Summary
Chromothripsis is a catastrophic cellular event recently described in cancer in which chromosomes undergo massive deletion and rearrangement. Here we report a case in which chromothripsis spontaneously cured a patient with WHIM syndrome, an autosomal dominant combined immunodeficiency disease caused by gain-of-function mutation of the chemokine receptor CXCR4. In this patient, deletion of the disease allele, CXCR4R334X, as well as 163 other genes from one copy of chromosome 2 occurred in a hematopoietic stem cell (HSC) that repopulated the myeloid but not the lymphoid lineage. In competitive mouse bone marrow (BM) transplantation experiments, Cxcr4 haploinsufficiency was sufficient to confer a strong long-term engraftment advantage of donor BM over BM from either wild-type or WHIM syndrome model mice, suggesting a potential mechanism for the patient’s cure. Our findings suggest that partial inactivation of CXCR4 may have general utility as a strategy to promote HSC engraftment in transplantation.
Keywords: CXCR4, immunodeficiency; genetic mosaic; genetic reversion; papillomavirus; bone marrow transplantation; hematopoietic stem cell
Introduction
WHIM syndrome is an autosomal dominant combined primary immunodeficiency disease caused by mutations in the chemokine receptor CXCR4 (Hernandez et al., 2003). The term ‘WHIM’ is an acronym for the main manifestations of the disease: Warts, Hypogammaglobulinemia, recurrent Infections and Myelokathexis; myelokathexis refers to impaired egress of mature neutrophils from bone marrow causing neutropenia (Wetzler et al., 1990; Zuelzer, 1964). Most patients with WHIM syndrome are actually panleukopenic, with severely reduced peripheral blood B cells but less severe reductions in peripheral blood T cells and monocytes (McDermott et al., 2011). The signature pathogen in WHIM syndrome is HPV, which causes warts that cannot be controlled with standard medical treatment and may progress to cancer (Al Ustwani et al., 2014; Beaussant Cohen et al., 2012; Dotta et al., 2011; Kawai and Malech, 2009). Recurrent bacterial infections also occur, mainly in the sinopulmonary tract, oral cavity, ear, skin and soft tissue, where chronic complications may arise, especially bronchiectasis and hearing loss. Prophylactic antibiotics, IVIg and G-CSF are often used to reduce the incidence of infections; however, their precise efficacy has not been established (Al Ustwani et al., 2014; Beaussant Cohen et al., 2012; Dotta et al., 2011; Kawai and Malech, 2009). In contrast, safety and preliminary evidence of clinical efficacy has recently been reported from Phase I studies of the specific CXCR4 antagonist plerixafor (Mozobil™, AMD3100) (Dale et al., 2011; McDermott et al., 2011; McDermott et al., 2014). Spontaneous remission or cure of WHIM syndrome has not been previously reported.
WHIM mutations of CXCR4 increase signaling because they disrupt negative regulatory elements in the carboxy-terminus, thereby exaggerating the normal hematopoietic functions of the receptor (Haribabu et al., 1997; Signoret et al., 1998; Venkatesan et al., 2003). CXCR4 is normally expressed by most leukocytes and has one ligand, CXCL12 (Bachelerie et al., 2014; Bleul et al., 1996), which is constitutively expressed at high levels by stromal cells in the bone marrow and normally mediates HSC retention in bone marrow niches (Broxmeyer et al., 2003b; Broxmeyer et al., 2005; Dar et al., 2006; Sugiyama et al., 2006). In addition, CXCR4 signaling promotes HSC quiescence, homing to bone marrow from blood and differentiation into committed myeloid progenitors (Broxmeyer et al., 2003a; Broxmeyer et al., 2003b; Kawai et al., 2007; Nie et al., 2008; Sugiyama et al., 2006).
Chromothripsis refers to multiple clustered genetic rearrangements and deletions affecting one or a few chromosomes (Stephens et al., 2011). The abnormalities are thought to occur all at once in a single cell, which then presumably either dies or acquires a growth advantage, depending on the genes affected (Stephens et al., 2011). Accordingly, chromothripsis was first identified by whole genome sequencing of cancer cell lines, and has been reported to affect approximately 2% of all cancers (Jones and Jallepalli, 2012), as well as one patient with a severe congenital cognitive syndrome (Kloosterman et al., 2011). Criteria for chromothripsis include 1) clustering of breakpoints in limited areas of one or several chromosomes with large intervening regions of normal sequence, 2) copy number states that suddenly oscillate between areas of normal heterozygosity and loss of heterozygosity, and 3) rearrangements affecting a single haplotype with multiple fragments rearranged in random orientation and order (Korbel and Campbell, 2013). Here we describe chromothriptic deletions of one copy of chromosome 2, including deletion of the disease allele CXCR4R334X, in a patient with WHIM syndrome that resulted in cure of the disease.
Results
Cure of a patient with WHIM syndrome
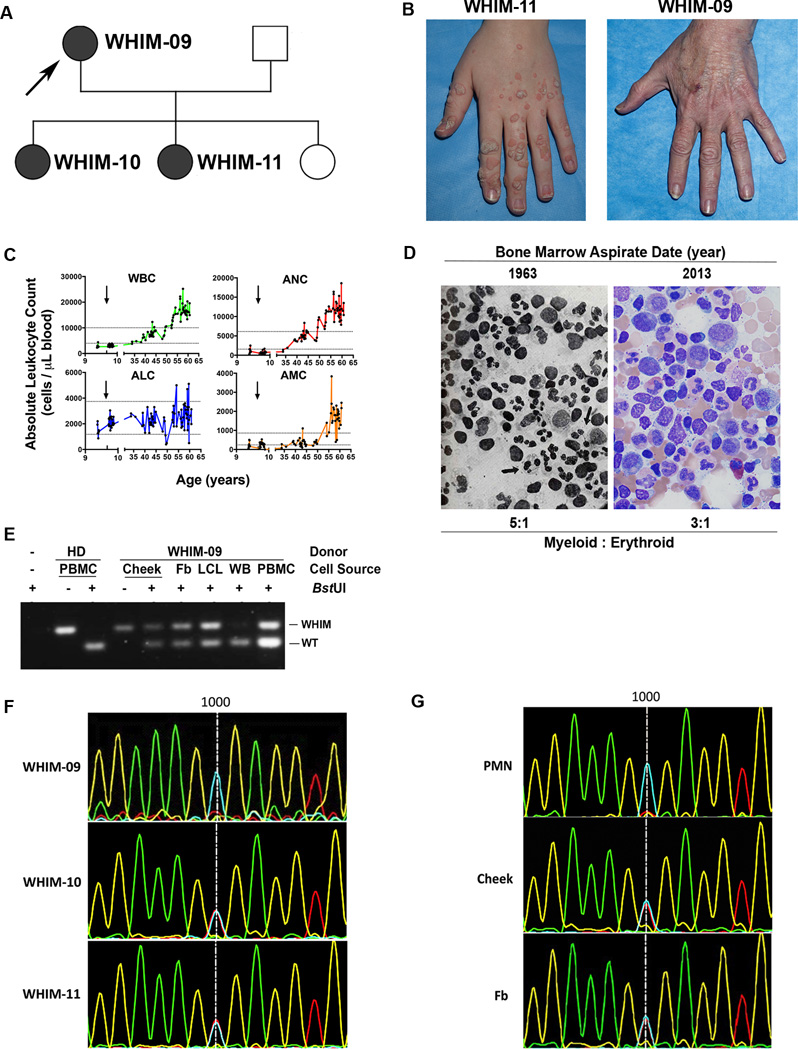
The index patient, designated WHIM-09, is a white female who presented at age 58 to the NIH requesting evaluation for herself and 2 of her 3 daughters, designated WHIM-10 (age 21) and WHIM-11 (age 23) (Figure 1A). Both daughters had a history of recurrent infections since early childhood, multiple cutaneous warts, panleukopenia and hypogammaglobulinemia, and therefore fulfilled all the clinical criteria for WHIM syndrome. WHIM-09, however, did not. Instead she reported that from childhood through age 38 she had had many serious infections, often requiring hospitalization, but then none in the 20 subsequent years, and that she had had confluent warts on her hands that spontaneously resolved also in her 30’s (Figure 1B). Moreover, we found that at the time of presentation WHIM-09 was not neutropenic, but instead had a mild leukocytosis, including an absolute neutrophil count (ANC) and absolute monocyte count (AMC) that were ~2-fold > the upper limit of normal; in contrast, the absolute lymphocyte count (ALC) was within normal limits. The past medical history revealed that WHIM-09 was in fact the first patient ever described with myelokathexis, the key hematopathologic feature in WHIM syndrome, reported in two articles published in The New England Journal of Medicine (NEJM) in 1964 (Krill et al., 1964; Zuelzer, 1964). She underwent a therapeutic splenectomy at age 9 for the possibility of autoimmune neutropenia, which was ineffective. There is no evidence that her parents or siblings had WHIM syndrome. Thus, the history and clinical evidence were compatible with a WHIM mutation occurring de novo in patient WHIM-09, autosomal dominant transmission to two of her three daughters, and spontaneous and durable complete clinical remission in WHIM-09 in her fourth decade of life (Figure 1A).
