Abstract
Nonalcoholic fatty liver disease (NAFLD) is a globally widespread disease of increasing clinical significance. The pathological progression of the disease from simple steatosis to nonalcoholic steatohepatitis (NASH) has been well defined, however, the contribution of altered branched chain amino acid metabolomic profiles to the progression of NAFLD is not known. The three BCAAs: leucine, isoleucine and valine are known to mediate activation of several important hepatic metabolic signaling pathways ranging from insulin signaling to glucose regulation. The purpose of this study is to profile changes in hepatic BCAA metabolite levels with transcriptomic changes in the progression of human NAFLD to discover novel mechanisms of disease progression. Metabolomic and transcriptomic data sets representing the spectrum of human NAFLD (normal, steatosis, NASH fatty, and NASH not fatty livers) were utilized for this study. During the transition from steatosis to NASH, increases in the levels of leucine (127 % of normal), isoleucine (139 %), and valine (147 %) were observed. Carnitine metabolites also exhibited significantly elevated profiles in NASH fatty and NASH not fatty samples and included propionyl, hexanoyl, lauryl, acetyl and butyryl carnitine. Amino acid and BCAA metabolism gene sets were significantly enriched among downregulated genes during NASH. These cumulative alterations in BCAA metabolite and amino acid metabolism gene profiles represent adaptive physiological responses to disease-induced hepatic stress in NASH patients.
Keywords: Branched-chain amino acids, Nonalcoholic fatty liver disease, Nonalcoholic steatohepatitis, Metabolomics, Transcriptomics
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a chronic, progressive liver disease of increasing significance worldwide. The incidence of NAFLD in many countries is similar to the 30–40 % prevalence estimates of NAFLD in the United States population (Ali and Cusi 2009; Bellentani et al. 2010; Koehler et al. 2012; Wong et al. 2012). NAFLD is currently recognized as the hepatic manifestation of the metabolic syndrome (Marchesini et al. 2001) and is linked to obesity, type 2 diabetes, and cardiovascular disease (Bonora 2006; Dam-Larsen 2004; Fabbrini et al. 2010). The pathological progression of NAFLD is classically described by the ‘two-hit’ hypothesis (Day and James 1998) in which a ‘first hit’ to the liver in the form of accumulating lipids results in simple steatosis. The excess lipid accumulation sensitizes hepatocytes to additional ‘second-hits’ in the form of increased oxidative stress and inflammation that can lead to the development of nonalcoholic steatohepatitis (NASH). NASH is considered the most severe stage of NAFLD and comprises two separate pathological designations: NASH fatty and NASH not fatty (Day 2002; McCullough 2006). Approximately, 5.7–17 % of the United States population is estimated to have some form of NASH and are at risk of progression to cryptogenic cirrhosis and hepatocellular carcinoma (HCC) (McCullough 2011; Rubinstein et al. 2008). NAFLD progression has been shown to acquire gene and protein expression alterations in phase I and II metabolism enzymes and transport proteins which put these patients at increased risk of hepatotoxicity (Hardwick et al. 2011; Lake et al. 2011; Merrell and Cherrington 2011). Metabolism and transport gene expression changes associated with amino acids and BCAAs have important roles in sustaining a healthy liver. These roles range from cancer stem cell suppression to inhibition of reactive oxygen species (ROS). BCAA supplementation is also reported to improve insulin resistance and can promote liver regeneration (Miyake et al. 2012; Nagao et al. 2012; Tajiri and Shimizu 2013; Yoshida et al. 2012). Determining the alterations in hepatic profiles of BCAA composition, with metabolism and signaling genes will provide important mechanistic information on the biochemical changes occurring in response to progression of this disease.
The catabolic pathway of BCAAs is reported to proceed through a reversible transamination reaction in extrahepatic organs by cytosolic branched chain amino acid aminotransferase 1 (BCAT1) or mitochondrial BCAT2 (Adeva et al. 2012; Suryawan et al. 1998). BCAT1 expression has previously been limited to the brain, placenta and ovaries (Suryawan et al. 1998; Sweatt et al. 2004), however, expression of the BCAT1 gene has also been shown in liver samples diagnosed as having NAFLD (Greco et al. 2008). Hepatic BCAA metabolite intermediates are the result of BCAT enzyme activity in extrahepatic tissues such as muscle and adipose tissue that are then shuttled to the liver in the blood (She et al. 2007). The second step in BCAA catabolism is an irreversible decarboxylative reaction of the alpha-keto acids that is mediated by the branched chain alpha-ketoacid dehydrogenase (BCKDH) complex which is regulated and inhibited by the branched chain ketoacid dehydrogenase kinase (BCKDK) (Adeva et al. 2012; Suryawan et al. 1998). BCAA catabolism and lipid processing produce acylcarnitine products in the mitochondria that have been shown to effect gluconeogenesis (Newgard et al. 2009; Newgard 2012). Furthermore, high lipid loads such as those associated with NAFLD may lead to an increase in acylcarnitine production in tissues according to previous studies (Schooneman et al. 2013).
The expression of transporters in the liver governs the distribution of BCAAs between the liver and blood. SLC16A10 (TAT1) functions as a uniporter in the transport of AAAs across the basolateral membrane of liver epithelial cells (Mariotta et al. 2012) while SLC43A1 (LAT3) is involved in the efflux of BCAAs from the liver to blood (Bodoy et al. 2012; Fukuhara et al. 2007). The transcriptomic profiling of alterations in amino acid metabolism and transporters during NAFLD progression will yield potential mechanistic clues for the biochemical alterations in BCAAs.
BCAA regulation of signaling pathways mediates an array of adaptive hepatic response mechanisms during disease. Leucine supplementation has been shown to activate the mammalian target of rapamycin (mTOR), a critical mediator of protein synthesis regulation, cellular proliferation, and insulin sensitivity regulation (Adeva et al. 2012; Newgard 2012). Overall, BCAAs have multiple roles in the liver that contribute to numerous biological functions including insulin signaling, the regulation of glucose and the protective inhibition of cancer development.
The purpose of this study is to characterize the hepatic profile of BCAAs and catabolism genes throughout the progressive stages of human NAFLD to provide mechanistic insight. Metabolomic and transcriptomic assays of hepatic samples representing the entire spectrum of human NAFLD were utilized to analyze gene expression changes of BCAA enzymes, transporters, and mTOR signaling components. Amino acids, BCAAs, carnitines, and other conventional metabolites of the liver were analyzed by a metabolomics profiling approach to determine the downstream biochemical consequences of transcriptomic changes in progressive NAFLD. This study will reveal unique profiles of hepatic amino acid composition and metabolism in the progression of human NAFLD with implications for adaptive stress response mechanisms.
Materials and methods
Human liver samples
Human liver tissue was acquired previously from the Liver Tissue Cell Distribution System (LTCDS) funded by the National Institutes of Health. The LTCDS is composed of the following institutions: the University of Minnesota, Virginia Commonwealth University, and the University of Pittsburgh. Clinical information and demographics on these same liver tissue samples has been previously described and published (Fisher et al. 2009). NAFLD activity scoring (NAS) of each sample for categorization was completed by an LTCDS medical pathologist (Kleiner et al. 2005). The samples were diagnosed by pathological analysis as normal (n = 19), steatosis (n = 10), NASH fatty livers (n = 9) and NASH not fatty livers (n = 7). The stage of steatosis was diagnosed as having >10 % fat deposition within hepatocytes and no accompanying inflammation or fibrosis. NASH fatty liver was characterized by >5 % fat deposition in the presence of inflammation and fibrosis. NASH not fatty liver was distinguished by <5 % fat deposition with inflammation and fibrosis. A portion of these liver samples were utilized for mRNA isolation and application to Affymetrix 1.0 ST GeneChip microarrays as described previously (Lake et al. 2011). Samples reserved for metabolomic analysis included fewer normal (n = 17) and steatosis (n = 4) samples. An increased sample size of NASH fatty liver (n = 14) and NASH not fatty liver samples (n = 23) were utilized and applied to the metabolomics assays.
