and hummingbird) has specialized regulation of axon guidance genes from the SLIT–ROBO molecular pathway. The SLIT1 ligand was differentially downregulated in the motor song output nucleus that makes the direct projection, whereas its receptor ROBO1 was developmentally upregulated during critical periods for vocal learning. Vocal nonlearning bird species and male mice, which have much more limited vocal plasticity and associated circuits, did not show comparable specialized regulation of SLIT–ROBO genes in their nonvocal motor cortical regions. These findings are consistent with SLIT and ROBO gene dysfunctions associated with autism, dyslexia, and speech sound language disorders and suggest that convergent evolution of vocal learning was associated with convergent changes in the SLIT–ROBO axon guidance pathway. J. Comp. Neurol. 523:892–906, 2015. © 2014 Wiley Periodicals, Inc.
Keywords: songbird, parrot, hummingbird, neural connectivity, axon guidance, vocal learning
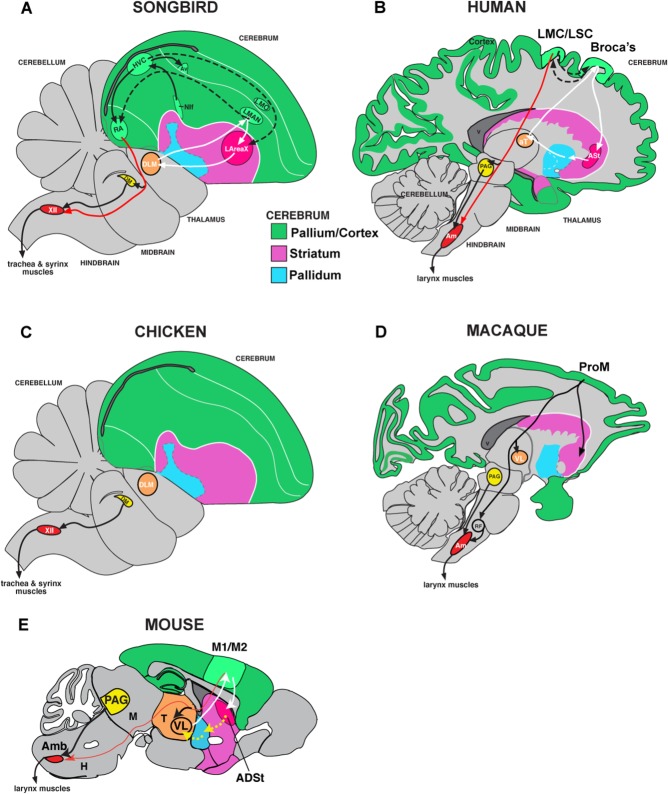
Vocal learning is the ability to imitate sounds, and is critical for spoken language. It is a rare trait found in several independent lineages of mammals (humans, cetaceans, elephants, pinnepeds, and bats) and birds (songbirds, parrots, and hummingbirds) (Petkov and Jarvis, 2012; Jarvis, 2013). Within birds, vocal learning is proposed to have evolved at least two to three independent times: in oscine songbirds and parrots, or their common ancestor, as they are sister lineages with only two vocal nonlearning lineages that separate them (New Zealand wrens and suboscine Passeriformes); and in hummingbirds, which are more distantly related to songbirds and parrots (Suh et al., 2011; Jarvis et al., 2014). The brains of all vocal learners studied to date (birds and humans) have forebrain song or speech control circuits not found in vocal nonlearners (including nonhuman primates) (Jarvis, 2004; Petkov and Jarvis, 2012), or found at very rudimentary levels in mice and suboscine Passeriformes closely related to songbirds (Arriaga et al., 2012; Arriaga and Jarvis, 2013; Liu et al., 2013) (Fig. 1). One of the relatively unique connections of this circuit is a robust direct projection from a vocal motor cortical region (the robust nucleus of the arcopallium [RA] in vocal learning birds; the laryngeal motor cortex [LMC] in humans) to the brainstem motor neurons that control the vocal muscles (syrinx in birds; larynx in humans; Fig. 1A,B) (Jurgens, 2002; Jarvis, 2004; Fitch et al., 2010). Direct projections from the cortex to motor neurons are rare, and when found they are often associated with fine motor skill learning and control (Lemon, 2008). Based on these findings, we hypothesized that brain regions with the direct motor projection may have unique convergent changes in expression of genes that control neural connectivity.
Figure 1.
Brain pathways for vocal behavior in vocal learning and vocal nonlearning birds and mammals. A: Drawing of a zebra finch male brain section showing connectivity of posterior (HVC, RA) and anterior (LMAN, area X) song pathways. B: Drawing of a human brain section showing proposed vocal pathway connectivity including the LMC/LSC and part of the anterior striatum (ASt) that shows convergence with songbird RA and area X (Pfenning et al., 2014). Black arrows, connections and regions of the posterior vocal motor pathway; white arrows, connections and regions of the anterior vocal pathway; dashed arrows, connections between the two pathways. Red arrows show the direct projections found only in vocal learners, from vocal motor cortex regions to brainstem vocal motor neurons. C: Connectivity of a vocal nonlearning bird, chicken, showing the absence of forebrain song nuclei. D: Connectivity of vocal nonlearning primates (macaque shown), which has forebrain regions that make indirect projections to the nucleus ambiguus (Amb), although they are not necessary for vocalizations. E: Proposed mouse vocal pathway connectivity, which has a very sparse direct projection to the Amb. For abbreviations, see list. Figure panels modified from Arriaga et al., 2012; Petkov and Jarvis, 2012; and Pfenning et al., 2014.
To test this hypothesis, we have been screening for potential differences in expression of genes in the vocal motor cortex and its projection targets, in humans and vocal learning birds, compared with control vocal nonlearning species. We previously found convergent molecular changes in two genes involved in neuroprotection (parvalbumin and DUSP1), which we hypothesized may be necessary for buffering highly active song and speech circuits (Hara et al., 2012; Horita et al., 2012). However, there have been no discoveries of convergent molecular changes of genes involved in neural connectivity beyond two vocal learning avian species (songbird and parrot) (Matsunaga et al., 2008). In a thesis study (Wang, 2011), we screened for protein coding sequence mutations in the genomes of vocal learning mammals and discovered that the ROBO1 axon guidance receptor has mutations enriched in mammalian vocal learners. In our related study (Pfenning et al., 2014), we performed genome-scale expression microarray analyses across vocal learning and vocal nonlearning primate and bird species, and discovered that the ligand of ROBO1, SLIT1, was among the top three most convergent differentially expressed genes in the human LMC and the RA analogs of vocal learning bird lineages. ROBO1 and SLIT1 are viable candidates for involvement in speech evolution and function, in that mutations in ROBO1 are associated with speech sound disorder and dyslexia (Hannula-Jouppi et al., 2005; Bates et al., 2011). Here we present a more detailed characterization of ROBO1 and SLIT1 mRNA expression, as well as other genes from the SLIT–ROBO family, in vocal learning and vocal nonlearning birds, and the putative LMC of mice. Our findings are consistent with the possibility that these genes play specialized roles in the repeated evolution and function of vocal learning circuits.
Materials and Methods
Animals
We collected fresh-frozen brains of 18 juvenile zebra finches (Taeniopygia guttata) from posthatch day (PHD) ∼20, ∼35, and ∼65 (n = 3 males and 3 females of each age), 6 male and 6 female adult (PHD > 90) zebra finches (a songbird), 4 male adult budgerigars (Melopsittacus undulates; a parrot), 3 male adult Anna's hummingbirds (Calypte anna), 4 male adult ring doves (Streptophilia risoria), 4 male adult quails (Coturnix cortunix japonica), 2 PHD2 male quails, and 2 male adult mice (Mus musculus; C57BL/6J). Song learning in zebra finch males occurs in three phases, beginning with a sensory phase of auditory learning and subsong production between ∼PHD 35 and 45, a sensory–motor plastic song phase at ∼PHD 45–75, and a crystallization song phase during which the imitated song becomes stereotyped at ∼PHD 90 (Immelmann, 1969; Tchernichovski et al., 2004). We chose males, as they are more often the vocal learning sex when sex differences exist in vocal learning abilities. The PHD20 and PHD35 zebra finch juveniles, and PHD2 quails, do not yet have gender-specific plumage, and so we used polymerase chain reaction (PCR) on the CHD (Chromo Helicase DNA-binding) gene to identify their gender following a protocol in Wada et al. (2006). The specific vocal nonlearning species were chosen for several reasons: 1) known experimental testing for absence of vocal learning (Nottebohm and Nottebohm, 1971; Deregnaucourt et al., 2001); and 2) phylogenetic relationships. For the latter, when we began the project, hummingbirds and pigeons (i.e., our ring dove) were considered closely related (Hackett et al., 2008). That proposed relationship has changed based on a new genome-scale phylogenetic tree, shown in our related study (Jarvis et al., 2014). The ring dove is now in a group of species more basal in Neoaves. Nevertheless, the species employed do represent logical choices to account for phylogenetic variation (Neoaves [ring dove] and a Galloanseres [quail] vocal nonlearning controls).