Figure 1. Long-term clinical remission of WHIM syndrome and evidence for somatic mosaicism in patient WHIM-09.
(A) Family Pedigree. Squares, males; circles, females; shaded symbols, WHIM syndrome; arrow, index patient WHIM-09.
(B) Spontaneous and complete remission of warts in patient WHIM-09. According to patient WHIM-09, through her fourth decade of life she had had extensive warts on her hands, similar to her daughters, (illustrated here for 24 year old daughter WHIM-11) that spontaneously resolved.
(C) Spontaneous sustained correction of neutropenia and monocytopenia in patient WHIM-09. WBC, white blood cell count; ANC, absolute neutrophil count; AMC, absolute monocyte count; ALC, absolute lymphocyte count. Arrows indicate age at splenectomy; horizontal lines indicate normal range for each cell type. Note, × axis is discontinuous to show pre/post splenectomy results more clearly.
(D) Normalization of bone marrow pathology in patient WHIM-09. A representative high magnification (500×) Wright-Giemsa stain of the bone marrow aspirate is shown for the index patient WHIM-09 in 1963 (Zuelzer, 1964) (reproduced with permission) and in 2013 at ages 9 and 59, respectively. Arrows in left image, eyeglass nuclei in neutrophils. (E) Polymerase chain reaction-BstUI restriction fragment length polymorphism (BstUI) analysis of genomic DNA. HD, healthy donor; WHIM-09, index patient; Cheek, cheek swab cells; PBMC, peripheral blood mononuclear cells; Fb, fibroblast; LCL, lymphoblastoid cell line; WB, whole blood; WT, wild-type allele; WHIM, CXCR4R334X allele that causes WHIM syndrome. (F) Sanger DNA sequencing analysis of whole blood DNA for affected family members in the region near nucleotide position 1000 (vertical line), the site of WHIM mutation CXCR4R334X (1000 C→T). Blue, C; Red, T; Fb, fibroblast; PMN, polymorphonuclear leukocyte (PMN). (G) Sanger DNA sequencing analysis of DNA from purified peripheral blood neutrophils, cells obtained from a buccal swab (cheek) and cultured skin fibroblasts for patient WHIM-09 in the same region as (F).
To evaluate potential mechanisms for clinical remission, we first graphed all available white blood cell counts for WHIM-09, including those previously published in the NEJM (Figure 1C). Consistent with the clinical history, this revealed severe neutropenia at least from age 9 that was unaffected by splenectomy but that began to resolve spontaneously early in the fourth decade of life, rising slowly over time to a new and stable baseline slightly above the upper limit of normal. The AMC followed the same time course, whereas, interestingly, the ALC did not, starting in the normal range for healthy individuals as a child then increasing inconsistently and only slightly as an adult. Nevertheless, when lymphocyte subsets were examined in detail, all B cell subsets and both naïve CD4+ and CD8+ T cell subsets were below the lower limit of normal (Table 1), as they were in both daughters and most other patients reported with WHIM syndrome. Consistent with this, WHIM-09 was slightly hypogammaglobulinemic at the time of presentation to NIH with IgG = 535 mg/dL (normal range, 642–1730 mg/dL). In contrast, memory CD4+ and CD8+ T cell subsets were elevated in WHIM-09, but deficient in both daughters. Unfortunately, WHIM-09’s archival lymphocyte subset values from the years when she fulfilled the clinical criteria for WHIM syndrome were not available.
Table 1.
Distribution of leukocyte subsets in the peripheral blood of index patient WHIM-09, in clinical remission from WHIM syndrome, and her two affected daughters (WHIM-10 and WHIM-11). Data are presented as absolute numbers of cells having the indicated immunophenotype per microliter of whole blood.
| Leukocyte subset | WHIM-09 | WHIM-10 | WHIM-11 | Normal adult reference range† |
|---|---|---|---|---|
| Neutrophil | 9980* | 380# | 530# | 1560–6130 |
| CD3+ T cell | 1736 | 382# | 218# | 714–2266 |
| CD4+ T cell | 1389 | 317# | 155# | 359–1565 |
| CD4+ CD45RA+ naïve T cell | 211# | 44# | 12# | 454–733 |
| CD4+ CD45RO+ memory T cell | 1178* | 273 | 143# | 219–1048 |
| CD8+ T cell | 222 | 41# | 45# | 178–853 |
| CD8+ CD45RA+ naïve T cell | 44# | 22# | 14# | 231–371 |
| CD8+ CD45RO+ memory T cell | 178* | 19# | 31# | 57–130 |
| CD3−CD56+ NK cell | 1270* | 32# | 51# | 126–729 |
| CD19+ B cell | 18# | 5# | 10# | 61–329 |
| CD19+ CD27+ memory B cell | 2# | 1# | 1# | 12–68 |
| CD19+ CD27−B cell | 16# | 4# | 9# | 90–176 |
| CD19+ CD27−IgD+ IgM+ transitional/naïve B cell | 12# | 3# | 4# | 42–85 |
| CD19+ CD27−IgD−IgM+ immature B cell | 4 | 1# | 1# | 2–10 |
| CD14+ CD16−Classical Monocyte | 1344* | 35# | 117# | 371–539 |
| CD14+ CD16+ Inflammatory Monocyte | 235* | 5# | 23 | 14–30 |
Value above the upper limit of normal;
Value below the lower limit of normal.
Based on the values of 11–40 healthy blood donors seen at the NIH Clinical Center.
We considered myeloid leukemia or a possible pre-leukemic state as a cause of her mild leukocytosis; however, the patient was clinically well over this ~20 year time span when her neutrophils and monocytes were increasing, and her blood smear and bone marrow histopathology at the NIH were not consistent with these diagnoses (Figure 1D); moreover, specific testing for B and T cell clonality as well as for BCR-ABL and JAK mutations were negative (see Extended Experimental Methods for details). Consistent with her apparent ~20 year complete remission of WHIM syndrome by clinical criteria, her bone marrow did not present the characteristic features of the disease (increased myeloid:erythroid ratio, neutrophil vacuolization, eyeglass nuclei in neutrophils), which were, however, all present in her bone marrow histopathology reported in the NEJM in 1964, shown again here for comparison, with permission, in Figure 1D (Zuelzer, 1964). Since the patient reported she had undergone several prior surgeries and blood transfusions, we tested her blood for evidence of allogeneic chimerism and found none (data not shown). Thus, although the patient appeared to be clinically cured, she was hematologically mosaic, with sustained spontaneous correction of neutropenia, monocytopenia and myelokathexis, and continued deficiency of B and naïve T lymphocytes in the blood.
Patient WHIM-09 is a genetic mosaic for CXCR4R334X, the most common mutation causing WHIM syndrome
We next genotyped whole blood DNA from family members using an established polymerase chain reaction-BstUI restriction fragment length polymorphism (PCR-RFLP) assay for the most common mutation in WHIM syndrome, CXCR4R334X. Both affected daughters (WHIM-10 and WHIM-11) tested positive, whereas the unaffected husband and third unaffected daughter tested negative. Surprisingly, two independent whole blood samples from WHIM-09, in which the leukocyte content was composed mostly of neutrophils and monocytes (Figure1E and 1F), also tested negative for the mutation. In contrast, DNA from WHIM-09 PBMCs, which were composed mostly of lymphocytes, as well as from a lymphoblastoid cell line generated from WHIM-09 PBMCs both tested positive for CXCR4R334X (Figure 1E). The mutation was also not detectable by direct sequencing of whole blood cell DNA from WHIM-09, whereas whole blood cell samples from both daughters, WHIM-10 and WHIM-11, were both positive (Figure 1F). In contrast, DNA samples from a cheek swab and fibroblasts cultured from a skin biopsy from WHIM-09 were both heterozygous for CXCR4R334X (Figure1E and 1G), defining her as a somatic genetic mosaic. A WHIM pedigree with germ-line/somatic genetic mosaicism, where the mother of 2 affected children was hematologically normal, has previously been described (Hernandez et al., 2003), but was unlikely to apply to WHIM-09 since she had been markedly symptomatic with severe neutropenia and myelokathexis as a child and cutaneous warts as an adult (Krill et al., 1964; Zuelzer, 1964). Therefore, we investigated whether a genetic reversion had occurred.