Affymetrix microarray data
Individual Affymetrix GeneChip Human 1.0 ST Arrays (Affymetrix, La Jolla, CA) were generated from purified mRNA isolated from each liver sample as described previously (Lake et al. 2011). Liver samples for normal (n = 19), steatosis (n = 10), NASH fatty (n = 9) and NASH not fatty livers (n = 7) were utilized. A total of 33,252 genes for each sample in four pathologically defined groups (normal, steatosis, NASH fatty and NASH not fatty) are available in this array data set. The array data have been made available to the public in the ArrayExpress repository for microarray data and are accessible under the accession number: E-MEXP-3291 (http://www.webcitation.org/5zyojNu7T).
Transcriptomic analysis
Lists of 184 amino acid genes, 53 BCAA genes, and 54 MTOR signaling genes were compiled using literature database searches and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/). These gene sets for BCAA genes, global amino acid genes, and MTOR signaling genes were analyzed by a gene set enrichment tests (Supplemental Tables 2, 3 and 4). Principal component analysis (PCA) was performed on the genes to determine differences among diagnosis groups according to gene expression. Hierarchical clustering of the liver samples with heatmaps of BCAA and MTOR signaling genes was also generated for the visualization of clustering patterns among these liver samples.
Gene expression analysis
The log2-transformed mRNA expression of selected BCAA genes was analyzed using the previously published human NAFLD microarray data set (Lake et al. 2011). Selected genes were graphed using bar plots for normal, steatosis, and NASH fatty and NASH not fatty samples. Values were normalized to the median value of normal samples. A one-way ANOVA with post hoc Tukey testing was performed in Graphpad Prism 5 Software (La Jolla, CA). Significance was determined by p values less than or equal to 0.05.
Analytical methods for LC/MS
A total of 17 healthy, 4 steatosis, 14 NASH fatty, and 23 NASH not fatty frozen liver samples were weighed (weights recorded between 60 and 140 mg) and homogenized in 10 times the tissue weight of ice-cold methanol solution with 0.1 % formic acid for 18–20 s using a polytron homogenizer over ice. Liver samples were kept frozen and on dry ice during all steps of the process. Samples were spun for 10 min at 4 °C at 10,000 RPM in a Beckman Coulter Allegra 25 centrifuge with a TA 10.250 rotor (Beckman Coulter Inc. Indianapolis, IN). Supernatants were transferred to new tubes, gently vortexed, and 200 µl aliquots of each sample were added to the corresponding positions in a 96 deep well polypropylene plate (Brand-Tech Scientific, Inc. Essex, CT). The 96 well plate was dried using a V&P Scientific Model VP-177 96 well plate manifold dryer (V&P Scientific Inc. San Diego, CA) with nitrogen gas for approximately 6 h prior to storage in a −80 °C freezer and processing for UHPLC high-resolution LC–MS analysis. Samples were reconstituted in a 90:10 water:methanol solution. The internal standard d5-hippurate was added to all samples which were then injected in randomized fashion onto a Thermo UHPLC Accela coupled to a Thermo Exactive high-resolution Orbitrap mass spectrometer. A total of 10 amino acid and 11 acylcarnitine metabolites were captured in negative and positive ion mode. Metabolite peak areas under the curve (AUC) LC–MS measurements were calculated using expedient data mining by Bristol-Myers Squibb scientists (Hnatyshyn et al. 2013). Principal component analysis of amino acids, acylcarnitines, and conventional metabolic metabolites was performed to determine how biochemical differences profile the different diagnosis groups of normal, steatosis, NASH fatty and NASH not fatty.
Statistical analysis of metabolomics data
The peak area under the curve (AUC) values for the LC–MS metabolite data were log transformed based upon a normal distribution approximation. After log transformation a one-way ANOVA with post hoc Tukey honest significant differences (HSD) testing was utilized for multiple comparisons of metabolites among the different pathology groups to test for mean differences. Significance was defined as p values ≤0.05. Percent of normal values was calculated using the raw area under the curve (AUC) metabolite data of the steatosis and NASH sample groups.
Immunoblot protein analysis
Western blots were performed to validate the protein expression of cytosolic BCAT1 and BCKDK in normal (n = 7), steatosis (n = 7), NASH fatty (n = 11), NASH not fatty (n = 13) cytosolic liver lysates. Cytosolic liver lysates were prepared from human liver tissue as previously described (Hardwick et al. 2012). Cytosolic lysates were utilized at a concentration of 20 µg of total protein. All samples were prepared in Laemmli sample dye (Bio-Rad Laboratories, Hercules, CA) with β-mercaptoethanol and boiled for 10 min. Proteins were separated on 10 % SDS polyacrylamide gels and transferred to methanol activated polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA) at 30 mA for 12.5 h. Polyclonal rabbit antibody (Abcam Inc., Cambridge, MA) for BCAT1 (ab107191) was utilized at a 1:3,000 dilution in 5 % nonfat dry milk dissolved in PBST. Rabbit polyclonal BCKDK antibody (NBP1-70814) was acquired (Novus Biologicals, Littleton, CO) and used at a concentration of 1:3,000 in 5 % nonfat dry milk dissolved in PBST. Secondary goat anti-rabbit HRP conjugate antibody (sc-2004, Santa Cruz Biotechnology Inc., Santa Cruz, CA) was utilized for each of the blots at a dilution of 1:55,000 in 5 % nonfat dry milk in PBST. Blots were imaged with Femto Chemiluminescence Substrate (Thermo Scientific, Rockford, IL). Relative protein expression was quantified using densitometric Image J software (National Institutes of Health, Bethesda, MD). Cytosolic liver lysate proteins were normalized to total ERK (C-16 and C-14, Santa Cruz Biotechnology Inc., Santa Cruz, CA) as per previous descriptions (Hardwick et al. 2012). GAPDH was not utilized as a control due to variations in GAPDH expression in human liver samples with inflammation (Boujedidi et al. 2012; Congiu et al. 2011). Statistical significance was determined by one-way ANOVA with Tukey post hoc analysis in GraphPad Prism 5 software (La Jolla, CA).
Results
Principal component analysis (PCA)
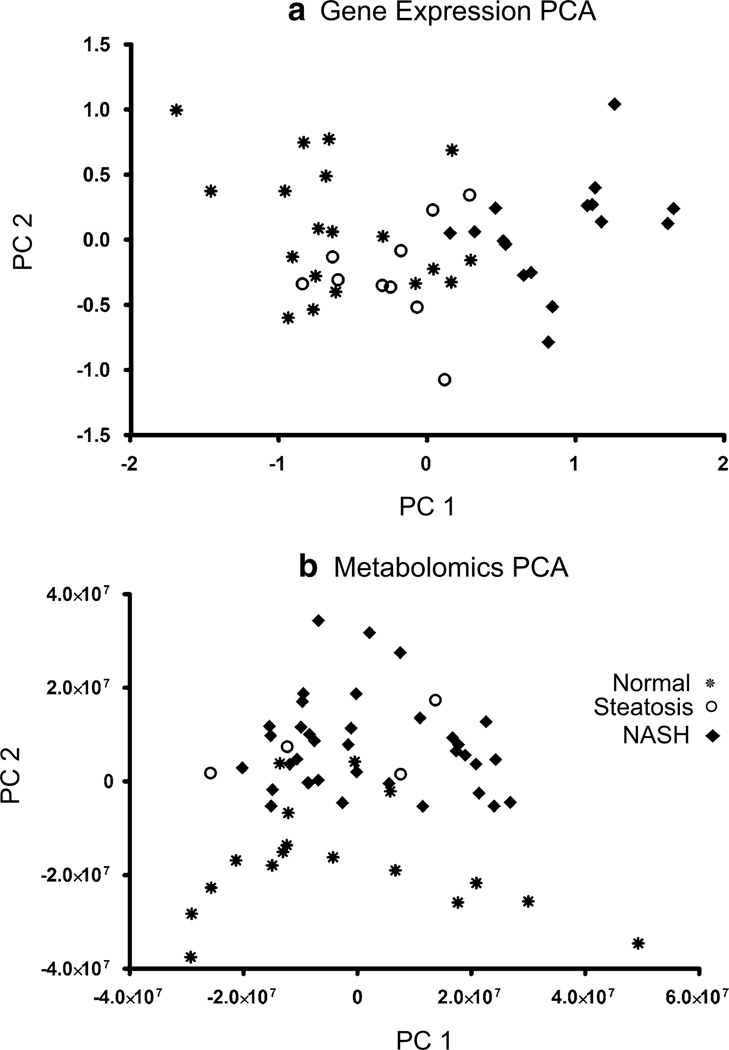
Gene expression data of amino acid metabolism, transport, and mTOR pathway signaling genes were analyzed by PCA to determine how these expression changes differentiate diagnosis in human NAFLD liver samples (Fig. 1a). Gene expression profiles differentiated NASH-diagnosed samples from normal or steatosis samples according to the first principal component. Amino acid, acylcarnitine lipid products, and conventional liver metabolites were also profiled by principal components to determine if the metabolomic profile was able to differentiate the different states of NAFLD (Fig. 1b). The metabolomic profiles differentiated diagnosis by the second principal component but not the first. Therefore, another parameter other than diagnosis is responsible for the primary differences in metabolomic profiles among these NAFLD liver samples.