All animals were obtained from our breeding colonies at the Duke University Medical Center (Durham, NC), except for the Anna's hummingbirds, which were obtained with the help of Dr. Douglas Altshuler at the University of California, Riverside for a previous study (Feenders et al., 2008). Brains were collected from quiet animals after an overnight period of silence in a sound isolation chamber (cooler box of ∼ 31 L × 13 W × 14 H inches insulated with soundproof foam) with no singing (determined by automated recording with an Avisoft recorder [http://www.avisoft.com]) or after overnight in the nest for PHD20 animals. This prevents detection of differences in gene expression due to neural activity associated with singing and movement (Feenders et al., 2008). All animal procedures were approved by the Duke University Institutional Animal Care and Use Committee.
In situ hybridization
In situ hybridizations were conducted on 12-µm coronal sections following our previously published protocol (Chen et al., 2012), at 65°C for the hybridization and washes for zebra finch brain sections, or at 60°C for all other species. Zebra finch clones for ROBO1, ROBO2, SLIT1, SLIT2, and SLIT3 were used to make 35S-radioactive riboprobes for hybridization to all avian species. We used zebra finch brain mRNA from a female in PCR reactions with primers generated against conserved sequences in the chicken, mouse, human, Xenopus (frog), and/or zebra fish genomes (Table1). The zebra finch PCR products were cloned into the T overhangs of the PGEM Teasy vector (Promega, Madison, WI), and both ends of the insert were sequenced with M13 forward and reverse primers to verify the identity of the gene. The SLIT1 zebra finch clone was completely sequenced and deposited into GenBank (accession no. KF738084). For riboprobes, the inserts were amplified in a PCR reaction with the M13 primers and purified, and the antisense strand was made with SP6 or T7 RNA polymerases (Table1). Mouse clones were from the mouse Mammalian Gene Collection (full-length cDNA clones) with accession numbers as follows: ROBO1 (CA326894), ROBO2 (BC055333), SLIT1 (BC057131), SLIT2 (BC059267), and SLIT3 (BU151959). The antisense strand was made with T3 RNA polymerase except for SLIT3, which was made with T7 RNA polymerase. The mouse sense strands were generated with T7 RNA polymerase except for SLIT3, which was generated with the SP6 RNA polymerase. The sense strand of each gene for each species did not result in any specific signal (data not shown).
Table 1.
Zebra Finch ROBO and SLIT Clones Generated for This Study1
| Gene | Clone ID | Foward primer | Reverse primer | Direction α-sense |
|---|---|---|---|---|
| Robo1 | 1–11-08 #03 | AGTCCCGTCTTTTACCTTCAC | CCCAGCCATTGATCATGGA | Minus SP6 |
| Robo2 | 1–25-08 #12 | GAGATGGAGGACTAATGAGCAA | ACAGAGCTGTCTAGATTGTCCAT | Minus SP6 |
| Slit1 | 1–31-08 #15 | AACCCNTTCAACTGCAACTGCCA | CACTTGVTGGGYTTYTCCAC | Plus T7 |
| Slit2 | 4–17-08 #13 | CAGATTGCCACAGACGAAGACAG | TTCCCCTCGACAAGAGATTTCT | Plus T7 |
| Slit3 | 1–31-08 #07 | CCATTTGTGTGCGACTGCCA | CTTATGCTGTTRTTGCTCA | Minus SP6 |
Listed are the clone IDs (date cloned and colony #), the forward (5′) and reverse (3′) primers used to amplify the cDNA product from zebra finch female brain, and the direction of the antisense and polymerase used to generate antisense riboprobes.
Quantification
Quantification of gene expression in identified forebrain regions was conducted by using a previously described method (Jarvis et al., 2013). In brief, in situ hybridization images from x-ray film or emulsion-dipped slides were digitized with a macrozoom microscope (Olympus DP Controller 3.2.1.276 software), transferred to Photoshop (version CS3, Adobe Systems, San Jose, CA), and converted to grayscale to reduce intensity differences created by differences in cresyl violet staining between experiments and to provide a comparable grayscale range among images. Regions of interest were outlined with the Lasso Tool (Photoshop), and the average pixel density was calculated by using the histogram function. The average adjacent background level on the glass slide without tissue was subtracted, and the resultant values were averaged across at least two different brain sections. For vocal learning species, the regions of interest included the RA analog and the medially adjacent intermediate arcopallium (Ai) that is active during movement (Feenders et al., 2008). The vocal nonlearning species do not have an RA analog, and thus we quantified two adjacent regions within the movement-activated Ai region, where the RA analog would be expected to be found.
For the brainstem regions, a more sensitive quantification method was needed due to sparsely spaced large motor neurons with large gene expression differences from other nearby cells. Thus, we took brightfield sections of the motor neurons at 63× magnification, and then used a threshold function in ImageJ v1.44 (http://imagej.nih.gov/ij/) to mark the maximum amount of grains without marking other tissue objects. Then the number of silver grains in the image was counted automatically (using the measure function), and that number was divided by the number of motor neurons (manually counted) in each image. The average adjacent background level on the glass slide without tissue was subtracted, and the resultant values were averaged across at least two different brain sections.
Figure processing
To represent the full color range of the in situ hybridization images in figures, the levels function of Photoshop was used to expand the image RGB levels to a full 0–250 range, by moving the black (0, left) and white (255, right) sliders to the edges of the brightness histogram values for the image in the RGB color setting.
Results
SLIT1 and ROBO1 are differentially regulated in the RA analog of vocal learners
We focused on the brain region considered to show the most specialized connection in vocal learning species, the RA analog (RA in songbird, AAC in parrot, and VA in hummingbird), which makes a direct projection to brainstem vocal motor neurons (nXIIts) (Paton et al., 1981; Wild, 1997; Gahr, 2000); the song nuclei of different vocal learning lineages have been given different names, because of the hypothesis that each lineage evolved their nuclei independently of a common ancestor (Striedter, 1994; Jarvis and Mello, 2000; Jarvis et al., 2000). We examined expression in the frontal plane, which allowed us to make expression comparisons directly adjacent to the more lateral nonvocal motor arcopallium in the same brain sections (Feenders et al., 2008).