Chromothripsis as the mechanism for genetic mosaicism in patient WHIM-09
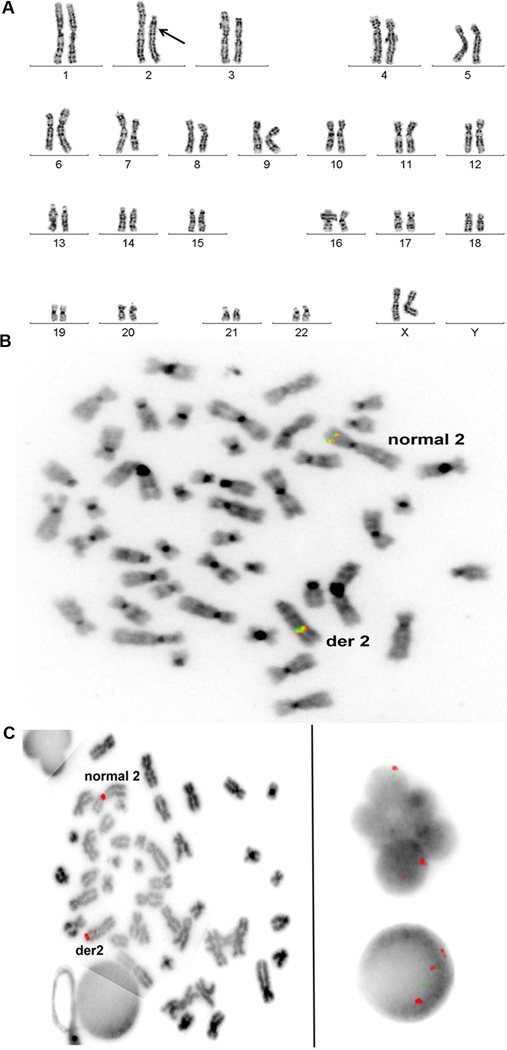
The PCR-RFLP assay and DNA sequencing results may both be explained by either reversion or deletion in patient leukocytes of the mutant nucleotide that defines the CXCR4R334X allele. To address this, we first performed cytogenetic analysis of bone marrow cells. In all 20 metaphase cells examined, one copy of chromosome 2, where CXCR4 is located, was acrocentric rather than submetacentric, and was significantly shorter than its normal homolog (Figure 2A). All other chromosomes appeared normal. In addition to deletions, the banding pattern of the abnormal chromosome 2 suggested the presence of inversions. Fluorescence in situ hybridization (FISH) revealed that the anaplastic lymphoma receptor tyrosine kinase (ALK) gene, at 2p23 on the normal chromosome 2, was on the long arm of the abnormal chromosome 2 (Figure 2B). FISH also demonstrated that the abnormal chromosome 2 had a portion of the centromere inverted into the long arm and the N-myc gene (MYCN), normally on 2p24, was absent (Figure 2C). Almost all of the interphase bone marrow cells with polymorphic nuclei, consistent with neutrophils, had the abnormal hybridization patterns of the abnormal chromosome 2 (Figure 2C), whereas cells with round nuclei had a significantly lower percentage of the abnormal chromosome (data not shown). An immunohistochemical stain of ALK activity on a bone marrow core biopsy section did not reveal abnormal activation of the enzyme despite its abnormal chromosomal location (Figure S1).
Figure 2. Massive deletion and rearrangement of one copy of chromosome 2, the location of CXCR4, in patient WHIM-09.
Cytogenetics and Fluorescence in-situ hybridization (FISH) analyses were performed on cultured bone marrow cells.
(A) Cytogenetics. Karyogram showing a short acrocentric chromosome 2 (arrow) with abnormal banding pattern suggesting deletions and inversions observed in all 20 metaphase cells analyzed from bone marrow.
(B) FISH with intact ALK break apart probe (Abbott Molecular) signal at 2p23 on normal chromosome 2 and on long arm of abnormal chromosome 2 (der 2).
(C) FISH with NMYC and CEP 2 probe set (Abbott Molecular). Left panel, Metaphase cell showing the normal chromosome 2 with intact centromere signal (red) and NMYC signal (green) at 2p24. The abnormal chromosome 2 has a portion of its centromere inverted into the long arm splitting the red signal; the green NMYC signal is absent. Right panel, Polymorphonuclear and round interphase nuclei showing the abnormal hybridization pattern with one green NMYC signal and three red CEP 2 signals. One of the red signals is smaller than the other two and is close to one of them.
To define the chromosomal abnormalities with greater resolution, microarray was performed using patient bone marrow cell and cultured fibroblast DNA. This revealed that the abnormal chromosome 2 was ~35 megabases shorter than usual due to 7 large deletions (Figure S2A), as demonstrated by loss of heterozygosity in these regions and by an abrupt change in the relative copy number from 2 to 1. One of the deletions involved the MYCN gene, confirming the FISH results. In addition, one of the deletions included the position of the CXCR4 gene (Figure S2B). This had made the patient hemizygous for CXCR4 in cells having the abnormal chromosome 2 with loss of the CXCR4R334X mutation as well as loss of one copy of 163 other annotated genes (Table S1). Thus, development of hemizygosity of CXCR4, with only the wildtype copy remaining, explained why the CXCR4R334X mutation was not detectable in neutrophil DNA by either DNA sequencing or PCR-RFLP. However, this potentially fortuitous deletion was clearly only a small part of a much larger and more complex genetic event.
We next performed whole genome sequencing (WGS) for single nucleotide definition of the event, comparing blood neutrophil and skin fibroblast DNA from WHIM-09 to each other and to the standard human genome sequence in Genbank (version hg19). We obtained an average of 40× coverage of both samples (Submitted to Genbank, accession number pending) and an analysis was performed to locate the DNA reads that had homology with multiple distinct areas of chromosome 2. This technique allowed precise base pair level identification of the inversions and deletions that had occurred on one copy of chromosome 2 (Table S2). No other large deletions or inversions were detected in the genome.
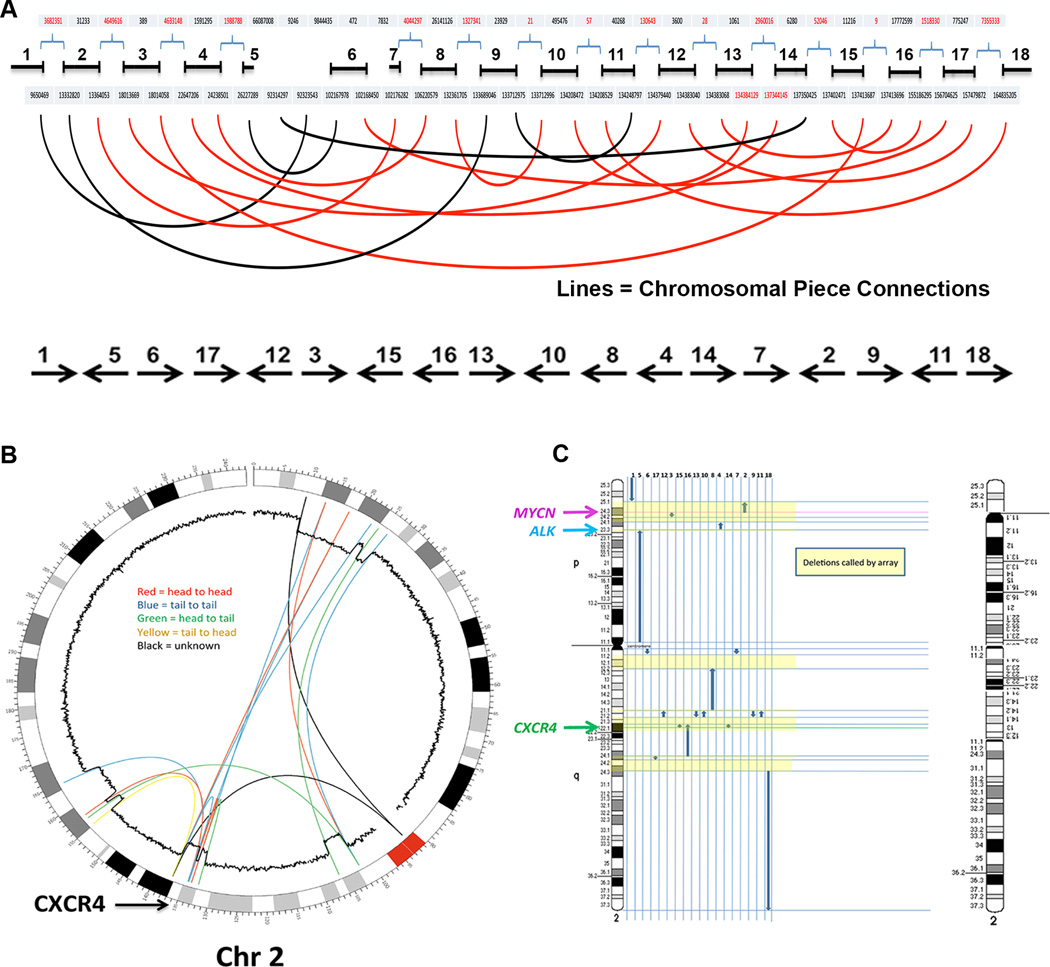
The changes seen in the microarray were confirmed by WGS but were actually more complex than had been initially suspected. The derivative chromosome was composed of 18 remaining pieces arranged in random orientation and in a random order, characteristic of chromothripsis (Figure 3A). A circular plot of the connections revealed by paired end WGS of the neutrophil DNA is shown in Figure 3B. The abnormal derivative chromosome, modeled using the breakpoints and connections (Figure 3C), revealed a predicted structural banding pattern that was identical to what was observed by cytogenetic analysis. We also developed primer pairs spanning 4 of the unique chromothriptic boundaries and demonstrated experimentally that they generated the predicted rearranged product from WHIM-09 neutrophil DNA but not from healthy donor DNA (data not shown). By analyzing the rearrangement breakpoints for the creation of novel fusion genes, we found two possibilities, fusions of MFSD2B with LOC285000 and POTEKP with NCKAP5 (Figure S3); however, these genes are unlikely to be expressed or functional because each lacks a promoter and transcription initiation site, and major regions of the potentially fused genes are deleted. Thus, the most likely explanation for the patient’s clinical improvement was haploinsufficiency for one or more of the 164 genes affected by chromothripsis.