Fig. 1.
Principal components analysis of amino acid genes and metabolites in human NAFLD. In a the first principal component distinguishes NASH from normal and steatosis amino acid gene expression. This plot demonstrates that gene expression differences among amino acid genes are able to differentiate between samples with a NASH diagnosis versus those designated as normal or steatosis. b The metabolomics PCA profile of amino acid and acylcarnitine metabolites distinguishes between sample diagnosis groups although diagnosis is shown to be correlated more with the second and not the first principal component in this plot
Hierarchical clustering of amino acid global genes, BCAA metabolism and MTOR signaling genes
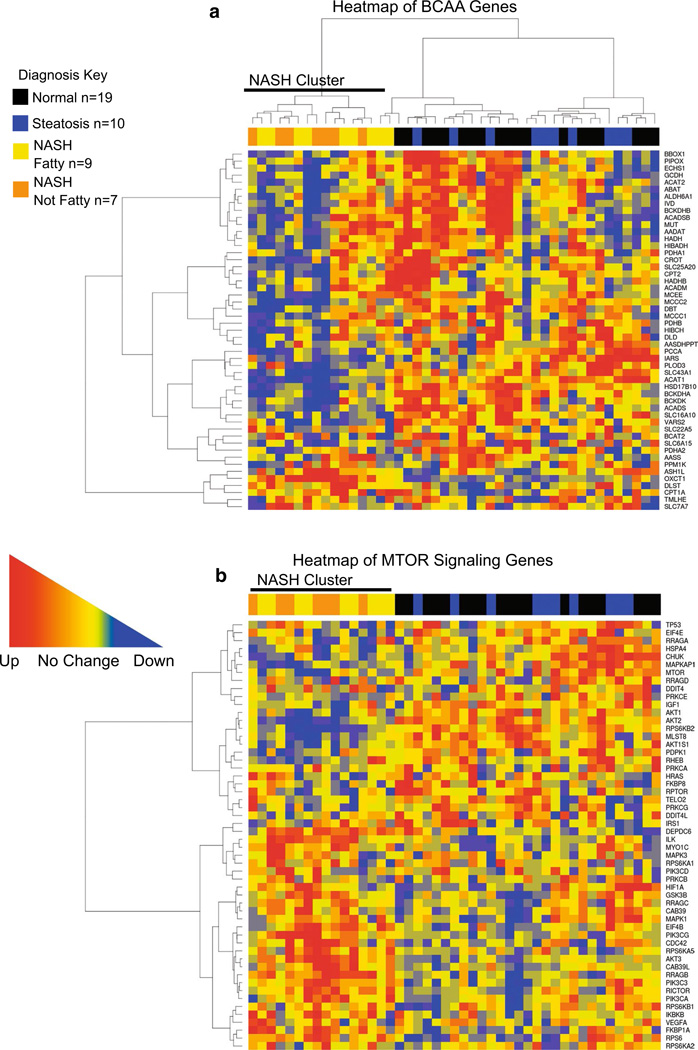
A hierarchical clustering analysis of each gene set was performed to demonstrate if liver samples clustered according to diagnosis. Heatmaps for global amino acid genes, BCAA associated genes, and MTOR signaling genes show that samples diagnosed as NASH clustered together. Steatosis and normal liver samples did not cluster with respect to gene expression but grouped together separately from NASH samples (Fig. 2a–c).
Fig. 2.
Hierarchical clustering analysis of BCAA and MTOR signaling genes. In a the heatmap shown for BCAA metabolism, transport, and signaling genes demonstrates the clustering of samples diagnosed as NASH fatty or NASH not fatty according to gene expression changes that distinguishes NASH liver samples as a whole from normal and steatosis samples. In b the MTOR signaling gene category also shows that samples diagnosed as NASH cluster together within the heatmap while steatosis and normal samples do not. Upregulation of genes is represented by red squares within the heat-map while downregulation is represented by blue squares and genes that are unchanged are represented by yellow. Diagnosis groups are represented along the tops of the heatmap as black (normal), blue (steatosis), yellow (NASH fatty), and orange (NASH not fatty) (color figure online)
Transcriptomic analysis
Gene set enrichment testing was performed on compiled lists of 183 global amino acid genes, 53 BCAA genes, and 54 MTOR pathway signaling genes (Supplemental Tables 2, 3 and 4). Global amino acid and BCAA gene categories were significantly enriched for downregulation of gene expression. No significance was found for either category in the tests for enrichment of upregulated gene expression (Table 1).
Table 1.