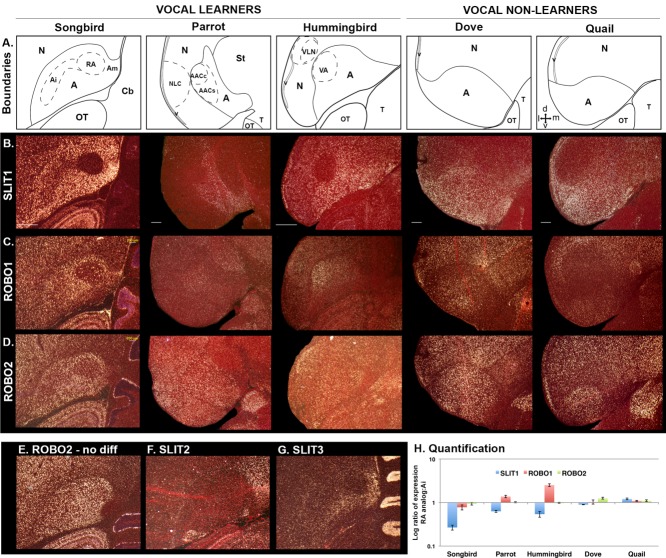
We found that the RA analog of all three vocal learning lineages showed striking SLIT1 mRNA downregulation relative to the surrounding arcopallium (Fig. 2A,B,H) (Pfenning et al., 2014). The directly adjacent lateral intermediate arcopallium (Ai), which does not make direct projections to brainstem motor neurons (Dubbeldam, 1998; Bottjer et al., 2000), had intermediate levels of SLIT1 relative to the remaining arcopallium, which projects to other forebrain regions (Wild, 1997; Bottjer et al., 2000). Furthermore, the AAC of parrots has a core subdivision (AAC core) that forms the direct projection to nXIIts vocal motor neurons and a surrounding ventrally skewed shell subdivision (AAC shell) unique to parrots that projects to other forebrain song nuclei (Durand et al., 1997; Chakraborty et al., in press). Only the parrot AAC core showed the differential SLIT1 downregulation (Fig. 2A,B). In hummingbirds, the pattern of SLIT1 downregulation in the RA analog (the VA) was similar to that in songbirds, which have only a core song nucleus that, like the songbird RA and parrot AAC core, makes a direct projection to brainstem vocal motor neurons (Gahr, 2000). No such large differential downregulation of SLIT1 was seen within the arcopallium of the two vocal nonlearning avian species tested, ring doves and quails (Fig. 2A,B,H).
Figure 2.
Brain expression of SLIT and ROBO genes in vocal learning and vocal nonlearning birds. A: Diagrams of the arcopallium region in coronal view in the species studied (zebra finch, a songbird; budgerigar, a parrot; Anna's hummingbird; ring dove; Japanese quail). Inset with arrows in the quail panel is a compass for orientation: d, dorsal; m, medial; v, ventral; l, lateral. B–D: mRNA expression patterns of SLIT1 (B), ROBO1 (C), and ROBO2 (D) in the RA analog and the surrounding arcopallium of avian vocal learners and vocal nonlearners. White silver grains show mRNA expression in the darkfield view; red label is cresyl violet stain. Images are expanded views of the in situ hybridizations shown in Pfenning et al. (2014), except that the zebra finch is from a different animal. E: Expression pattern of ROBO2 from another zebra finch that did not show a differential expression in RA. F,G: Expression pattern of SLIT2 (F) and SLIT3 (G) in the zebra finch. H. Quantification of SLIT1, ROBO1, and ROBO2 mRNA expression in the RA analog in vocal learners or the intermediate arcopallium in vocal nonlearners versus adjacent motor intermediate arcopallium in each species. Only SLIT1 was significantly differentially expressed in the same direction in all vocal learners versus vocal nonlearners (P < 0.05; paired t-tests; n= 3–6 per species). Error bars, SEM. For abbreviations, see list. Scale bar = 500 µm in second row (applies to rows below). Scale bar in panel B, zebra finch, applies to E–G.
Expression of the SLIT1 receptor, ROBO1, was high only in isolated cells in songbird RA, whereas it was high in most cells in the surrounding arcopallium (Fig. 2C). In contrast to songbirds, ROBO1 expression in the RA analog of parrot (the AAC core portion) and hummingbird (the VA) was higher than in the surrounding arcopallium (Fig. 2C,H). As in songbirds, there was higher ROBO1 expression in the Ai adjacent to the RA analog relative to the rest of the arcopallium. However, such localized differential ROBO1 expression was also seen in the Ai of the two vocal nonlearning species (Fig. 2C), and thus was not unique to vocal learners.
Expression of the sister receptor gene ROBO2 in the songbird RA was variable among animals, with some (∼50%) showing lower expression in the RA relative to the surrounding arcopallium (Fig. 2D) and others showing no difference (Fig. 2E). This difference among animals was not related to immediate singing behavior, as the animals examined were taken after an overnight period of silence. There was no detectable difference in ROBO2 expression for all animals within the RA analogs of the parrot and the hummingbird or within the central arcopallium of the two vocal nonlearners, dove and quail, relative to the surrounding arcopallium (Fig. 2D, H).
In contrast to SLIT1, expression of the sister genes SLIT2 and SLIT3 was low throughout most of the songbird arcopallium, including the RA (Fig. 2F,G), and thus we did not test them further in other species. The most medial part of the arcopallium (Am; a limbic region; Reiner et al., 2004) had higher patches of expression of all three SLIT genes, but this did not result in the RA analog having specialized expression for SLIT2 and SLIT3 relative to the rest of the arcopallium.
Intriguingly, based on enrichment of SLIT1 expression in the arcopallium, we noted that the arcopallium in all three vocal learners is positioned more medially in the forebrain than in the two vocal nonlearning species tested. In the vocal learners, the adjacent nidopallium is both dorsal and lateral to the arcopallium, whereas in the two vocal nonlearners, the nidopallium is all dorsal. Future experiments on other vocal nonlearners will be necessary to determine whether this is a trait specific to vocal learners.
Overall, these findings demonstrate convergent differential regulation specific to SLIT1 and ROBO1 in the RA analog of vocal learning avian species. Unlike ROBO1, the SLIT1 gene is differentially regulated in the same direction across all vocal learning avian species.
Other song nuclei and broader brain expression profiles
We qualitatively examined expression of the SLIT and ROBO genes in other telencephalic song nuclei, particularly in the zebra finch, a songbird. We did not note specialized expression of SLIT1 or ROBO1 in the HVC analogs of vocal learners (Figs. 2B,C, 3A,B); in zebra finches all song nuclei can all be seen in the same plane in sagittal sections (Fig. 3), whereas the HVC analogs in parrots (the NLC) and hummingbirds (the VLN) is directly adjacent to the RA analog in frontal sections (Fig. 2A–D). In contrast to these two genes, ROBO2 appeared to have convergent upregulation in the HVC analogs (Figs. 2D, 3C). We examined only the anterior song nuclei in zebra finches (using sagittal sections) and noted differential downregulation of SLIT1 and ROBO1 in the LMAN and Area X, but no difference for ROBO2 (Fig. 3A-C). We did not note any differential expression of these genes in the smaller songbird song nuclei of the mesopallium (the Av and MO) and of the nidopallium (Nif; Figs. 1A, 3).
Figure 3.