Figure 3. Chromothripsis as the mechanism for loss of CXCR4R334X in patient WHIM-09.
Purified neutrophil and cultured skin fibroblast DNA from patient WHIM-09 was isolated and subjected to whole genome sequencing with paired end analysis.
(A) Linear non-proportional plot of the abnormal copy of chromosome 2 in patient WHIM-09 labeled from the p arm telomere (0) to the q arm telomere (~240) in megabases with the 18 remaining pieces arranged in their numeric order (top). Connections between these pieces are depicted by the curved lines. Note that some connections were poorly defined because of the involvement of repetitive centromeric sequence (black lines). The order and orientation of the 18 remaining pieces in the derivative chromosome are indicated at the bottom.
(B) Circos plot of chromosome 2 and its normal Giemsa cytogenetic banding pattern labeled from the p arm telomere (0) to the q arm telomere (~240) in megabases. Large pieces of chromosome 2 were missing from patient WHIM-09 neutrophil DNA and the 18 remaining pieces were arranged in random order. Connections between these pieces and their orientation (inset) are depicted by the colored lines (See Figure S2 and Tables S1 and S2 for additional details). Note that 2 connections were poorly defined because of the involvement of repetitive centromeric sequence. The inner circular trace is the copy number variation data derived from microarray analysis (Figure S2A and S2B). Note that the sites of connections derived from the paired end sequencing analysis closely match the sites where copy number variation abruptly falls from 2 to 1. The location of CXCR4 is indicated at lower left.
(C) Derivative chromothriptic chromosome 2. Ideogram of intact chromosome 2 (left) and a model of the Giemsa cytogenetic banding pattern and the order and orientation of the 18 pieces with the deletions called by microarray shaded in yellow (right) are shown. The resultant remaining chromosome 2 banding pattern predicted by whole genome sequencing closely matches that seen by cytogenetic analysis (see Figure 3). Note the location of CXCR4 at 2q22.1 in one of the deleted segments.
Myeloid/lymphoid mosaicism established by chromothripsis in an HSC from patient WHIM-09
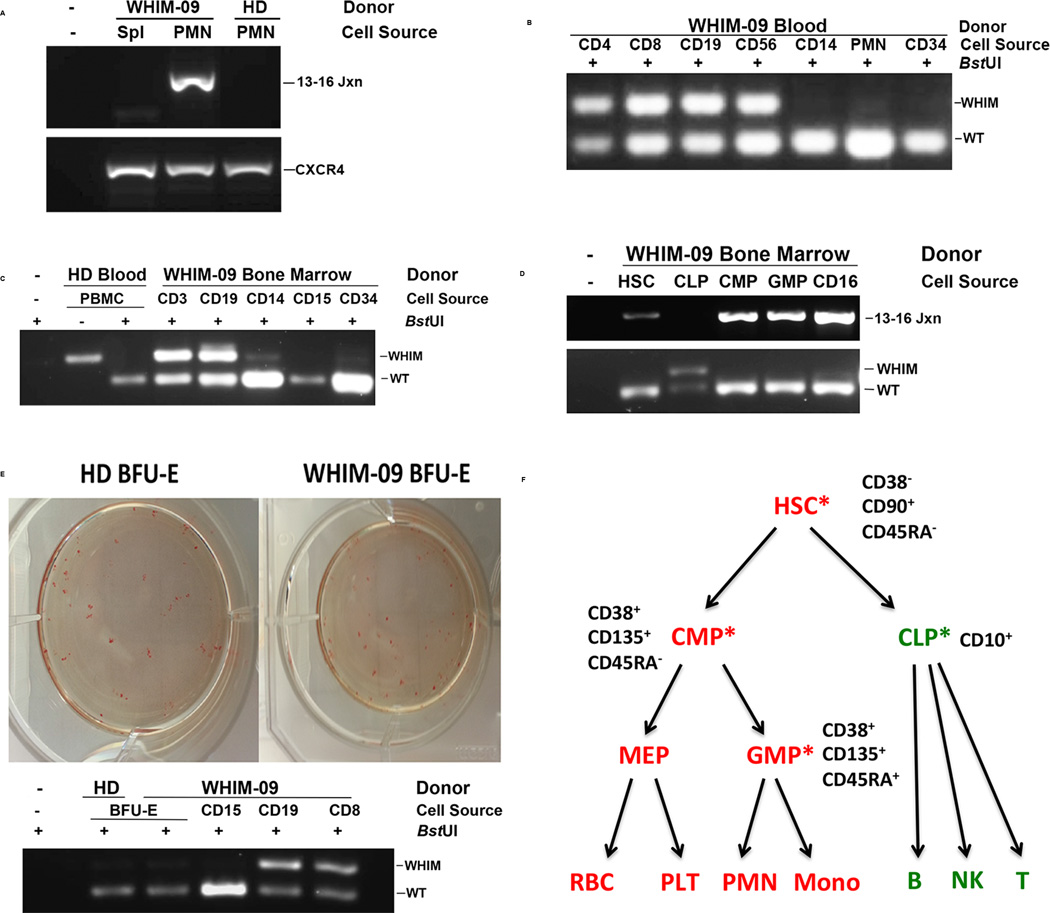
Chromothripsis-specific primers did not generate a detectable product by PCR when DNA prepared from the archival spleen sample from patient WHIM-09 was amplified, suggesting that the chromothriptic event occurred after age 9, when splenectomy was performed, but before age 35, when the rise in WBC was already underway (Figure 4A). The emergence of uniformly chromothriptic neutrophils, as suggested by WGS, PCR and bone marrow cytogenetics and FISH, implied that at least some patient HSCs must also be chromothriptic. We verified this with CD34+ leukocytes cultured from peripheral blood, as well as with CD38−CD90+CD45RAHSCs FACS-sorted from patient bone marrow (Figure S4 and Figures 4B and 4C), which both appeared to be markedly deficient in or lack the CXCR4R334X allele by the BstUI PCR-RFLP assay. Consistent with this, CD38+CD135+CD45RA− common myeloid precursor cells (CMP) and CD38+CD135+CD45RA+ granulocyte-monocyte precursor cells (GMP) sorted from patient bone marrow also lacked the CXCR4R334X allele by the PCR-RFLP assay, whereas FACS-sorted CD45+ CD34+ CD38+ CD10+ common lymphoid precursors (CLP) remained CXCR4+/R334X (Figure 4D, lower panel). This pattern was confirmed by PCR using chromothripsis-specific primers and DNA from HSCs, CMPs and CLPs (Figure 4D, upper panel). Consistent with this, we found that all mature myeloid cell types tested in both blood and bone marrow lacked or were markedly deficient in the CXCR4R334X WHIM allele, whereas all mature lymphoid cell types tested in peripheral blood and bone marrow were heterozygous CXCR4+/R334X (Figures 4B and C). This aligns with the cytogenetic results presented in Figure 2C. Further, we found 100% of EBV-transformed B cell lines (n=10) prepared from patient blood lacked the chromothriptic chromosome (data not shown). As expected, erythroid precursors, which derive from CMPs, also lacked the CXCR4R334X allele as demonstrated by BstUI PCR-RFLP analysis of Burst Forming Unit-Erythroid (BFU-E) colonies generated from patient CD34+ cells cultured from PBMCs ex vivo (Figure 4E). Thus, the combined evidence suggests that an HSC underwent chromothripsis and selectively repopulated the myeloid lineage (including the erythroid lineage), but not the lymphoid lineage (Figure 4F).