Gene set enrichment tests of amino acid gene categories
| Gene category | p value | q value | Gene set size |
|---|---|---|---|
| Tests for gene downregulation | |||
| BCAA genes | 0.0002 | 0.0006 | 53 |
| Global amino acid GENES | 0.0046 | 0.0069 | 183 |
| MTOR signaling genes | 0.831 | 0.831 | 54 |
| Tests for gene upregulation | |||
| BCAA genes | 1.000 | 1.000 | 53 |
| Global amino acid genes | 0.995 | 1.000 | 183 |
| MTOR signaling genes | 0.169 | 0.507 | 54 |
Gene set enrichment analysis. The gene set enrichment test results for down and upregulation of the gene categories for BCAA genes, global amino acid genes, and MTOR signaling genes are shown. Global p values and FDR-corrected p values (q value) of significance are reported for each gene set category
Metabolomics analysis
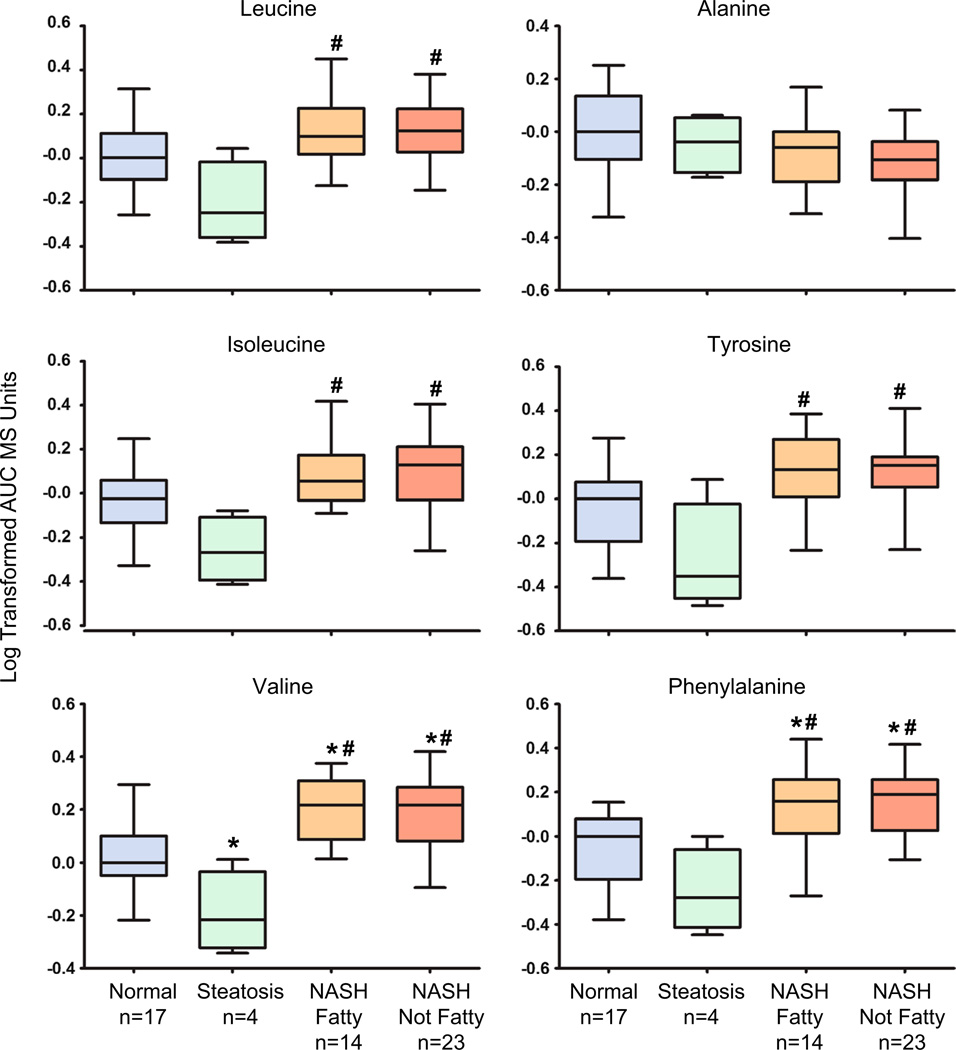
A total of 33 metabolites measured by LC–MS in normal, steatosis, NASH fatty and NASH not fatty human liver samples were utilized to profile hepatic composition changes throughout the different stages of NAFLD (Supplemental Table 1). The metabolomics profile included conventional amino acids and BCAAs (leucine, isoleucine and valine), together with functional liver metabolites (glucose, glutathione) and mitochondrial lipid products (acylcarnitines) (Supplemental Table 1). BCAAs were significantly changed in hepatic composition with progression to NASH. Leucine (127 % of normal), isoleucine (139 % of normal), and valine (147 % of normal) were significantly elevated in both NASH fatty and NASH not fatty samples (Fig. 3; Table 2). Aromatic amino acids (AAAs) including tyrosine and phenylalanine were also significantly elevated in both types of NASH. Tyrosine was significantly elevated (139 % of normal) in NASH samples when compared to steatosis, while phenylalanine was significantly increased (158 % of normal) in NASH when compared to both normal and steatosis samples (Fig. 3; Table 2). Hepatic levels of alanine exhibited no change across the spectrum of NAFLD (Fig. 3; Table 2). Several essential and non-essential amino acids exhibited altered metabolomics profiles in NASH samples (Supplemental Table 1).
Fig. 3.
Metabolomic composition of hepatic amino acids in progressive stages of human NAFLD. The metabolomics profiling results for the BCAAs leucine, isoleucine and valine with the amino acids tyrosine, alanine, and phenylalanine are shown as log-transformed area under the curve (AUC) mass spectrometry units normalized to the median value of the normal samples ± the standard deviation. Significance from normal is shown by asterisk and significance from steatosis is represented by pound. Significance set at p ≤ 0.05
Table 2.
Percent of normal values for BCAA and acylcarnitine metabolites
| Metabolite | % Of normal in steatosis |
% Of normal in NASH |
|---|---|---|
| Acetylcarnitine | 343* | 286* |
| Propionylcarnitine | 149 | 266* |
| Lauryl carnitine | 441* | 403* |
| Butyryl carnitine | 368* | 353* |
| Hexanoyl carnitine | 39* | 349* |
| Stearoyl carnitine | 125 | (163* NASH not fatty) |
| Palmitoyl carnitine | 230* | 149 |
| Isovaleryl carnitine | 135 | 115 |
| Isobutyryl carnitine | 89 | 176 |
| Linoleoyl carnitine | 209 | 133 |
| Oleoyl carnitine | 248 | 178 |
| Valine | 60* | 147*# |
| Isoleucine | 60 | 139# |
| Leucine | 60 | 127# |
| Alanine | 90 | 80 |
| Phenylalanine | 163 | 158*# |
| Tyrosine | 70 | 139# |
Percent of normal values for metabolomics analysis. The percent of normal values for each metabolite measured in the metabolomics profiling analysis of the liver samples diagnosed as steatosis and NASH are shown.
Significantly changed metabolites are shown by asterisk (*) for significance of normal and pound (#) for significance from steatosis. Significance is set at p ≤ 0.05
Fatty acid and carnitine metabolite levels
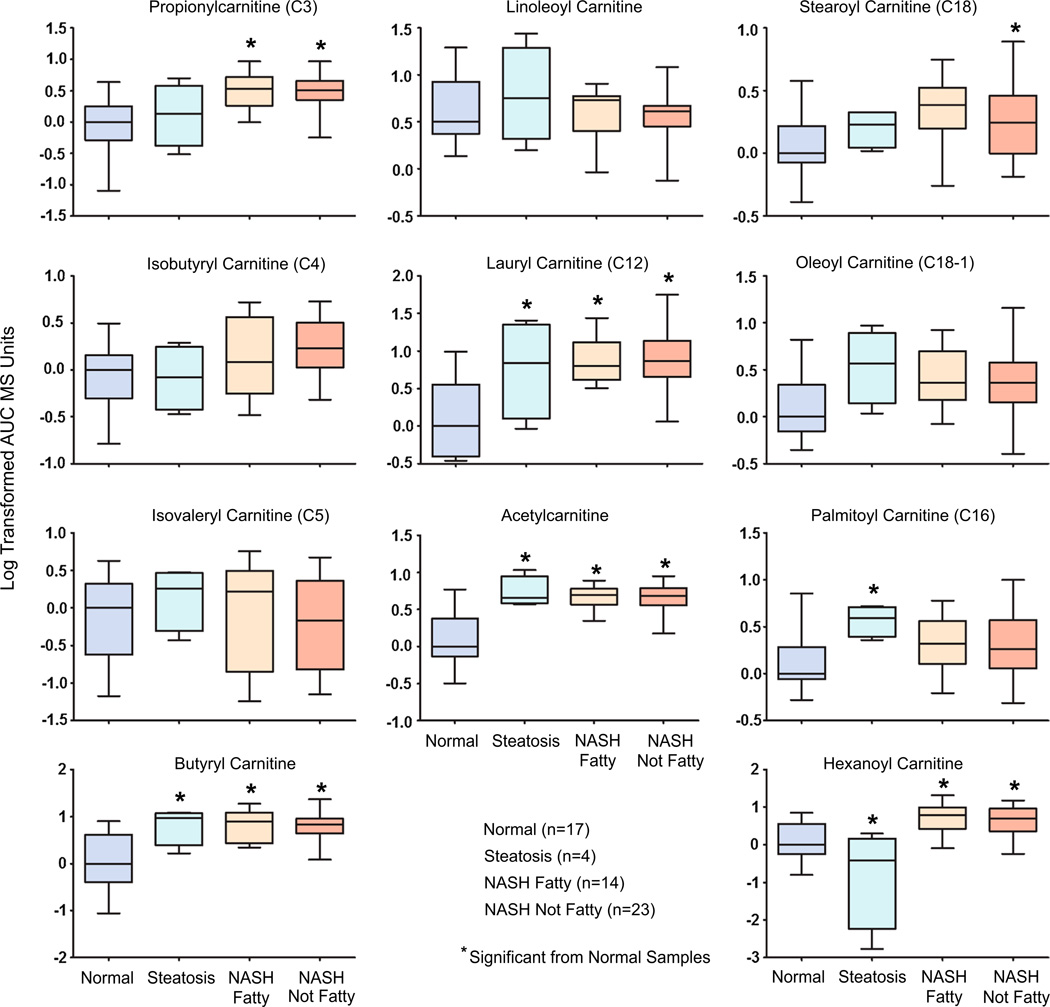
The fatty acids linoleic acid (C18:2), oleic acid (C18:1), and arachidonic acid (C20:4) were also analyzed by metabolomics profiling. Linoleic and arachidonic acid were significantly elevated from normal levels in both types of NASH. Oleic acid was only significantly elevated in NASH not fatty Samples (Supplemental Table 1). Several acylcarnitine products of lipid processing and BCAA catabolism by the mitochondria were significantly changed with progression of NAFLD to steatosis and NASH (Fig. 4; Supplemental Table 1). Lauryl carnitine (403 % of normal), acetylcarnitine (286 % of normal), hexanoyl carnitine (349 % of normal), and butyryl carnitine (353 % of normal) levels were all significantly elevated from normal in NASH fatty and NASH not fatty specimens. These same metabolites were also significantly elevated in steatosis with the exception of hexanoyl carnitine which was significantly decreased in steatosis samples. Propionylcarnitine (266 % of normal) was significantly increased in NASH fatty and NASH not fatty while stearoyl carnitine (C18) was only significantly elevated in NASH not fatty samples (195 % of normal). Palmitoyl carnitine (C16) was significantly increased from normal in steatosis samples (230 % of normal) but not NASH (Fig. 4). Isovaleryl carnitine (C5), isobutyryl carnitine (C4), linoleoyl carnitine, and oleoyl carnitine (C18-1) were not significantly altered with the progression of NAFLD (Fig. 4).