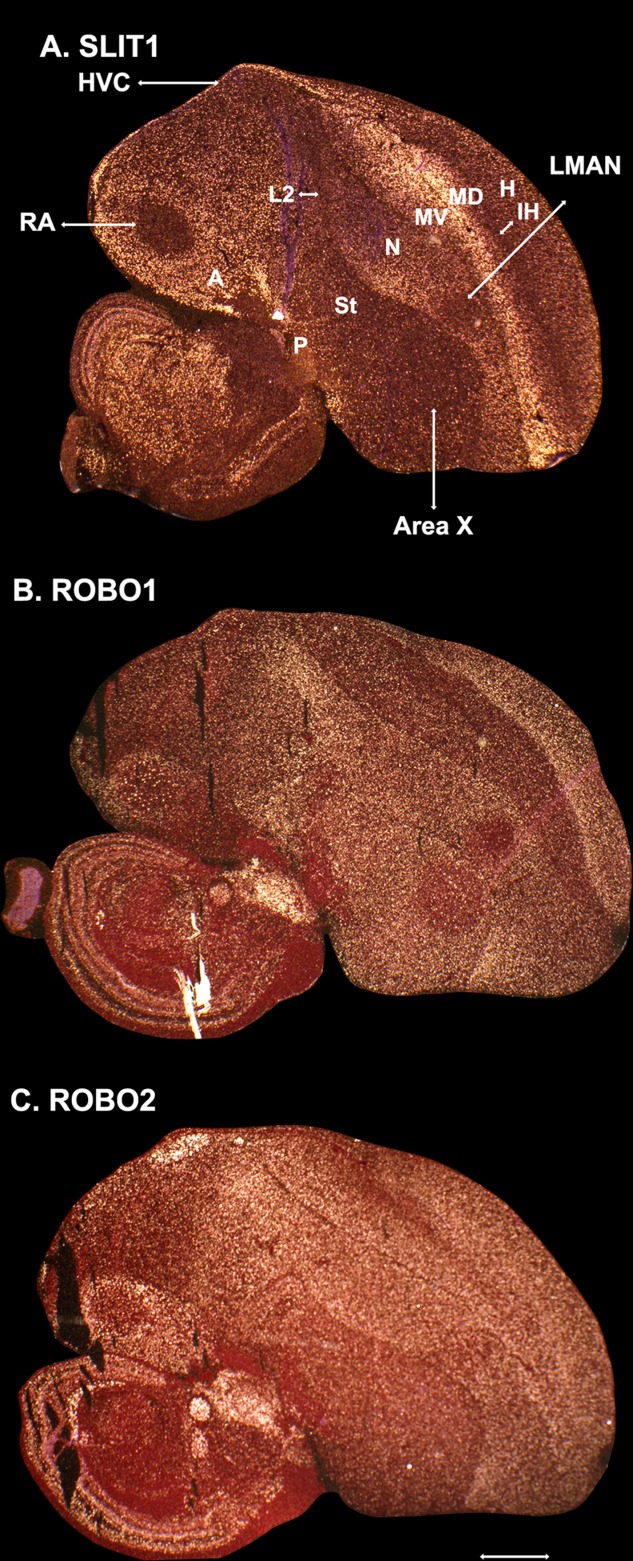
Expression of SLIT and ROBO genes that have prominent expression in the telencephalon, in the sagittal plane in the male zebra finch brain. A: SLIT1 mRNA expression in a plane of section in which all major song nuclei are present (highlighted with lines). B: ROBO1 mRNA expression showing a mostly complementary pattern to SLIT1, except in song nuclei. The white streak is in the midbrain. C: ROBO2 mRNA expression in an adjacent section. For telencephalic subdivision abbreviations in A, see list. Scale bar = 1.0 mm in C (applies to A–C).
In terms of the overall expression patterns of these genes in the telencephalon, we noted that SLIT1 was highest in the arcopallium (quaternary pallium), and had a higher expression gradient toward the lamina between the dorsal (MD) and ventral (MV) mesopallium (tertiary pallium), intermediate levels of expression in the nidopallium and hyperpallium (secondary pallium), and the lowest expression levels in the intercalated pallium (auditory L2 and visual IH; primary pallium), which receives the primary sensory input into the telencephalon; expression levels were lower as well in the striatum (Fig. 3A). ROBO1 had a complementary pattern, with highest expression levels in the intercalated pallium, intermediate levels in the nidopallium plus the hyperpallium and striatum, and the lowest levels in the mesopallium (Fig. 3B). Even in the arcopallium, ROBO1 appeared to be lower where SLIT1 was highest. ROBO2 did not have a complementary pattern to either SLIT1 or ROBO1, and was expressed at comparably high levels in most telencephalic areas except for the striatum, pallidum, and caudal nidopallium (Fig. 3C).
Overall, these findings suggest that, as has been noted in the rat brain (Marillat et al., 2002), SLIT1 and ROBO1 in the avian brain have mostly complementary expression profiles. The findings also support the recent revisions to our understanding of avian brain organization, with semi-mirror organization of the telencephalic cell populations around the mesopallium lamina and lateral ventricle (Jarvis et al., 2013). However, the song nuclei display some striking exceptions to this complementary pattern, most notably for ROBO1 in the RA and the downregulation of both genes in the LMAN and Area X.
Brainstem vocal motor neurons show specialized regulation of ROBO2 in both vocal learning and vocal nonlearning species
We next compared expression of the ROBO and SLIT genes in the target of the RA analog, the nXIIts, with the nearby supraspinal (SSp) neck motor neurons in the same coronal sections. Unlike the nXIIts, the SSp is thought to not receive a direct projection from the arcopallium, but rather an indirect projection from the Ai to the reticular formation (RF) around the SSp, and then from the RF to the SSp in both vocal learners and vocal nonlearners (Dubbeldam, 1998). Both vocal learning and nonlearning species have vocal, neck, and other brainstem and spinal motor neurons, which are the final stations for generation of both innate and learned movements (Wild, 1997; Dubbeldam, 1998).
Similar to the forebrain, we found a complementary pattern of SLIT1 and ROBO1 expression, whereby SLIT1 was low in motor nuclei and high in the surrounding RF (Fig. 4A), and ROBO1 was higher in the motor nuclei (Fig. 4C). SLIT2 was not expressed in this brainstem region (not shown), and SLIT3 was specifically high in the motor nuclei (Fig. 4B); unlike the other genes, ROBO2 was specifically enriched in the nXIIts (Fig. 3D). Brightfield images at high magnification verified these differences, and further revealed that these genes were expressed in the large motor neurons (Fig. 4E,F).
Figure 4.
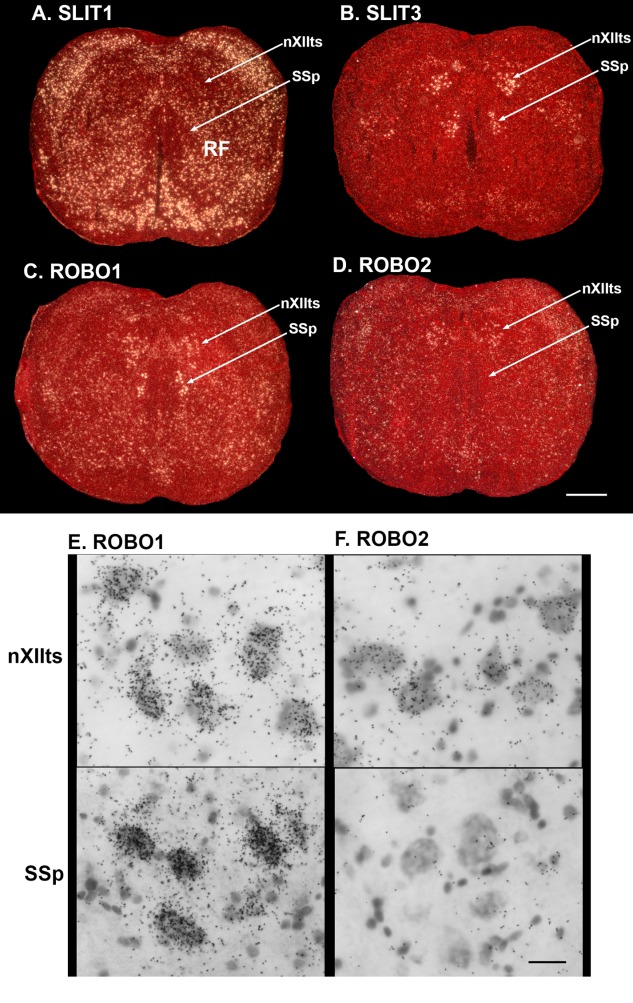
Expression of SLIT and ROBO genes in brainstem vocal (nXIIts) and neck (SSp) motor neurons. A–D: Example in situ hybridization expression patterns of SLIT1 (A), SLIT3 (B), ROBO1 (C), and ROBO2 (D) in the zebra finch brainstem motor neurons. E,F: Brightfield high magnification of ROBO1 (E) and ROBO2 (F) mRNA expression in nXIIts and SSp, showing differences of mRNA levels (measured by black silver grains) in the motor neurons. For abbreviations, see list. Scale bar = 500 µm in D (applies to A–D); 30 µm in F (applies to E,F).
We quantified the ratios of gene expression between the nXIIts and SSp across species, and found that, unlike in the forebrain where specialized expression was restricted to vocal learners, ROBO2 had higher expression in the nXIIts and was nearly absent in the SSp (Figs. 4F, 5) and other motor neurons (not shown) in all vocal learning and vocal nonlearning species examined. For the other genes, most species had lower expression levels of one or more of them in the nXIIts compared with the SSp (Fig. 5). We found no detectable differences between zebra finch males and females (not shown). These findings indicate that the SLIT–ROBO genes have differential expression in the vocal motor neurons, but, unlike their input to the forebrain, the differences are not specific to vocal learners.