Figure 4. Chromothriptic CXCR4-haploinsufficient HSC replacement of the myeloid lineage, but not the lymphoid lineage, is associated with clinical remission in patient WHIM-09.
(A–E) Representative results from a BstUI PCR-restriction fragment length polymorphism assay (BstUI), designed to distinguish the wild type CXCR4 allele (WT) from the CXCR4R334X WHIM allele (WHIM), as well as from a PCR assay specific for the chromothriptic chromosome (13–16 Jxn). PCR was performed on DNA obtained from the indicated donor leukocyte subsets purified either from blood using magnetic bead purification (B, E) or from a bone marrow aspirate using flow cytometric sorting (C, D). DNA was also prepared from archived WHIM-09 spleen and compared with peripheral blood PMN DNA (A), as well as from Burst-forming Unit-Erythroid colonies and compared with blood leukocyte subsets (E). WHIM-09, index patient; HD, healthy donor; Spl, spleen; PMN, polymorphonuclear leukocytes; 13–16 Jxn, PCR product specific for the chromothriptic junction between segments 13 and 16 of the chromothriptic chromosome of patient WHIM-09; PBMC, peripheral blood mononuclear cells; CD4, purified CD4+ T cells; CD8, purified CD8+ T cells; CD19 purified CD19+ B cells; CD56, purified CD56+ natural killer cells; CD14, purified CD14+ monocytes; CD34, purified CD34+ hematopoietic cells; CD3, purified CD3+ T cells; CD15, purified CD15+ neutrophils; CD16, purified CD16+ neutrophils; HSC, hematopoietic stem cells; CLP, common lymphoid precursor; CMP, common myeloid precursor; GMP, granulocyte/monocyte precursor MEP, megakaryocyte-erythroid precursor; BFU-E, Burst-forming Unit-Erythroid; CXCR4, CXCR4 amplicon not digested with BstUI. (F) Summary of myeloid/lymphoid mosaicism for CXCR4R334X in patient WHIM-09. The immunophenotype used to purify each cell type from enriched CD34+CD45+ cells is summarized next to each cell type shown. Red, positive for CXCR4R334X; green, negative for CXCR4R334X; asterisks, purified cell types directly analyzed by PCR-RFLP for the WHIM mutation.
Cxcr4 haploinsufficiency enhances engraftment during bone marrow transplantation
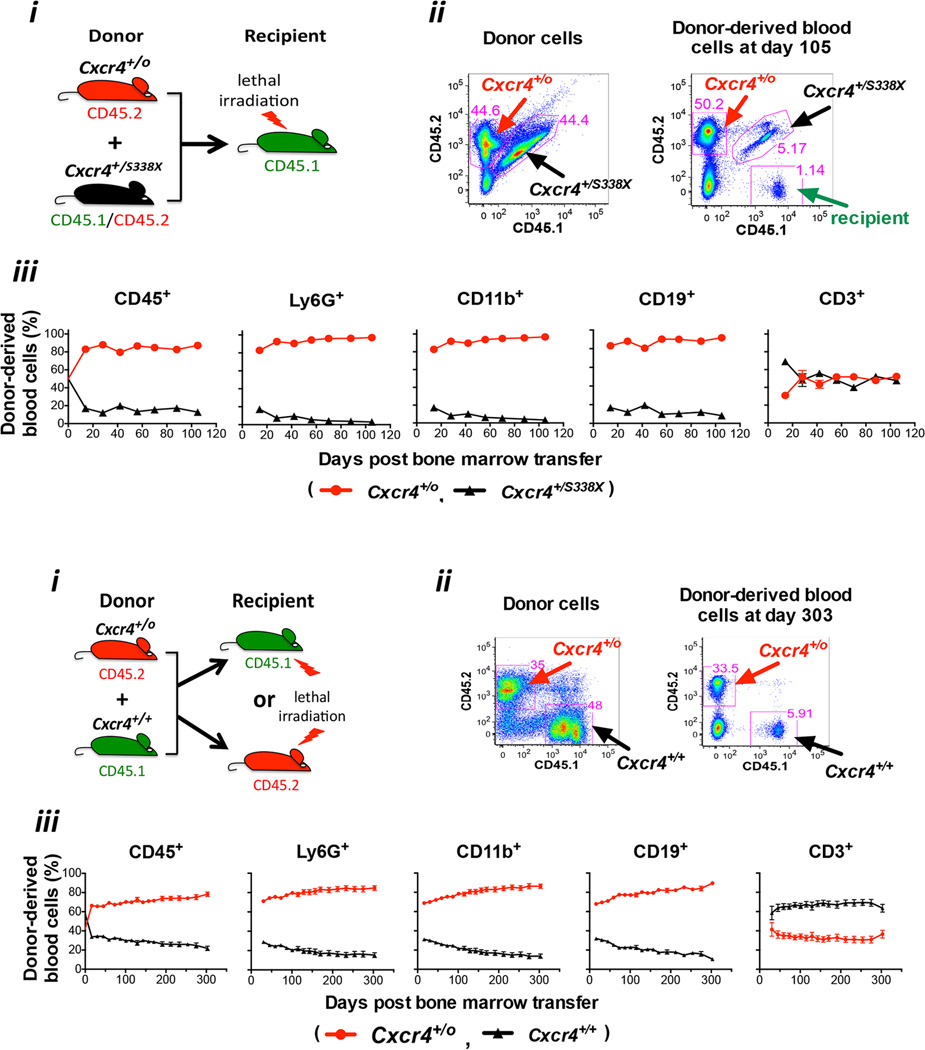
To test whether deletion of the disease allele CXCR4R334X alone, and not any of the other 163 genes deleted by chromothripsis, would be sufficient to confer a selective advantage to an HSC in the context of WHIM syndrome, we performed competitive repopulation experiments using a mouse model of WHIM syndrome (Cxcr4+/S338X), hemizygous Cxcr4 mice (Cxcr4+/o, the same as the HSC and myeloid cell CXCR4 genotype of patient WHIM-09) and wild type (Cxcr4+/+) mice as donors and recipients. Leukocytes from these strains were specifically marked with either CD45.1 or CD45.2 or both, allowing tracking of transplanted donor cell fate in vivo by FACS. When ~2.5 × 106 total donor bone marrow cells from both Cxcr4+/S338X and Cxcr4+/o mice were mixed together in equal proportions (~50%:50%) and transplanted into lethally irradiated Cxcr4+/+ recipients, by day 105 post-transplantation the percentage of Cxcr4+/S338X neutrophils, monocytes and B cells detectable in the peripheral blood had declined to ~5%, whereas the percentage of each of the corresponding Cxcr4+/o leukocyte subsets had increased to ~95% (Figure 5A). The rate at which the proportion of donor-derived blood cells diverged in frequency from the input ratio of 50:50 occurred in two phases, a rapid early phase within two weeks and a slow late phase evident by two weeks after transplantation. The inflection point could be earlier than two weeks, since this is the first time point when data were collected to allow for recovery after transplantation. Importantly, there was no difference in the baseline HSC frequency in bone marrow for Cxcr4+/o and Cxcr4+/S338X donor mice, so the number of HSCs transplanted was similar (Figure S5). Since the ANC in the peripheral blood of Cxcr4+/S338X donor mice is only ~50% less than the corresponding value in both Cxcr4+/+ mice (Balabanian et al., 2012, and J-L Gao et al., data not shown) and Cxcr4+/o mice (J-L Gao, data not shown), the extreme skewing of these transplantation results in the blood suggested that hemizygous Cxcr4 (Cxcr4+/o) HSCs may have a selective advantage over WHIM (Cxcr4+/S338X) HSCs for engraftment in this system. Consistent with this, in the bone marrow, as in the blood, we observed a greater content of mature Cxcr4+/o leukocytes compared to mature Cxcr4+/S338X leukocytes (data not shown). No difference was observed for the frequencies of Cxcr4+/o and Cxcr4+/S338X T cells in the blood, presumably owing to homeostatic proliferation of mature T cells present in donor bone marrow transferred into irradiated hosts.
Figure 5. Cxcr4 haploinsufficiency enhances HSC engraftment during mouse bone marrow transplantation.
Two types of competitive bone marrow transplantation experiments were performed: (A) Cxcr4+/o vs. Cxcr4+/S338X (mouse model of WHIM syndrome); and (B) Cxcr4+/o vs. Cxcr4+/+. For panels A and B: i) Experimental design; ii) Representative flow cytometry plots demonstrating the relative contributions of CD45 congenic markers in mixed donor bone marrow prior to transplantation (left panel) and in blood after bone marrow transplantation (right panel) in a single mouse; iii) Cell frequency data for the leukocyte subsets indicated at the top of each panel, presented as the mean ± SEM percentage (%) of total donor-derived cells for each subset (n=10 mice per data point). SEM was < 5% of the mean in all cases, and therefore is not visible for most data points. Results were verified in one and two additional independent experiments for panels A and B, respectively.