Fig. 4.
Metabolomic composition of acylcarnitine levels in progressive stages of human NAFLD. Short, medium, and long chain hepatic acylcarnitine levels are shown as log-transformed AUC mass spectrometry units normalized to the median value of the normal samples ± the standard deviation. Significance from normal is shown by asterisk and significance from steatosis is represented by pound. Significance was set at p ≤ 0.05
Microarray gene expression
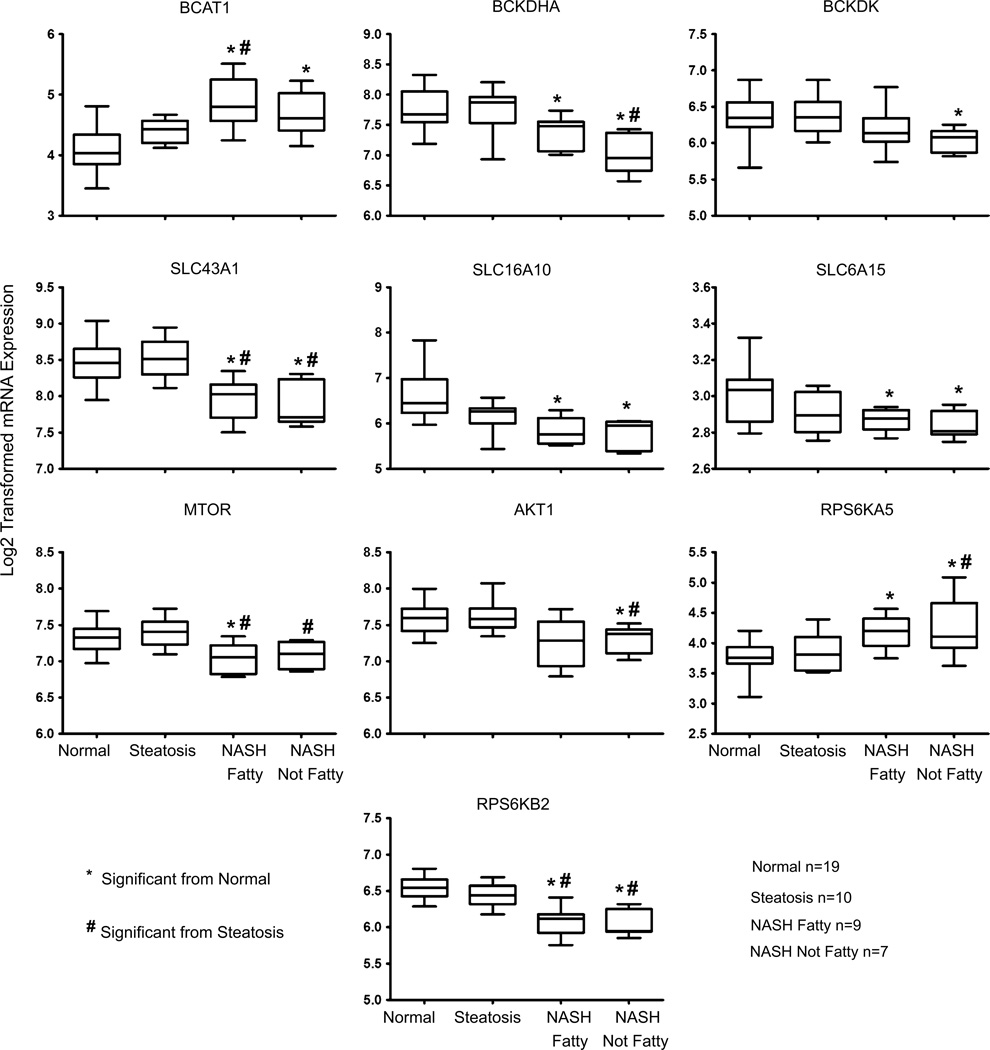
BCAA catabolism, transporter, and MTOR signaling genes were analyzed using a previously validated set of human NAFLD microarray data (Lake et al. 2011). The cytosolic BCAT1 enzyme showed significantly increased expression in NASH fatty and NASH not fatty compared to normal samples (Fig. 5). The BCKDHA complex gene was significantly decreased in expression for both types of NASH. The BCKDH complex inhibiting kinase, BCKDK gene, was significantly decreased in expression in NASH not fatty tissues but no change was evident in steatosis or NASH fatty samples (Fig. 5). Two uptake amino acid transporter genes of the solute carrier transporter family (SLC), SLC6A15 and SLC16A10, were both significantly downregulated in both NASH fatty and NASH not fatty samples, while the efflux transporter SLC43A1 was also significantly decreased when compared to both steatosis and normal samples (Fig. 5). In addition to the transcriptional changes in metabolism and transport genes, mTOR and AKT gene expression was also analyzed. The gene expression of mTOR exhibited significantly decreased levels in both NASH fatty and NASH not fatty (Fig. 5). AKT1 exhibited decreased expression in NASH not fatty samples (Fig. 5). The gene for ribosomal protein S6 kinase alpha 5 (RPS6KA5) a positive regulator of MTORC1 and MTORC2 pathways was significantly increased in both types of NASH, while gene expression for another positive regulator of MTOR (RPS6KB2) was significantly decreased in NASH (Fig. 5).
Fig. 5.