Figure 5.
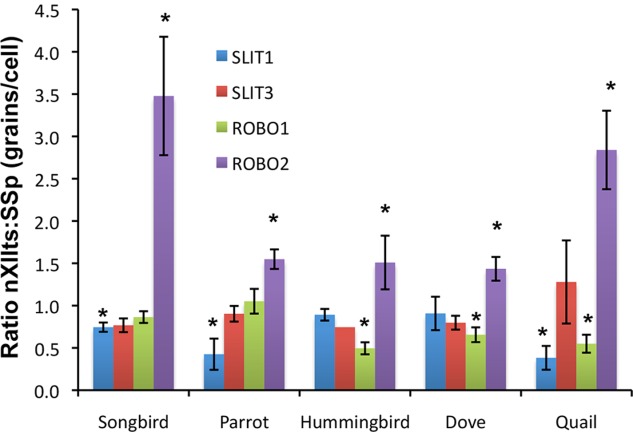
Quantification of nXIIts:SSp gene expression ratios across species. Shown are quantifications of relative differences of four genes from the SLIT-ROBO family in the vocal motor (nXIIts) and neck motor (SSp) neurons of three vocal learning (songbird, parrot, hummingbird) and two vocal nonlearning (dove and quail) species. * P < 0.05; paired t-test between nXIIts and SPP of the same animals, indicating significantly greater or less than a ratio of 1; n = 3–4 per species. Error bars, SEM. For abbreviations, see list.
ROBO1 and SLIT1 specializations in RA are formed during critical periods for vocal learning
In zebra finches, the RA to nXIIts projection is already formed in both sexes by PHD18, but sex differences emerge around PHD30, at which time the RA neuron population and projection continue to grow and develop in males but shrink significantly with a developmental loss of RA neurons in females (Johnson and Sellix, 2000). We found that both zebra finch males and females showed differential downregulation of SLIT1 expression in RA throughout all measured posthatch stages into adulthood, except female adults (Fig. 6A,C). In constrast, males and females exhibited similarly strong ROBO1 expression in the arcopallium with no differential expression in RA by PHD20 (Fig. 6B,C). Significant differential upregulation of ROBO1 expression occurred in the RA of PHD35 males, followed by differential downregulation excepted in isolated cells by PHD65 only in males (Fig. 6B,C). These findings suggest that the mechanism for the specialized regulation of the receptor (ROBO1) and ligand (SLIT1) in the RA are not directly coupled.
Figure 6.
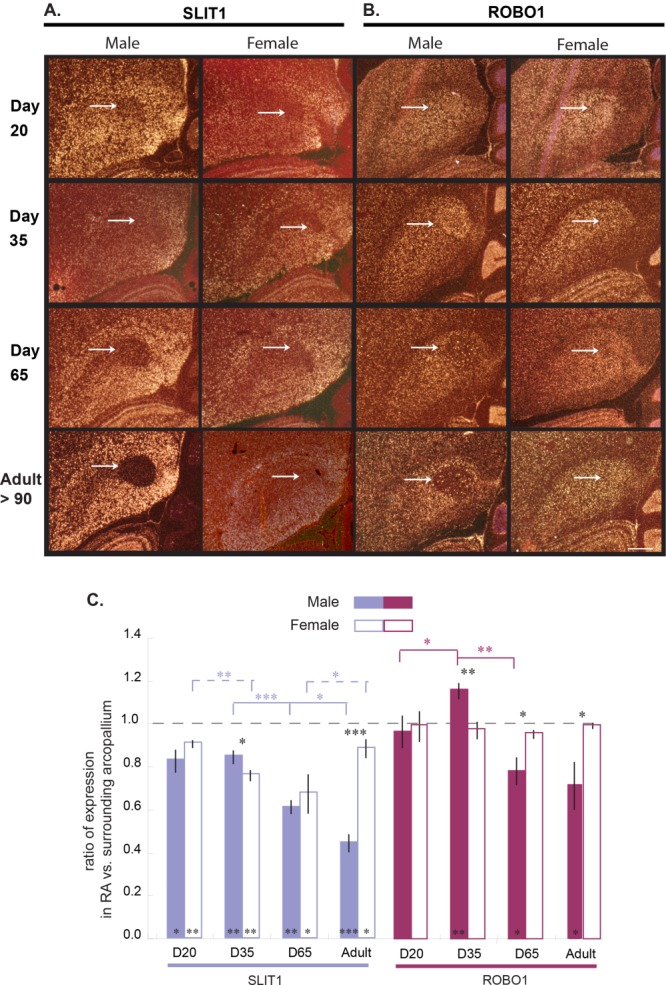
Expression of SLIT1 and ROBO1 in zebra finch RA during song development. A,B: In situ hybridization in darkfield view showing expression of (A) SLIT1 and ROBO1 (B) mRNA (white) in RA of male and female zebra finches during juvenile development: days 20, 35, and 65 post hatch relative to adult (>90). C: Quantification of expression in RA versus adjacent intermediate arcopallium in male (filled bars) and female (open bars) zebra finches. * P < 0.05; ** P < 0.01; *** P < 0.001 (one-tailed t-tests; n = 3 animals/group). Error bars, SD. Scale bar = 500 µm in B (applies to A,B).
We examined SLIT1 expression in 2-day-old juvenile quails (equivalent to ∼PHD20 zebra finches in terms of feeding and walking), to test whether a vocal nonlearning species would show an early posthatch difference that would have been missed in adults. However, we did not observe a distinct downregulation of SLIT1 in the juvenile quail arcopallium (Fig. 7A). We also did not observe a posthatch developmental difference in the XIIts motor neurons of quail (Fig. 7B) or of male zebra finch (Fig. 7C,D). These findings suggest that the specialized expression of SLIT1 in the RA is restricted to vocal learners at an early developmental stage, and is probably not the result of a posthatch difference disappearing in adult vocal nonlearners
Figure 7.
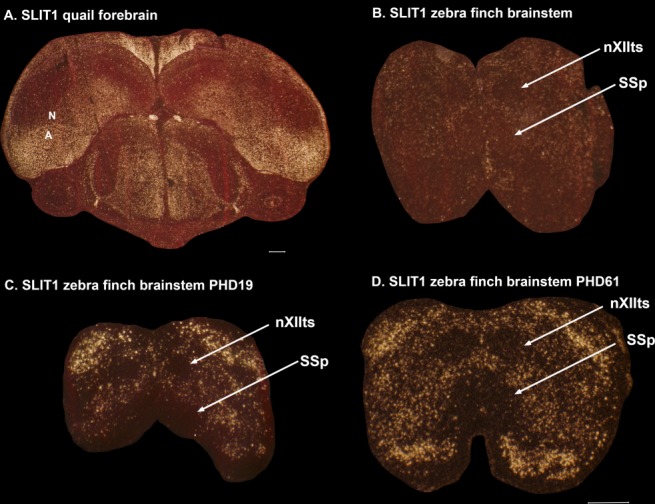
Expression of SLIT1 in quail forebrain and zebra finch brainstem at earlier ages. A: Coronal section through a 2-day-old male quail brain, showing no region of strong differential downregulation of SLIT1. B: Expression of SLIT1 in the brainstem of the same 2-day-old quail, showing that there is no difference from that seen in the motor neurons of zebra finches. C: Expression of SLIT1 in the brainstem at in a PHD (posthatch day) 19 zebra finch. D: Expression of SLIT1 in the brainstem of a PHD61 zebra finch. For abbreviations, see list. Scale bars = 500 μm.
The putative laryngeal motor cortex of mice has SLIT1 and ROBO1 expression patterns similar to those of vocal nonlearning birds
We next examined expression of SLIT and ROBO genes in a recently discovered putative mouse LMC, which has a much sparser projection from its cortical layer 5 neurons to brainstem vocal motor neurons (nucleus ambiguous) compared with the RA analog of vocal learning birds and the LMC of humans (Arriaga et al., 2012; Arriaga and Jarvis, 2013). We found that, similar to vocal nonlearning birds, the mouse layer 5 cells of the putative LMC had high SLIT1 expression in isolated cells and high SLIT2 expression similar to the adjacent cortex (Fig. 8A,B). Also similar to the nonvocal arcopallium of both vocal learning and vocal nonlearning birds, ROBO1 expression was higher in layer 5 cells of the primary (M1) and secondary (M2) motor cortices relative to other regions (Fig. 8C,F), and this was not specific to the putative LMC. Similar to a more general distributed pattern throughout the avian pallium, mouse ROBO2 expression was high in all cortical layers (Fig. 8D). Thus, the mouse brain has a SLIT–ROBO family gene expression pattern in its motor cortex that is more similar to that of vocal nonlearning birds.