To test whether there might be a general selective advantage of Cxcr4 hemizygosity in transplantation, we performed competitive repopulation experiments by mixing ~2.5 × 106 total donor bone marrow cells each from Cxcr4+/+ and Cxcr4+/o mice and transplanting the mixture into lethally irradiated Cxcr4+/+ recipients. The baseline HSC frequency in bone marrow was the same for Cxcr4+/o and Cxcr4+/+ donor mice (Figure S5). In this case, the input ratio was skewed slightly in favor of the Cxcr4+/+ cells over the Cxcr4+/o cells (58%:42%). Nevertheless, by day 303 post-transplantation, when the animals were sacrificed, the percentage of Cxcr4+/+ neutrophils, monocytes and B cells detectable in the peripheral blood had declined to less than ~15%, whereas the percentage of each of the corresponding Cxcr4+/o leukocyte subsets had increased to ~85% with similar kinetics as for the competition with Cxcr4+/S338X bone marrow (Figure 5B). The same effect was observed whether the irradiated recipient mouse was CD45.1 or CD45.2 or Cxcr4+/o (data not shown), and whether the donor bone marrows were depleted of lineage-positive cells (Figure S6).
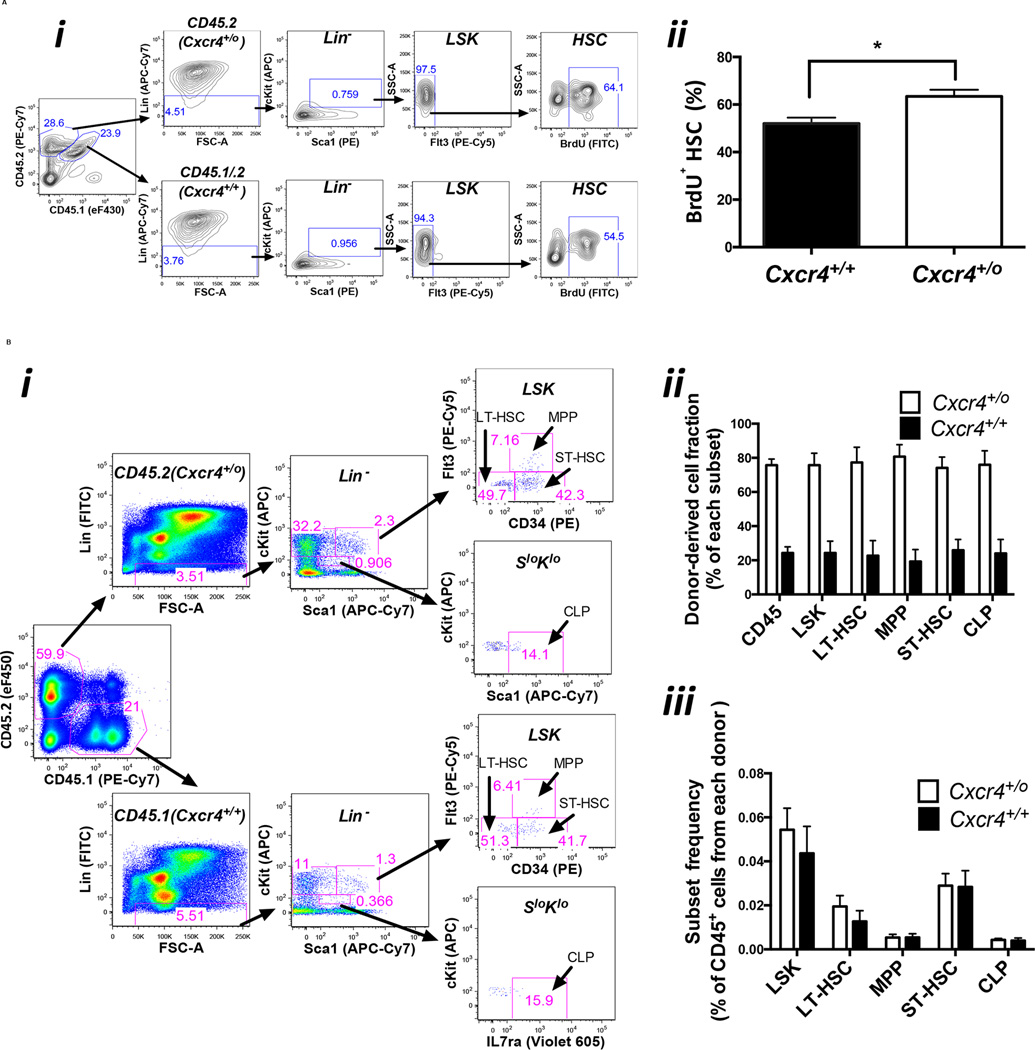
To investigate the mechanism for the apparent competitive advantage of Cxcr4+/o over Cxcr4+/+ bone marrow cells for reconstituting the blood, we first measured the proliferative status of the corresponding HSCs in vivo by BrdU incorporation early after transplantation (day 7) (Figure 6A). The results indicated ~20% greater frequency of BrdU+ Cxcr4+/o HSCs as compared to Cxcr4+/+ HSCs suggesting a proliferative advantage of Cxcr4+/o HSCs in the bone marrow. To test whether differential retention of leukocytes in the bone marrow might also contribute to the skewed distribution of mature leukocytes in the blood, we sacrificed mice at day 303 after competitive transplantation with Cxcr4+/o and Cxcr4+/+ bone marrow cells (the same combination of donor bone marrow cells as were analyzed in the proliferation experiments). The results showed that Cxcr4+/o HSCs, HPCs (hematopoietic progenitor cells) and total CD45+ cells (mostly mature leukocytes) predominated in the bone marrow by the same 4:1 ratio over the corresponding Cxcr4+/+ cells as was found for mature leukocytes in the blood. Thus, the predominance of mature Cxcr4+/o over Cxcr4+/+ leukocytes in the blood of competitively transplanted mice cannot be simply explained by low retention of mature Cxcr4+/o leukocytes relative to the retention of mature Cxcr4+/+ leukocytes in the bone marrow. Moreover, the results at this very late time point clarify that enhanced Cxcr4+/o HSC proliferation does not result in long term depletion of HSCs. The frequency distribution of stem and progenitor subtypes was the same for each genotype, indicating no block to progenitor cell differentiation (Figure 6B).
Figure 6. Cxcr4 haploinsufficiency enhances HSC proliferation and engraftment in a mouse bone marrow transplantation model.
(A) Proliferation. Donor bone marrow cells with a Cxcr4+/o genotype on a homozygous CD45.2 background were mixed with donor bone marrow cells with a Cxcr4+/+ genotype on a heterozygous CD45.1/CD45.2 background (47:53) and injected intravenously into a lethally irradiated CD45.1 recipient mouse. Six days after bone marrow transplantation, each mouse was given 1.25 mg of BrdU I.P. Twenty hours later, the mice were euthanized for HSC proliferation analysis. i) Gating scheme for BrdU+ HSCs. Bone marrow cells were first gated with CD45.2 (Cxcr4+/o) and CD45.1/CD45.2 (Cxcr4+/+), then HSCs were gated as Flt3− Lin− Sca1+ c-Kit+ (Flt3−LSK), which includes long-term and short-term HSCs, and BrdU+ cells were quantitated. ii) Percentage of BrdU+ HSCs in each donor. Data are expressed as mean ± SEM from four mice. The experiment was repeated once with similar results.
(B) Long term engraftment and differentiation. Bone marrow cells from donor mice with a Cxcr4+/o genotype on a CD45.2 background were mixed with bone marrow cells from donors with a Cxcr4+/+ genotype on a CD45.1 background (42:58) and the mixed cells were injected intravenously into lethally irradiated recipient mice. 303 days later bone marrow was harvested. i) Gating scheme for long term HSC (LT-HSC: CD34−Flt3− Lin− Sca1+ c-Kit+), short term HSC (ST-HSC: CD34+Flt3− Lin− Sca1+ c-Kit+), multipotent progenitors (MPP: CD34+Flt3+ Lin− Sca1+ c-Kit+), and common lymphoid progenitors (CLP: IL7ra+ Lin− Sca1low c-Kitlow). ii) Long-term engraftment. iii) Differentiation. The distribution frequency of bone marrow cell subsets is similar for Cxcr4+/o compared to Cxcr4+/+ donor-derived cell populations. Each data point represents 10 mice presented as the mean ± SEM.
Finally, when Cxcr4+/o bone marrow was used in competitive repopulation assays with either Cxcr4+/+ or Cxcr4+/S338X, we found in both cases that the total white blood cell and differential counts in the blood post-transplantation were not significantly altered as compared to reference values for C57Bl/6J mice (phenome.jax.org), suggesting that Cxcr4+/o genotype does not predispose to myelodysplasia or leukemia in the setting of this transplantation model (Figure S7). Moreover, there is no evidence that Cxcr4+/o mice have a constitutive predisposition to myelodysplasia (data not shown).