BCAA metabolizing enzyme, transporter, and mTOR signaling genes in progressive human NAFLD. Gene expression values of BCAT1, BCKDK, SLC6A15, SLC16A10, SLC43A1, MTOR, AKT1, RPS6KA5, and RPS6KB2 are shown for normal, steatosis, NASH fatty and NASH not fatty samples. Log2-transformed values ± the standard deviation are normalized to the median of the normal samples. Significance from normal is shown by asterisk and significance from steatosis is represented by pound. Significance was set at p ≤ 0.05
Immunoblot analysis
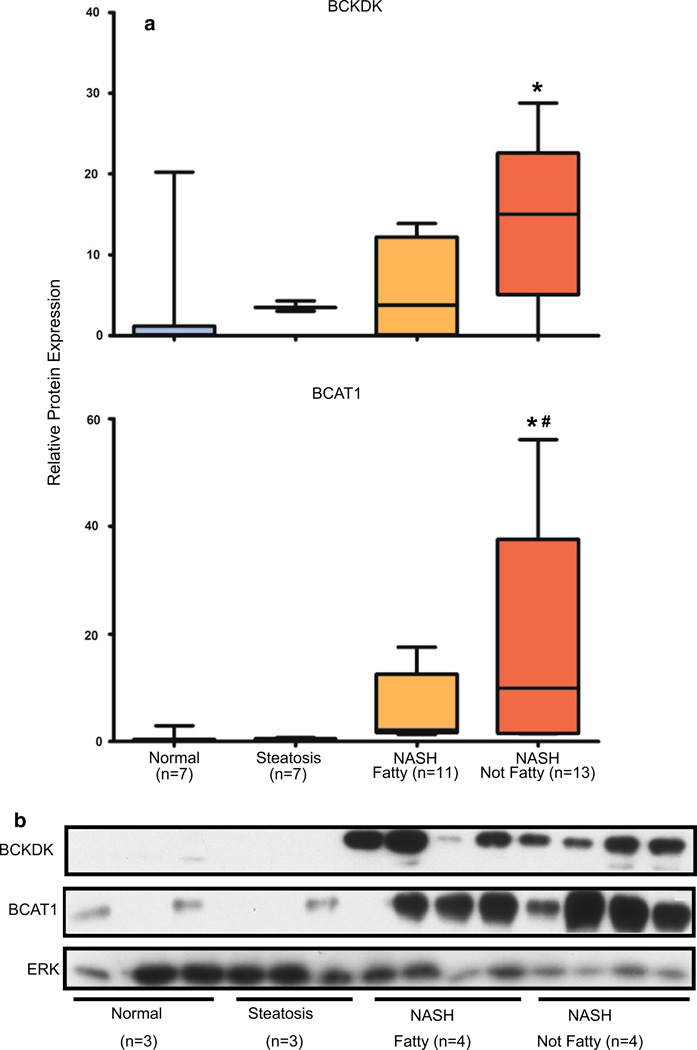
Protein expression of the BCAA enzyme BCAT1 and the kinase BCKDK that is responsible for inhibiting the BCKDH complex were assessed against ERK control protein in cytosolic human liver preparations of normal, steatosis, NASH fatty and NASH not fatty samples. The protein levels of BCAT1 and BCKDK exhibited significant increases in expression for NASH not fatty samples (Fig. 6a, b) but expression was not observed in normal and steatosis samples and was also not significant for NASH fatty samples.
Fig. 6.
Hepatic BCAA enzyme protein expression in progressive human NAFLD. Relative protein expression levels ± the standard deviation of BCKDK and BCAT1 are shown for normal (n = 7), steatosis (n = 7), NASH fatty (n = 11), and NASH not fatty (n = 13) samples. Representative blots of normal (n = 3), steatosis (n = 3), NASH fatty (n = 4), and NASH not fatty (n = 4) samples are shown. Total ERK was utilized as control protein. Significance from normal is shown by asterisk and significance from steatosis is represented by pound. Significance was set at p ≤ 0.05
Discussion
This study revealed significant changes in hepatic BCAA composition and transcriptomic metabolism profiles during the progression of human NAFLD. Significant elevations of hepatic BCAAs in NASH together with altered profiles for several essential and non-essential amino acids demonstrate the perturbations of systemic amino acid homeostasis that occur in NAFLD patients. Results from previous studies in the literature have also shown increased levels of BCAAs (leucine, isoleucine and valine) in plasma samples of NASH patients that are comparable to the changes in BCAA alterations observed in NASH liver in the present study (Kalhan et al. 2011). Similarly, rodent models of obesity have shown increases in BCAA levels in addition to elevations of other critical amino acids such as tyrosine and phenylalanine which were also elevated in our NASH samples (She et al. 2007, 2013). Importantly, systemic amino acid increases are reported to function as adaptive responses against disease (Adams 2011; Kalhan et al. 2011; Newgard et al. 2009; She et al. 2007). In light of recent discoveries demonstrating the clinical effectiveness of BCAA supplementation in liver disease and cancer prevention (Hagiwara et al. 2012; Hayaishi et al. 2011; Miyake et al. 2012), the present study provides important information for NASH researchers and clinicians.
Elevated mRNA and protein levels of cytosolic BCAT1 were observed in liver samples diagnosed as NASH in the present study, while no expression of BCAT1 was shown in normal or steatosis samples. Whether or not this observation is due to increased systemic BCAT1 circulating to the liver or if this elevation is isolated to the liver in NASH, this finding is indicative of increased BCAA metabolism occurring in NASH. Elevated BCAT1 gene expression has been shown previously in NAFLD liver tissue (Greco et al. 2008) and BCAT activity levels have been reported for human cirrhotic livers which support our findings in the present study. However, BCAT activity and expression was thought to be due to other enzymatic sources in extrahepatic tissues (Suryawan et al. 1998; Sweatt et al. 2004).
The irreversible enzymatic step of the alpha-ketoacid intermediate breakdown is mediated by the BCKDH complex. Gene expression levels for this complex are significantly downregulated in our NASH liver samples while other studies have reported on the alterations of this complex in rodent models of obesity (She et al. 2013). Decreased gene expression of BCKDK, the kinase which inhibits BCKDH function was observed in NASH not fatty livers in our study, however, BCKDK protein levels, conversely, are significantly increased in the NASH not fatty samples. Obese animal models have exhibited elevated hepatic BCKDK protein levels. The high plasma insulin levels in these obese rodent models are thought to contribute to stabilization of protein BCKDK (She et al. 2007) which may also occur in humans with NASH due to the close association of NAFLD with insulin resistance and obesity. Overall, the results we have shown are indicative of increased hepatic metabolism of BCAAs in NASH patients which may be indicators of a hepatic stress response mechanism.
Increased levels of hepatic acylcarnitine metabolites were observed in the metabolomic profile analysis of our NASH samples. Alterations in carnitine levels have been reported in the literature to be a result of high lipid loads, mitochondrial lipotoxicity, and altered lipid metabolism (Schooneman et al. 2013). Increased levels of acylcarnitines in the plasma of NASH patients have been previously reported (Kalhan et al. 2011) which are comparable to the hepatic elevations observed in the present study. An increase in short chain acylcarnitines (C3, C4 and C5) in plasma studies that are associated with obesity and insulin resistance (Kalhan et al. 2011; Schooneman et al. 2013) is also present in our NASH liver samples. Accumulation of the C3 chain propionylcarnitine is thought to be due to increased isoleucine and valine catabolism, while increased C5 acylcarnitines in NASH (butyryl and isovaleryl carnitine) may be a result of increases in both leucine and isoleucine catabolism (Newgard et al. 2009). The elevated levels of long-chain acylcarnitines in our steatosis and NASH liver samples in the present study parallel elevated levels of long-chain acylcarnitines observed in other studies centered on obesity and insulin resistance (Schooneman et al. 2013). These findings also indicate the presence of complications such as excessive fatty acid oxidation and mitochondrial lipid metabolism (Schooneman et al. 2013). Thus, the metabolomics profile of hepatic acylcarnitines in our samples may be a highly sensitive indicator of mitochondrial overload amid increased lipotoxicity in NAFLD progression.