Figure 8.
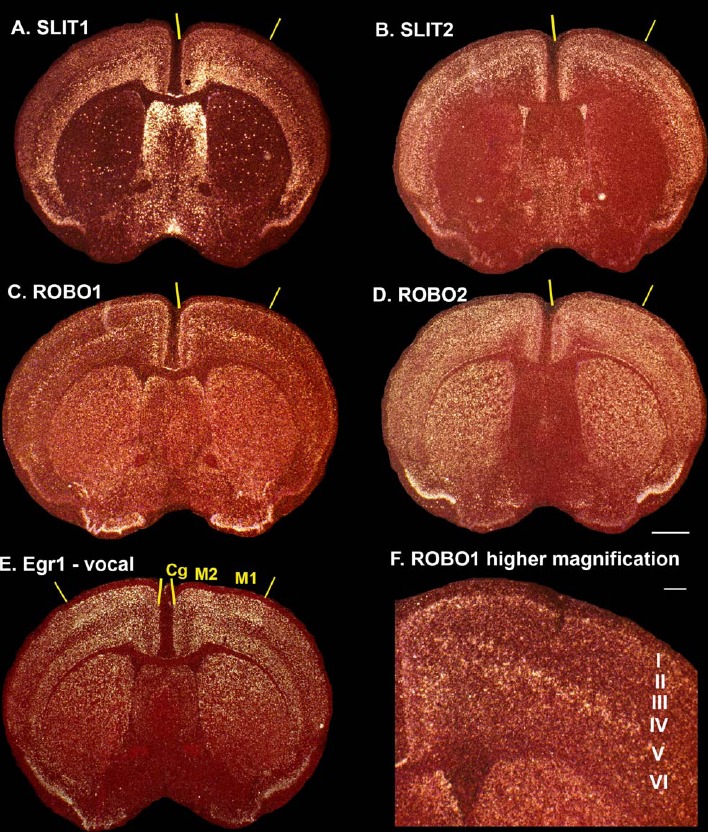
Expression of SLIT and ROBO genes in mouse putative LMC. A–D: In situ hybridization expression patterns of SLIT1 (A), SLIT2 (B), ROBO1 (C), and ROBO2 (D) in sections containing putative LMC. E: Expression of singing-driven EGR1 gene expression in a comparable section from Arriaga et al. (2012) to help identify the vocally active part of the mouse cortex (used with permission). F: Higher magnification of ROBO1 expression in layer 5 cells of the M1+M2 LMC region. For abbreviations, see list. Scale bar = 1 mm in D (applies to A–E); 200 µm in F.
Discussion
In this study we found convergent downregulation of the SLIT1 gene in the RA song production analog of all vocal learning avian lineages. We also found differential up- or downregulation of its receptor, ROBO1, although the upregulation was not as specific to the RA, because the intermediate arcopallium of vocal nonlearners also has some ROBO1 upregulation relative to the surrounding arcopallium. The ROBO1 differential expression in the songbird RA relative to the surrounding arcopallium was transiently and developmentally upregulated during critical periods of song learning. We also found suggestive evidence of convergent upregulation of the ROBO2 receptor in the HVC analogs. Along with our finding of differential downregulation of SLIT1 in the laryngeal motor cortex and adjacent laryngeal somatosensory cortex in humans (Pfenning et al., 2014), these results suggest that differential regulation of the SLIT–ROBO pathway could be critical for the evolution of vocal learning circuits.
This suggestion is consistent with prior discoveries in ROBO1 and SLIT1 genes. Some mutations in the ROBO1 gene in humans are associated with dyslexia, autism, and speech sound disorder language deficits (Hannula-Jouppi et al., 2005; Bates et al., 2011). The mRNA expression of one ROBO1 splice variant, ROBO1a, is enriched in the human temporal auditory and/or temporal association cortex, and another variant, ROBO1b is enriched in the prefrontal cortex, where Broca's speech area develops (Johnson et al., 2009). SLIT1 is a direct target of FOXP2, and human FOXP2 modulates stronger upregulation of SLIT1 than chimpanzee FOXP2 (Konopka et al., 2009). The SLIT–ROBO GTPase activating protein 2 (SRGAP2) is uniquely duplicated three times in the human genome, and one of the partial duplicated copies partially suppresses SLIT protein activity, thereby causing slower forebrain dendritic pruning, leaving longer and more dendrites (Charrier et al., 2012; Dennis et al., 2012); one interpretation of this finding is that the duplicated SRGAP2 results in the human adult brain remaining in a more juvenile-like state, thereby allowing humans greater cognitive flexibility as adults, more so than other animals (Charrier et al., 2012; Dennis et al., 2012). Thus, our results add to a growing body of studies converging on the SLIT–ROBO pathway genes as candidates for developing circuits for specialized complex behaviors.
Historically, SLIT ligands and ROBO receptors were found to control commissural guidance of axons across midlines of the two brain hemispheres during development, by way of a repulsive mechanism (Brose and Tessier-Lavigne, 2000); binding of secreted SLIT proteins from cell bodies to ROBO receptors on axons was found to mediate repulsion of the axons from the midline. When one or the other molecule was not present, the axons could cross. However, subsequent studies have revealed a more complex story for these genes, including involvement in cell proliferation and migration of neuronal and non-neuronal cells, and axon pathfinding and dendritic branching of neurons (Brose and Tessier-Lavigne, 2000; Lopez-Bendito et al., 2007; Ypsilanti et al., 2010; Kolodkin and Tessier-Lavigne, 2011; Blockus and Chédotal, 2014; Ypsilanti and Chedotal, 2014). Furthermore, the different SLITs can bind to the different ROBO receptors in various combinations, including among different splice variants, leading to either repulsive or attractive interactions between neurons (Blockus and Chédotal, 2014; Ypsilanti and Chedotal, 2014). Although we find developmental changes in ROBO1 expression in the RA during critical periods of vocal learning, the overall pattern in adulthood across species, the expression specializations that last into adulthood (including high ROBO1 in parrot and hummingbird RA analogs), and the nonvocal pathway expression patterns in adults all suggest that these genes have continuing important roles in adults for maintaining or modulating general as well as specialized connections. It is unlikely that their primary roles in the avian and thereby mammalian telencephalon would be in commissural crossings, as there is no corpus callosum in the avian telencephalon and the expression profiles in the avian telencephalon parallel some of those found in the mammalian brain, within analogous cell types.
Based on the expression profiles presented here, the repulsive interactions between SLIT1 and ROBO1 under some conditions, and the unique connectivity of song nuclei in vocal learners, we propose that these genes serve as viable candidate axon guidance molecules that could allow for the specialized direct projection of the RA to brainstem vocal motor neurons. Since the RA analog makes significantly less SLIT1 mRNA, its axons would theoretically not be repelled by ROBO1 or ROBO2 receptor expression in the nXIIts, whereas the surrounding arcopallium axons would be repelled from this and other motor neurons (Fig. 9). Conversely, because the reticular formation surrounding the motor neurons does not express high levels of ROBO1 or ROBO2, such neurons would be receptive to projections from the surrounding arcopallium with high SLIT1 expression (Fig. 9). It is also possible that the specialized expression of SLIT1 in the RA influences the connectivity of input from other song nuclei, such as the HVC and LMAN (Figs. 1A, 9). However, the connectivity of song nuclei within the forebrain is not that different from the connectivity of adjacent nonvocal motor or sensory neurons (Iyengar and Bottjer, 2002; Feenders et al., 2008), and we did not note convergent specialized expression of SLIT1 or ROBO1 for the HVC. Upregulation of ROBO2 in the HVC analogs could be a contributing factor, although this difference in expression was relatively minor compared with the striking downregulation of SLIT1 in the RA analogs of all vocal learners. This overall hypothesis would be viable if, as opposed to common belief (Blockus and Chédotal, 2014; Ypsilanti and Chedotal, 2014), SLIT1 protein is released from descending axons from the arcopallium neurons and ROBO1 protein is on the dendrites or cell bodies on brainstem motor neurons (Fig. 9). This hypothesis can be tested with immunolabel and high-resolution microscopy, as well as protein–protein interaction assays.