Discussion
Our analysis of this unprecedented experiment of nature in a single patient provides the first evidence that chromothripsis, a newly described form of genetic instability, may result in clinical benefit, in particular, cure of an inherited disease. This patient also provides the second example of chromothripsis occurring in a stem cell still capable of terminal differentiation, (Kloosterman et al., 2011) as well as one of the few examples of spontaneous genetic cure of a Mendelian condition (Hirschhorn, 2003).
We use the word ‘cure’ to refer to the patient’s clinical outcome because she had met all four of the acronymic clinical criteria diagnostic for WHIM syndrome through her 4th decade of life, but has fulfilled none except for mild hypogammaglobulinemia since then (~20 years, to date). Moreover, the disease allele CXCR4R334X is no longer detectable in key cell types that drive the disease: HSCs, neutrophils and monocytes. To our knowledge, patient WHIM-09's spontaneous long-term complete remission of warts without treatment is unprecedented in WHIM syndrome (Al Ustwani et al., 2014; Beaussant Cohen et al., 2012; Kawai and Malech, 2009; Tassone et al., 2009; Wetzler et al., 1990). Since she remains lymphopenic (B and naïve T lymphocytes), this suggests that the myeloid arm of the immune system, probably through monocytes or monocyte-derived cells, plays an essential role in HPV clearance.
The combined evidence suggests that an HSC in WHIM-09 underwent chromothripsis between the second and fourth decade of life and selectively repopulated the myeloid lineage, but not the lymphoid lineage. This pattern of hematologic mosaicism implies that the chromothriptic changes precluded differentiation of HSCs to CLPs but not to CMPs. Thus, the mechanism for maintenance of the B cell lineage, which is WHIM in the patient, is unclear. Possibilities include differentiation from a small population of persistent WHIM HSCs below the level of detection of our assays, or by differentiation of self-sustaining or very long-lived WHIM CLPs. In contrast, the T cell lineage may simply have been maintained by homeostatic proliferation of the pre-chromothriptic WHIM T cell repertoire.
Chromothripsis is a complex chromosomal catastrophe that is thought to occur all at once in one cell (Stephens et al., 2011). If the affected cell dies, chromothripsis is clinically silent and undetectable, therefore the true frequency with which it occurs cannot be determined. If the chromothriptic cell acquires a strong selective advantage, it may emerge as a readily detectable, clinically apparent clonal population harboring a single pattern of deletions and rearrangements, resulting for example either in cancer, as previously reported (Jones and Jallepalli, 2012), or if the location of the event is fortuitous, in cure of a genetic condition, as occurred in patient WHIM-09. Although it is possible that deletion of the disease allele CXCR4R334X in the original chromothriptic HSC of patient WHIM-09, which rendered the cell CXCR4 haploinsufficient (CXCR4+/o), may have been sufficient to repopulate the myeloid lineage--as suggested by the strong HSC engraftment advantage of Cxcr4 haploinsufficient mouse bone marrow cells in our competitive bone marrow transplantation experiments--other factors, particularly haploinsufficiency for one or more of the 163 other genes that were deleted by chromothripsis, may also have contributed. In this regard, at least three of the other 163 deleted genes, DNMT3A (Challen et al., 2012), MYCN (Laurenti et al., 2008), and IL1R (Orelio et al., 2009), have been reported to regulate hematopoiesis (Table S1).
That Cxcr4 haploinsufficiency should provide a competitive engraftment advantage to HSCs is somewhat counterintuitive since Cxcr4 has been well-documented to promote HSC homing to and retention in bone marrow (Broxmeyer et al., 2003a; Broxmeyer et al., 2003b; Broxmeyer et al., 2005; Dar et al., 2006; Kawai et al., 2007; Nie et al., 2008; Sugiyama et al., 2006). However, it has also been reported that CXCL12 secreted by CXCL12-abundant reticular (CAR) cells that contact HSCs in the bone marrow niche is able to enhance HSC quiescence by stimulating CXCR4 signaling (Nie et al., 2008; Sugiyama et al., 2006). The overall effect on engraftment of reducing signaling via CXCR4 haploinsufficiency has not been previously tested in vivo and cannot be confidently predicted given the diverse and contrasting roles of the receptor on HSCs as well as on mature leukocytes. Our mouse data confirmed in vivo that Cxcr4 haploinsufficiency enhanced HSC proliferation while maintaining long-term hematopoiesis, therefore a competitive advantage acquired from enhanced proliferation and potentially other mechanisms by these cells may supercede any potential disadvantage they may have at the level of bone marrow retention and homing.
Additional work using more stringent engraftment protocols in mice and ultimately gene editing/transplantation trials in humans will be needed to precisely determine the impact of CXCR4 haploinsufficiency on HSC engraftment as well as on each of the parameters that regulate the physiologic steady state levels of mature leukocytes and hematopoietic stem and progenitor cells in blood, bone marrow and other hematopoietic compartments. Such studies may further elucidate precisely how patient WHIM-09 was cured, which may point to general applications in transplantation and gene therapy.
Experimental Procedures
Patients
Consistent with the Declaration of Helsinki, all human subjects signed informed consent to participate in NIAID Institutional Review Board-approved clinical protocols. All subjects were studied at the NIH Clinical Center.
Leukocyte analysis
Leukocytes were isolated from blood and bone marrow by Ficoll-Hypaque centrifugation and either magnetic bead selection (Miltenyi Biotec, Cambridge, MA) for blood samples, or cell sorting with a FACS Aria II cytometer (BD Biosciences, San Jose, CA) for bone marrow. PBMCs and bone marrow cells were cultured ex vivo to expand CD34+ hematopoietic progenitor cells and to generate BFU-E colonies. PBMCs were used to create lymphoblastoid cell lines (LCL) from Epstein-Barr virus (EBV)-transformed B cells.
Genetic analysis
BstUI PCR-RFLP analysis was performed as previously described (Hernandez et al., 2003) using patient genomic DNA. The genome was also analyzed by PCR using primers spanning chromothriptic junctions of the derivative chromosome 2 of patient WHIM-09; by Sanger method sequencing; by standard cytogenetics and fluorescence in situ hybridization (FISH) of metaphase chromosomes (Quest Diagnostics, Chantilly, VA) using Abbott Molecular probes (Abbott Molecular, Des Plaines, IL); by microarray analysis using the Affymetrix Cytoscan HD array (Affymetrix, Santa Clara, CA); and by whole genome sequencing (WGS). For WGS, paired-end patient WHIM-09 fibroblast and neutrophil DNA libraries were prepared using the TruSeq DNA protocol (Illumina Inc., San Diego, CA) and were sequenced using a HiSeq 2000 next generation sequencer (Illumina) generating ~380 million reads per library of 2 × 100 bp paired-ends. Reads were mapped to build hg19 of the human genome to identify fusion junctions present in the neutrophil but not in the fibroblast samples.
Transplantation Experiments
Cxcr4 floxed mice (Strain 008767, B6.129P2-Cxcr4tm2Yzo/J) and EIIa promoter driven Cre recombinase transgenic mice (Strain 003724, B6.FVB-Tg(EIIa-cre)C5379Lmgd/J) were obtained from The Jackson Laboratory (Bar Harbor, ME) and bred together to generate Cxcr4+/o mice on a homozygous CD45.2 background. Cxcr4+/+ mice were obtained from Jackson Laboratory on the homozygous CD45.1 and homozygous CD45.2 backgrounds. Creation of WHIM knockin mice bearing a heterozygous Cxcr4S338X mutation has been previously described (Balabanian et al., 2012) and sperm from these mice were used for in vitro fertilization at NIH to impregnate female C57BL/6 mice from Taconic Farms (Hudson, NY) to produce Cxcr4+/S338X on a homozygous CD45.2 background. Both Cxcr4+/o and Cxcr4+/S338X mice on the homozygous CD45.2 background were bred to Cxcr4+/+ mice on a homozygous CD45.1 background to produce Cxcr4+/o and Cxcr4+/S338X mice on a heterozygous CD45.1/CD45.2 background. In this way, differentially marked donor bone marrow cells could be tracked in vivo during competitive transplantation experiments. Five million bone marrow cells (~50% from each donor) were transferred via tail vein injection into recipient mice that had undergone lethal irradiation (900 rads) 8 hours prior to transplant. The mice were fed with neomycin water for four weeks post irradiation. All animal experiments were performed using an NIAID Animal Care and Use Committee (ACUC) approved protocol in approved and certified facilities. For additional details see Extended Experimental Procedures.