While it is known that many compounds are transported between the liver and the blood, BCAA transport is a critical component of this process affected by disease status of the liver. We have previously reported changes in the expression several metabolizing enzymes and transporters in the same set of human liver samples (Fisher et al. 2009; Hardwick et al. 2011). An enrichment of downregulated uptake drug transporter genes was reported previously for these NASH samples (Lake et al. 2011). The gene expression of three important amino acid transporters SLC6A15, SLC16A10, and SLC43A1 from the NAFLD array data is included in this study. Gene expression of the efflux transporter, SLC43A1, was decreased in NASH liver samples and is potentially indicative of impaired BCAA efflux from liver to blood during NASH. SLC16A10 was also downregulated in NASH. In previous studies, deletion of SLC16A10 in rodent models resulted in an accumulation of plasma AAAs and decreased metabolism of AAA in the liver (Mariotta et al. 2012). Thus, decreased gene expression of SLC16A10 may indicate a potential mechanism for the elevation of tyrosine and phenylalanine levels in our NASH samples. A significant decrease in hepatic gene expression of SLC6A15 in NASH samples was observed while this transporter has been previously observed primarily in brain tissue (Takanaga et al. 2005). Overall, these findings indicate that uptake (SLC16A10, SLC6A15) and efflux (SLC43A1) transporter gene expression is compromised in NASH.
BCAAs significantly contribute to and help regulate mTOR signaling (Dodd and Tee 2012; Hagiwara et al. 2012; Newgard et al. 2009). BCAAs have been reported to potentially promote insulin resistance and glucose dysregulation through activation of the mTOR signaling pathway (Dodd and Tee 2012; Newgard et al. 2009). Other studies have shown that BCAAs are protective and beneficial to patients with advanced liver disease (Hagiwara et al. 2012; She et al. 2007). Importantly, chronic activation of mTOR signaling induced by excess fatty nutrition and BCAA supplementation is capable of exacerbating insulin resistance (Newgard et al. 2009). Gene expression levels for MTOR, AKT1, RPS6KA5 (positive regulator of MTOR) and RPS6KB2 (positive regulator of MTOR) were analyzed in this study to determine the status of mTOR activation at the transcriptional level in hepatic NAFLD samples. A significant decrease in mTOR gene expression among NASH samples was observed, while AKT1 was only significantly decreased in NASH not fatty samples. mTOR pathway signaling has several branches and roles that may be critical for hepatoprotection (Hagiwara et al. 2012). The mTORC1 signaling branch is activated by the BCAA leucine, which is thought to help drive negative feedback of the mTOR signaling pathway and inhibit development of cancer (Hagiwara et al. 2012). The influence of altered hepatic BCAA profiles upon MTOR signaling in human NAFLD progression is a critical aspect of this disease that will require future mechanistic studies.
The altered hepatic amino acid composition and BCAA catabolism profile results reveal important biochemical changes in human NASH. These findings contribute an improved understanding of the role of amino acids in progressive human NAFLD with implications for adaptive response mechanisms to lipotoxicity. Elevated hepatic BCAAs and acylcarnitines during NAFLD progression are indicative of a hepatic stress response to the increased inflammation and oxidative stress which characterize this disease. Overall, this study provides a strong basis for future mechanistic studies of amino acid homeostasis in human NAFLD.
Supplementary Material
Acknowledgments
We extend our sincere gratitude to Dr. Walter T. Klimecki for his valued scientific advice and contribution to the development of the human NAFLD microarray data set. We also thank Jose Munoz-Rodriguez and the Genomics Core Facility at the University of Arizona Cancer Center for the processing, archiving, and data acquisition of the microarrays. The authors also thank the National Institutes of Health (NIH)-sponsored Liver Tissue Cell Distribution System members for their help in the acquisition of human liver tissue at the University of Minnesota, Virginia Commonwealth and the University of Pittsburgh. This work was supported by the National Institutes of Health Grants, [AI083927], [HD062489], [ES006694]; the National Institute of Environmental Health Science Toxicology Training Grant [ES007091]; the Academy of Sciences of the Czech Republic with institutional support [RVO:60077344], and the Liver Tissue Cell Distribution System National Institutes of Health Contract [NO1-DK-7-0004/HHSN267200700004C].
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00726-014-1894-9) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that there are no conflicts of interest, financial or otherwise that would influence the performance or presentation of work in this manuscript.
Contributor Information
April D. Lake, Department of Pharmacology and Toxicology, The University of Arizona, 1703 East Mabel St, PO Box 210207, Tucson, AZ 85721, USA
Petr Novak, Biology Centre ASCR, Institute of Plant Molecular Biology, České Budějovice, Czech Republic.
Petia Shipkova, Bristol-Myers Squibb Co, Pharmaceutical Candidate Optimization, Princeton, NJ, USA.
Nelly Aranibar, Bristol-Myers Squibb Co, Pharmaceutical Candidate Optimization, Princeton, NJ, USA.
Donald G. Robertson, Bristol-Myers Squibb Co, Pharmaceutical Candidate Optimization, Princeton, NJ, USA
Michael D. Reily, Bristol-Myers Squibb Co, Pharmaceutical Candidate Optimization, Princeton, NJ, USA
Lois D. Lehman-McKeeman, Bristol-Myers Squibb Co, Pharmaceutical Candidate Optimization, Princeton, NJ, USA
Richard R. Vaillancourt, Department of Pharmacology and Toxicology, The University of Arizona, 1703 East Mabel St, PO Box 210207, Tucson, AZ 85721, USA
Nathan J. Cherrington, Email: cherrington@pharmacy.arizona.edu, Department of Pharmacology and Toxicology, The University of Arizona, 1703 East Mabel St, PO Box 210207, Tucson, AZ 85721, USA.
References
- Adams SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv Nutr Int Rev J. 2011;2(6):445–456. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeva MM, Calvino J, Souto G, Donapetry C. Insulin resistance and the metabolism of branched-chain amino acids in humans. Amino Acids. 2012;43(1):171–181. doi: 10.1007/s00726-011-1088-7. [DOI] [PubMed] [Google Scholar]
- Ali R, Cusi K. New diagnostic and treatment approaches in non-alcoholic fatty liver disease (NAFLD) Ann Med. 2009;41(4):265–278. doi: 10.1080/07853890802552437. [DOI] [PubMed] [Google Scholar]
- Bellentani S, Scaglioni F, Marino M, Bedogni G. Epidemiology of non-alcoholic fatty liver disease. Dig Dis. 2010;28(1):155–161. doi: 10.1159/000282080. [DOI] [PubMed] [Google Scholar]
- Bodoy S, Fotiadis D, Stoeger C, Kanai Y, Palacin M. The small SLC43 family: facilitator system L amino acid transporters and the orphan EEG1. Mol Aspects Med. 2012;12:S0098–S2997. doi: 10.1016/j.mam.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Bonora E. The metabolic syndrome and cardiovascular disease. Ann Med. 2006;38:64–80. doi: 10.1080/07853890500401234. [DOI] [PubMed] [Google Scholar]
- Boujedidi H, Bouchet-Delbos L, Cassard-Doulcier AM, Njike-Nakseu M, Maitre S, Prevot S, Dagher I, Agostini H, Voican CS, Emilie D, Perlemuter G, Naveau S. Housekeeping gene variability in the liver of alcoholic patients. Alcohol Clin Exp Res. 2012;36(2):258–266. doi: 10.1111/j.1530-0277.2011.01627.x. [DOI] [PubMed] [Google Scholar]
- Congiu M, Slavin JL, Desmond PV. Expression of common housekeeping genes is affected by disease in human hepatitis C virus-infected liver. Liver Int. 2011;31(3):386–390. doi: 10.1111/j.1478-3231.2010.02374.x. [DOI] [PubMed] [Google Scholar]
- Dam-Larsen S. Long-term prognosis of fatty liver: risk of chronic liver disease and death. Gut. 2004;53:750–755. doi: 10.1136/gut.2003.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP. Pathogenesis of steatohepatitis. Best Pract Res Clin Gastroenterol. 2002;16(5):663–678. doi: 10.1053/bega.2002.0333. [DOI] [PubMed] [Google Scholar]