Figure 9.
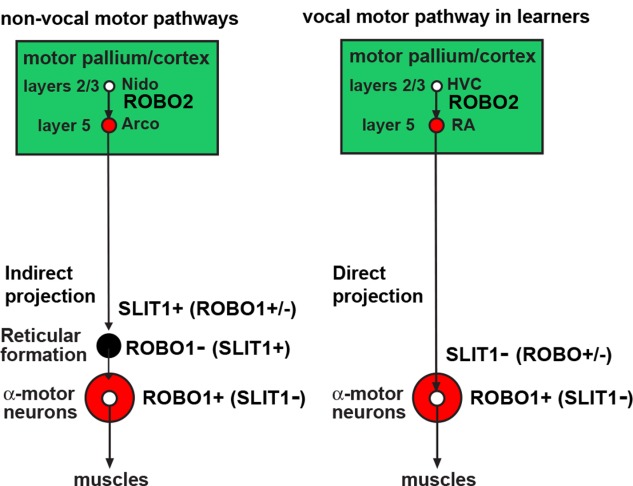
Summary of ROBO and SLIT gene expression in forebrain motor pathway projection neurons. Shown is a simplified diagram of the cell populations that express SLIT1, ROBO1, and ROBO2 in the songbird (left) and mouse (right) brains. The ligands and receptors are labeled along the axons that express the mRNAs. We do not yet know whether the expression of these genes is in the projection neurons of each brain region or the local cells.
At the other end of the spectrum, our results lead to additional questions on convergence of gene regulation in the brain across species. The NEUROD6 transcription factor is known to regulate SLIT1 expression (Blockus and Chédotal, 2014), and we found in our related study (Pfenning et al., 2014) that NEUROD6 is also differentially downregulated in the RA analog of vocal learning birds and human LMC. A possible alternative or complementary mechanism could involve enhancers. We recently found that histone 3 acetylation activity at lysine 27 (H3K27ac) differs in the enhancer and promoter regions among region-specific singing-regulated genes in the songbird RA and Area X in a manner that predicts which genes can be regulated in which brain regions (Whitney et al., 2014). It is possible that a similar mechanism could be involved in the control of differential regulation of axon guidance genes within the vocal learning nuclei relative to the surrounding cells.
Addressing these hypotheses about the consequences and causes of differential expression of SLIT–ROBO axon guidance genes will require further characterization of the genomes of individual cell types within and outside of vocal learning systems, as well as functional investigations that manipulate these genes in the vocal pathways of different species. The findings of the present study should help guide the testing of such hypotheses.
Acknowledgments
We thank Gustavo Arriaga for providing mouse brain sections.
Glossary
- A
Arcopallium
- AAc
Central nucleus of the anterior arcopallium
- AAcc
AAc core
- AAcs
AAc shell
- Am
Medial arcopallium
- Ai
Intermediate arcopallium
- Cb
Cerebellum
- Cg
Cingulate cortex
- H
Hyperpallium
- HVC
high vocal center
- IH
Intercalated hyperpallium
- LMAN
Lateral magnocellular nucleus of the anterior nidopallium
- LMC
Laryngeal motor cortex
- M1
Primary motor cortex
- M2
Secondary motor cortex
- MD
Dorsal mesopallium
- MV
Ventral mesopallium
- N
Nidopallium
- NLC
Central nucleus of the lateral nidopallium
- nXIIts
12th motor neurons, tracheosyringeal part
- OT
Optic tectum
- P
Pallidum
- SSp
Supraspinal motor nucleus
- St
Striatum
- T
Thalamus
- V
Ventricle
- VA
Vocal nucleus of the arcopallium
- VLN
Vocal nucleus of the lateral nidopallium
Conflict of Interest Statement
The authors have no conflicts of interest.
Role of Authors
All authors had full access to all the data in the study and take full responsibility for the integrity of the data and the accuracy of the data analysis. RW, CCC, EH, MR PLR, JTH, MC, and JNA performed experiments. RW and EDJ performed analyses and wrote the paper, and EDJ supervised the project.
Literature Cited
- Arriaga G, Jarvis ED. Mouse vocal communication system: are ultrasounds learned or innate? Brain Lang. 2013;124:96–116. doi: 10.1016/j.bandl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga G, Zhou EP, Jarvis ED. Of mice, birds, and men: the mouse ultrasonic song system has some features similar to humans and song-learning birds. Plos One. 2012;7:e46610. doi: 10.1371/journal.pone.0046610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TC, Luciano M, Medland SE, Montgomery GW, Wright MJ, Martin NG. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet. 2011;41:50–57. doi: 10.1007/s10519-010-9402-9. [DOI] [PubMed] [Google Scholar]
- Blockus H, Chédotal A. The multifaceted roles of Slits and Robos in cortical circuits: from proliferation to axon guidance and neurological diseases. Curr Opin Neurobiol. 2014;27:82–88. doi: 10.1016/j.conb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Bottjer SW, Brady JD, Cribbs B. Connections of a motor cortical region in zebra finches: relation to pathways for vocal learning. J Comp Neurol. 2000;420:244–260. [PubMed] [Google Scholar]
- Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr Opin Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- Chakraborty M, Walløe S, Nedergaard Sg, Fridel EE, Dabelsteen T, Pakkenberg B, Bertelson MF, Dorrestein GM, Brauth SE, Durand SE, Jarvis ED. Core and shell song systems unique to the parrot brain. PLoS ONE. doi: 10.1371/journal.pone.0118496. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, Polleux F. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wada K, Jarvis ED. Radioactive in situ hybridization for detecting diverse gene expression patterns in tissue. J Vis Exp. 2012;62:3764. doi: 10.3791/3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, Curry CJ, Shafer S, Shaffer LG, de Jong PJ, Wilson RK, Eichler EE. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deregnaucourt S, Guyomarc'h J, Richard V. Classification of hybrid crows in quail using artificial neural networks. Behav Processes. 2001;56:103–112. doi: 10.1016/s0376-6357(01)00188-7. [DOI] [PubMed] [Google Scholar]
- Dubbeldam JL. The neural substrate for 'learned' and 'nonlearned' activities in birds: a discussion of the organization of bulbar reticular premotor systems with side-lights on the mammalian situation. Acta Anat (Basel) 1998;163:157–172. doi: 10.1159/000046494. [DOI] [PubMed] [Google Scholar]
- Durand SE, Heaton JT, Amateau SK, Brauth SE. Vocal control pathways through the anterior forebrain of a parrot (Melopsittacus undulatus. J Comp Neurol. 1997;377:179–206. doi: 10.1002/(sici)1096-9861(19970113)377:2<179::aid-cne3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS ONE. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WT, Huber L, Bugnyar T. Social cognition and the evolution of language: constructing cognitive phylogenies. Neuron. 2010;65:795–814. doi: 10.1016/j.neuron.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahr M. Neural song control system of hummingbirds: comparison to swifts, vocal learning (songbirds) and nonlearning (suboscines) passerines, and vocal learning (budgerigars) and nonlearning (dove, owl, gull, quail, chicken) nonpasserines. J Comp Neurol. 2000;426:182–196. [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RC, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kaariainen H, Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Rivas MV, Ward JM, Okanoya K, Jarvis ED. Convergent differential regulation of parvalbumin in the brains of vocal learners. PLoS ONE. 2012;7:e29457. doi: 10.1371/journal.pone.0029457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita H, Kobayashi M, Liu WC, Oka K, Jarvis ED, Wada K. Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLoS One. 2012;7:e42173. doi: 10.1371/journal.pone.0042173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immelmann K. Song development in the zebra finch and other estridid finches. In: Hinde RA, editor. Bird vocalizations. Cambridge, UK: Cambridge University Press; 1969. pp. 64–74. [Google Scholar]