Supplementary Material
Acknowledgements
This work was supported by the Division of Intramural Research (DIR) of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) and was funded in part with federal funds from the National Cancer Institute, NIH under Contract number HHSN261200800001E. We thank Hongwei Zhang for flow cytometry advice, Nancy Murphy and Carolyn Boscia for help with graphics, and the research subjects for their participation in the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, 2 Tables and 7 Figures and can be found with the article online at http:\\xxx.
Conflict of Interest
A provisional patent on CXCR4 knock down as a method to enhance HSC engraftment has been filed by the US government with DHM, QL, MS, JG, HLM, and PMM as inventors. The authors confirm that there are no other conflicts of interest.
Author Contributions
Patient recruitment and care was provided by: DHM, DV, MMM, NK, SP, MAR, KRC, IM, RD, and PMM. Experimental design: Q.L., DHM, JG and PMM. Generation and analysis of experimental data: JG, QL, MS, CM, PJ, CRB, ZD, ES, EY, NT, and DALP with supervision and analysis by DHM, SFP, HLM, KLH, DBK, and PMM. KB and FB provided the WHIM knockin mice. All authors participated, but DHM, JG and PMM were principally responsible for writing the manuscript.
References
- Al Ustwani O, Kurzrock R, Wetzler M. Genetics on a WHIM. British journal of haematology. 2014;164:15–23. doi: 10.1111/bjh.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH, Locati M, Luster AD, et al. International Union of Pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacological reviews. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanian K, Brotin E, Biajoux V, Bouchet-Delbos L, Lainey E, Fenneteau O, Bonnet D, Fiette L, Emilie D, Bachelerie F. Proper desensitization of CXCR4 is required for lymphocyte development and peripheral compartmentalization in mice. Blood. 2012;119:5722–5730. doi: 10.1182/blood-2012-01-403378. [DOI] [PubMed] [Google Scholar]
- Beaussant Cohen S, Fenneteau O, Plouvier E, Rohrlich PS, Daltroff G, Plantier I, Dupuy A, Kerob D, Beaupain B, Bordigoni P, et al. Description and outcome of a cohort of 8 patients with WHIM syndrome from the French Severe Chronic Neutropenia Registry. Orphanet journal of rare diseases. 2012;7:71. doi: 10.1186/1750-1172-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) The Journal of experimental medicine. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE, Cooper S, Kohli L, Hangoc G, Lee Y, Mantel C, Clapp DW, Kim CH. Transgenic expression of stromal cell-derived factor-1/CXC chemokine ligand 12 enhances myeloid progenitor cell survival/antiapoptosis in vitro in response to growth factor withdrawal and enhances myelopoiesis in vivo. J Immunol. 2003a;170:421–429. doi: 10.4049/jimmunol.170.1.421. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Kohli L, Kim CH, Lee Y, Mantel C, Cooper S, Hangoc G, Shaheen M, Li X, Clapp DW. Stromal cell-derived factor-1/CXCL12 directly enhances survival/antiapoptosis of myeloid progenitor cells through CXCR4 and G(alpha)i proteins and enhances engraftment of competitive, repopulating stem cells. Journal of leukocyte biology. 2003b;73:630–638. doi: 10.1189/jlb.1002495. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Orschell CM, Clapp DW, Hangoc G, Cooper S, Plett PA, Liles WC, Li X, Graham-Evans B, Campbell TB, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. The Journal of experimental medicine. 2005;201:1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JK, Alper HS. The genome editing toolbox: a spectrum of approaches for targeted modification. Current opinion in biotechnology. 2014;30C:87–94. doi: 10.1016/j.copbio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Dale DC, Bolyard AA, Kelley ML, Westrup EC, Makaryan V, Aprikyan A, Wood B, Hsu FJ. The CXCR4 antagonist plerixafor is a potential therapy for myelokathexis, WHIM syndrome. Blood. 2011;118:4963–4966. doi: 10.1182/blood-2011-06-360586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A, Kollet O, Lapidot T. Mutual, reciprocal SDF-1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in NOD/SCID chimeric mice. Experimental hematology. 2006;34:967–975. doi: 10.1016/j.exphem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Dotta L, Tassone L, Badolato R. Clinical and genetic features of Warts, Hypogammaglobulinemia, Infections and Myelokathexis (WHIM) syndrome. Current molecular medicine. 2011;11:317–325. doi: 10.2174/156652411795677963. [DOI] [PubMed] [Google Scholar]
- Haribabu B, Richardson RM, Fisher I, Sozzani S, Peiper SC, Horuk R, Ali H, Snyderman R. Regulation of human chemokine receptors CXCR4. Role of phosphorylation in desensitization and internalization. The Journal of biological chemistry. 1997;272:28726–28731. doi: 10.1074/jbc.272.45.28726. [DOI] [PubMed] [Google Scholar]
- Hernandez PA, Gorlin RJ, Lukens JN, Taniuchi S, Bohinjec J, Francois F, Klotman ME, Diaz GA. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nature genetics. 2003;34:70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. Journal of medical genetics. 2003;40:721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MJ, Jallepalli PV. Chromothripsis: chromosomes in crisis. Developmental cell. 2012;23:908–917. doi: 10.1016/j.devcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nature reviews Molecular cell biology. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Choi U, Cardwell L, DeRavin SS, Naumann N, Whiting-Theobald NL, Linton GF, Moon J, Murphy PM, Malech HL. WHIM syndrome myelokathexis reproduced in the NOD/SCID mouse xenotransplant model engrafted with healthy human stem cells transduced with C-terminus-truncated CXCR4. Blood. 2007;109:78–84. doi: 10.1182/blood-2006-05-025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Malech HL. WHIM syndrome: congenital immune deficiency disease. Curr Opin Hematol. 2009;16:20–26. doi: 10.1097/MOH.0b013e32831ac557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Human molecular genetics. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Krill CE, Jr, Smith HD, Mauer AM. Chronic Idiopathic Granulocytopenia. The New England journal of medicine. 1964;270:973–979. doi: 10.1056/NEJM196405072701902. [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell stem cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DH, Liu Q, Ulrick J, Kwatemaa N, Anaya-O'Brien S, Penzak SR, Filho JO, Priel DA, Kelly C, Garofalo M, et al. The CXCR4 antagonist plerixafor corrects panleukopenia in patients with WHIM syndrome. Blood. 2011;118:4957–4962. doi: 10.1182/blood-2011-07-368084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DH, Liu Q, Velez D, Lopez L, Anaya-O'Brien S, Ulrick J, Kwatemaa N, Starling J, Fleisher TA, Priel DA, et al. A phase 1 clinical trial of long-term, low-dose treatment of WHIM syndrome with the CXCR4 antagonist plerixafor. Blood. 2014;123:2308–2316. doi: 10.1182/blood-2013-09-527226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Han YC, Zou YR. CXCR4 is required for the quiescence of primitive hematopoietic cells. The Journal of experimental medicine. 2008;205:777–783. doi: 10.1084/jem.20072513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelio C, Peeters M, Haak E, van der Horn K, Dzierzak E. Interleukin-1 regulates hematopoietic progenitor and stem cells in the midgestation mouse fetal liver. Haematologica. 2009;94:462–469. doi: 10.3324/haematol.13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley interdisciplinary reviews Systems biology and medicine. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoret N, Rosenkilde MM, Klasse PJ, Schwartz TW, Malim MH, Hoxie JA, Marsh M. Differential regulation of CXCR4 and CCR5 endocytosis. J Cell Sci. 1998;111(Pt 18):2819–2830. doi: 10.1242/jcs.111.18.2819. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- Tassone L, Notarangelo LD, Bonomi V, Savoldi G, Sensi A, Soresina A, Smith CI, Porta F, Plebani A, Badolato R. Clinical and genetic diagnosis of warts, hypogammaglobulinemia, infections, and myelokathexis syndrome in 10 patients. The Journal of allergy and clinical immunology. 2009;123:1170–1173. 1173 e1171–e1173. doi: 10.1016/j.jaci.2008.12.1133. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Venkatesan S, Rose JJ, Lodge R, Murphy PM, Foley JF. Distinct mechanisms of agonist-induced endocytosis for human chemokine receptors CCR5 and CXCR4. Molecular biology of the cell. 2003;14:3305–3324. doi: 10.1091/mbc.E02-11-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzler M, Talpaz M, Kleinerman ES, King A, Huh YO, Gutterman JU, Kurzrock R. A new familial immunodeficiency disorder characterized by severe neutropenia, a defective marrow release mechanism, and hypogammaglobulinemia. Am J Med. 1990;89:663–672. doi: 10.1016/0002-9343(90)90187-i. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Human molecular genetics. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- Zuelzer WW. "Myelokathexis"--a New Form of Chronic Granulocytopenia. Report of a Case. The New England journal of medicine. 1964;270:699–704. doi: 10.1056/NEJM196404022701402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.