- Day CP, James OFW. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114(4):842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- Dodd KM, Tee AR. Leucine and mTORC1: a complex relationship. Am J Physiol Endocrinol Metabol. 2012;302(11):E1329–E1342. doi: 10.1152/ajpendo.00525.2011. [DOI] [PubMed] [Google Scholar]
- Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, Cherrington NJ. Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos. 2009;37(10):2087–2094. doi: 10.1124/dmd.109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara D, Kanai Y, Chairoungdua A, Babu E, Bessho F, Kawano T, Akimoto Y, Endou H, Yan K. Protein characterization of Na+-independent system L amino acid transporter 3 in mice—a potential role in supply of branched-chain amino acids under nutrient starvation. Am J Pathol. 2007;170(3):888–898. doi: 10.2353/ajpath.2007.060428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco D, Kotronen A, Westerbacka J, Puig O, Arkkila P, Kiviluoto T, Laitinen S, Kolak M, Fisher RM, Hamsten A, Auvinen P, Yki-Jarvinen H. Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol. 2008;294(5):G1281–G1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- Hagiwara A, Nishiyama M, Ishizaki S. Branched-chain amino acids prevent insulin-induced hepatic tumor cell proliferation by inducing apoptosis through mTORC1 and mTORC2-dependent mechanisms. J Cell Physiol. 2012;227(5):2097–2105. doi: 10.1002/jcp.22941. [DOI] [PubMed] [Google Scholar]
- Hardwick RN, Fisher CD, Canet MJ, Scheffer GL, Cherrington NJ. Variations in ATP-binding cassette transporter regulation during the progression of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39(12):2395–2402. doi: 10.1124/dmd.111.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick RN, Ferreira DW, More VR, Lake AD, Lu Z, Manautou JE, Slitt AL, Cherrington N. Altered UDP-glucuronosyltransferase (UGT) and sulfotransferase (SULT) expression and function during progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2012;41(3):554–661. doi: 10.1124/dmd.112.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayaishi S, Chung H, Kudo M, Ishikawa E, Takita M, Ueda T, Kitai S, Inoue T, Yada N, Hagiwara S, Minami Y, Ueshima K. Oral branched-chain amino acid granules reduce the incidence of hepatocellular carcinoma and improve event-free survival in patients with liver cirrhosis. Dig Dis. 2011;29(3):326–332. doi: 10.1159/000327571. [DOI] [PubMed] [Google Scholar]
- Hnatyshyn S, Shipkova P, Sanders M. Expedient data mining for nontargeted high-resolution LC-MS profiles of biological samples. Bioanalysis. 2013;5(10):1195–1210. doi: 10.4155/bio.13.86. [DOI] [PubMed] [Google Scholar]
- Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60(3):404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- Koehler EM, Schouten JNL, Hansen BE, van Rooij FJA, Hofman A, Stricker BH, Janssen HLA. Prevalence and risk factors of non-alcoholic fatty liver disease in the elderly: results from the Rotterdam study. J Hepatol. 2012;57(6):1305–1311. doi: 10.1016/j.jhep.2012.07.028. [DOI] [PubMed] [Google Scholar]
- Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, Cherrington NJ. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39(10):1954–1960. doi: 10.1124/dmd.111.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesini G, Brizi M, Bianchi G, Tomassetti S, Bugianesi E, Lenzi M, McCullough AJ, Natale S, Forlani G, Melchionda N. Nonalcoholic fatty liver disease—a feature of the metabolic syndrome. Diabetes. 2001;50(8):1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- Mariotta L, Ramadan T, Singer D, Guetg A, Herzog B, Stoeger C, Palacin M, Lahoutte T, Camargo SMR, Verrey F. T-type amino acid transporter TAT1 (Slc16a10) is essential for extracellular aromatic amino acid homeostasis control. J Physiol. 2012;590(24):6413–6424. doi: 10.1113/jphysiol.2012.239574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AJ. Pathophysiology of nonalcoholic steatohepatitis. J Clin Gastroenterol. 2006;40:S17–S29. doi: 10.1097/01.mcg.0000168645.86658.22. [DOI] [PubMed] [Google Scholar]
- McCullough AJ. Epidemiology of the metabolic syndrome in the USA. J Dig Dis. 2011;12(5):333–340. doi: 10.1111/j.1751-2980.2010.00469.x. [DOI] [PubMed] [Google Scholar]
- Merrell MD, Cherrington NJ. Drug metabolism alterations in nonalcoholic fatty liver disease. Drug Metab Rev. 2011;43(3):317–334. doi: 10.3109/03602532.2011.577781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Abe M, Furukawa S, Tokumoto Y, Toshimitsu K, Ueda T, Yamamoto S, Hirooka M, Kumagi T, Hiasa Y, Matsuura B, Onji M. Long-term branched-chain amino acid supplementation improves glucose tolerance in patients with nonalcoholic steatohepatitis-related cirrhosis. Intern Med. 2012;51(16):2151–2155. doi: 10.2169/internalmedicine.51.7578. [DOI] [PubMed] [Google Scholar]
- Nagao Y, Kawaguchi T, Ide T, Sata M. Effect of branched-chain amino acid-enriched nutritional supplementation on interferon therapy in Japanese patients with chronic hepatitis C virus infection: a retrospective study. Virol J. 2012;9(1):282. doi: 10.1186/1743-422X-9-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard C. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15(5):606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy J, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E, Lavine JE, Schwimmer JB. Hepatic, cardiovascular, and endocrine outcomes of the histological subphenotypes of nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28(4):380–385. doi: 10.1055/s-0028-1091982. [DOI] [PubMed] [Google Scholar]
- Schooneman MG, Vaz FM, Houten SM, Soeters MR. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62(1):1–8. doi: 10.2337/db12-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Van Horn C, Reid T, Hutson SM, Cooney RN, Lynch CJ. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metabol. 2007;293(6):E1552–E1563. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Olson KC, Kadota Y, Inukai A, Shimomura Y. Leucine and protein metabolism in obese Zucker rats. PLoS One. 2013;8(3):e59443. doi: 10.1371/journal.pone.0059443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawan A, Hawes JW, Harris RA, Shimomura Y, Jenkins AE, Hutson SM. A molecular model of human branched-chain amino acid metabolism. Am J Clin Nutr. 1998;68(1):72–81. doi: 10.1093/ajcn/68.1.72. [DOI] [PubMed] [Google Scholar]
- Sweatt AJ, Wood M, Suryawan A, Wallin R, Willingham MC, Hutson SM. Branched-chain amino acid catabolism: unique segregation of pathway enzymes in organ systems and peripheral nerves. Am J Physiol Endocrinol Metabol. 2004;286(1):E64–E76. doi: 10.1152/ajpendo.00276.2003. [DOI] [PubMed] [Google Scholar]
- Tajiri K, Shimizu Y. Branched-chain amino acids in liver diseases. World J Gastroenterol. 2013;19(43):7620–7629. doi: 10.3748/wjg.v19.i43.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanaga H, Mackenzie B, Peng JB, Hediger MA. Characterization of a branched-chain amino-acid transporter SBAT1 (SLC6A15) that is expressed in human brain. Biochem Biophys Res Commun. 2005;337(3):892–900. doi: 10.1016/j.bbrc.2005.09.128. [DOI] [PubMed] [Google Scholar]
- Wong VWS, Chu WCW, Wong GLH, Chan RSM, Chim AML, Ong A, Yeung DKW, Yiu KKL, Chu SHT, Woo J, Chan FKL, Chan HLY. Prevalence of non-alcoholic fatty liver disease and advanced fibrosis in Hong Kong Chinese: a population study using proton-magnetic resonance spectroscopy and transient elastography. Gut. 2012;61(3):409–415. doi: 10.1136/gutjnl-2011-300342. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Yagi T, Sadamori H, Matsuda H, Shinoura S, Umeda Y, Sato D, Utsumi M, Nagasaka T, Okazaki N, Date A, Noguchi A, Tanaka A, Hasegawa Y, Sakamoto Y, Fujiwara T. Branched-chain amino acid-enriched nutrients improve nutritional and metabolic abnormalities in the early post-transplant period after living donor liver transplantation. J Hepato Biliary Pancreat Sci. 2012;19(4):438–448. doi: 10.1007/s00534-011-0459-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.