- Iyengar S, Bottjer SW. Development of individual axon arbors in a thalamocortical circuit necessary for song learning in zebra finches. J Neurosci. 2002;22:901–911. doi: 10.1523/JNEUROSCI.22-03-00901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Evolution of brain pathways for vocal learning in birds and humans. In: Bolhuis JJ, Everaert M, editors. Birdsong, speech, and language. Cambridge, MA: MIT Press; 2013. pp. 63–107. [Google Scholar]
- Jarvis ED, Mello CV. Molecular mapping of brain areas involved in parrot vocal communication. J Comp Neurol. 2000;419:1–31. doi: 10.1002/(sici)1096-9861(20000327)419:1<1::aid-cne1>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Ribeiro S, da Silva ML, Ventura D, Vielliard J, Mello CV. Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature. 2000;406:628–632. doi: 10.1038/35020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Yu J, Rivas MV, Horita H, Feenders G, Whitney O, Jarvis SC, Jarvis ER, Kubikova L, Puck AEP, Siang-Bakshi C, Martin S, McElroy M, Hara E, Howard J, Pfenning A, Mouritsen H, Chen C-C, Wada K. Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J Comp Neurol. 2013;521:3614–3665. doi: 10.1002/cne.23404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Simon HYW, Faircloth BC, Nabholz B, Howard JT, Suh A, Weber CC, da Fonseca RR, Li J, Zhang F, Li H, Zhou L, Narula N, Liu L, Ganapathy G, Boussau C., Md Bayzid S, Zavidovych V, Subramanian S, Gabaldón T, Capella-Gutiérrez S, Huerta-Cepas J, Rekepalli B, Munch K, Schierup M, Lindow B, Warren WC, Ray D, Green RE, Bruford M, Zhan X, Dixon A, Li S, Li N, Huang Y, Derryberry EP, Bertelsen MF, Sheldon FH, Brumfield RT, Mello CV, Lovell PV, Wirthlin M, Cruz Schneider MP, Prosdocimi F, Samaniego JA, Vargas Velazquez AM, Alfaro-Núñez A, Campos PF, Petersen B, Sicheritz-Ponten T, Pas A, Bailey T, Scofield T, Bunce M, Lambert DM, Zhou Q, Perelman P, Driskell AC, Shapiro B, Xiong Z, Zeng Y, Liu S, Li Z, Liu B, Wu K, Xiao J, Yinqi X, Zheng Q, Zhang Y, Yang H, Wang J, Smeds L, Rheindt FE, Braun M, Fjeldsa J, Orlando L, Barker K, Jønsson KA, Johnson W, Koepfli KP, O'Brien SJ, Haussler D, Ryder OA, Rahbek C, Willerslev E, Graves GR, Glenn TC, McCormack J, Burt D, Ellegren H, Alström P, Edwards SV, Stamatakis A, Mindell DP, Cracraft J, Braun EL, Warnow T, Jun W, Gilbert MTP, Zhang G. Whole genome analyses resolve the early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. (6215): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F, Sellix M. Reorganization of a telencephalic motor region during sexual differentiation and vocal learning in zebra finches. Brain Res Dev Brain Res. 2000;121:253–263. doi: 10.1016/s0165-3806(00)00052-3. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens U. Neural pathways underlying vocal control. Neurosci Biobehav Rev. 2002;26:235–258. doi: 10.1016/s0149-7634(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Tessier-Lavigne M. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol. 2011;3:a001727. doi: 10.1101/cshperspect.a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon RN. Descending pathways in motor control. Annu Rev Neurosci. 2008;31:195–218. doi: 10.1146/annurev.neuro.31.060407.125547. [DOI] [PubMed] [Google Scholar]
- Liu W-C, Wada K, Jarvis ED, Nottebohm F. Rudimentary substrates for vocal learning in a suboscine. Nat Commun. 2013;4:2082. doi: 10.1038/ncomms3082. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Flames N, Ma L, Fouquet C, Di Meglio T, Chedotal A, Tessier-Lavigne M, Marin O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J Neurosci. 2007;27:3395–3407. doi: 10.1523/JNEUROSCI.4605-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillat V, Cases O, Nguyen-Ba-Charvet KT, Tessier-Lavigne M, Sotelo C, Chédotal A. Spatiotemporal expression patterns of slit and robo genes in the rat brain. J Comp Neurol. 2002;442:130–155. doi: 10.1002/cne.10068. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Kato M, Okanoya K. Comparative analysis of gene expressions among avian brains: a molecular approach to the evolution of vocal learning. Brain Res Bull. 2008;75:474–479. doi: 10.1016/j.brainresbull.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Nottebohm ME. Vocalizations and breeding behaviour of surgically deafened ring doves (Streptopelia risoria. Anim Behav. 1971;19:313–327. doi: 10.1016/s0003-3472(71)80012-x. [DOI] [PubMed] [Google Scholar]
- Paton JA, Manogue KR, Nottebohm F. Bilateral organization of the vocal control pathway in the budgerigar, Melopsittacus undulatus. J Neurosci. 1981;1:1279–1288. doi: 10.1523/JNEUROSCI.01-11-01279.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov CI, Jarvis ED. Birds, primates, and spoken language origins: behavioral phenotypes and neurobiological substrates. Front Evol Neurosci. 2012;4:12. doi: 10.3389/fnevo.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning AR, Hara E, Whitney O, Rivas MR, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, Mountcastle J, Moseley MA, Thompson JW, Soderblum EJ, Iriki A, Kato M, Gilbert MTP, Zhang G, Bakken T, Bongarts A, Bernard A, Lein E, Mello CV, Hartemink AJ, Jarvis ED. Convergent transcriptional specializations in the brains of humans and song learning birds. Science. 2014;346(6215):1333. doi: 10.1126/science.1256846. & 1256846-1–1256846-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, Wild M, Ball GF, Durand S, Guturkun O, Lee DW, Mello CV, Powers A, White SA, Hough G, Kubikova L, Smulders TV, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter GF. The vocal control pathways in budgerigars differ from those in songbirds. J Comp Neurol. 1994;343:35–56. doi: 10.1002/cne.903430104. [DOI] [PubMed] [Google Scholar]
- Suh A, Paus M, Kiefmann M, Churakov G, Franke FA, Brosius J, Kriegs JO, Schmitz J. Mesozoic retroposons reveal parrots as the closest living relatives of passerine birds. Nat Commun. 2011;2:443. doi: 10.1038/ncomms1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernichovski O, Lints TJ, Deregnaucourt S, Cimenser A, Mitra PP. Studying the song development process: rationale and methods. Ann N Y Acad Sci. 2004;1016:348–363. doi: 10.1196/annals.1298.031. [DOI] [PubMed] [Google Scholar]
- Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, Horita H, Patterson MA, White SA, Scharff C, Haesler S, Zhao S, Sakaguchi H, Hagiwara M, Shiraki T, Hirozane-Kishikawa T, Skene P, Hayashizaki Y, Carninci P, Jarvis ED. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci U S A. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Dissecting the genetic basis of convergent complex traits based on molecular homoplasy. Durham, NC: Duke University; 2011. [Google Scholar]
- Whitney O, Pfenning AR, Howard JT, Blatti CA, Liu F, Ward JM, Wang R, Audet J-N, Kellis M, Mukherjee S, Sinha S, Hartemink AJ, West AE, Jarvis ED. Core and region enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346(6215):1333. doi: 10.1126/science.1256780. & 1256846-1–1256846-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM. Neural pathways for the control of birdsong production. J Neurobiol. 1997;33:653–670. doi: 10.1002/(sici)1097-4695(19971105)33:5<653::aid-neu11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Ypsilanti AR, Chedotal A. Roundabout receptors. Adv Neurobiol. 2014;8:133–164. doi: 10.1007/978-1-4614-8090-7_7. [DOI] [PubMed] [Google Scholar]
- Ypsilanti AR, Zagar Y, Chédotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:1939–1952. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]