Abstract
Plants produce hydroxycinnamic acid defense compounds (HCAs) to combat pathogens, such as the bacterium Ralstonia solanacearum. We showed that an HCA degradation pathway is genetically and functionally conserved across diverse R. solanacearum strains. Further, a Δfcs (feruloyl-CoA synthetase) mutant that cannot degrade HCAs was less virulent on tomato plants. To understand the role of HCA degradation in bacterial wilt disease, we tested the following hypotheses: HCA degradation helps the pathogen (1) grow, as a carbon source; (2) spread, by reducing physical barriers HCA-derived; and (3) survive plant antimicrobial compounds. Although HCA degradation enabled R. solanacearum growth on HCAs in vitro, HCA degradation was dispensable for growth in xylem sap and root exudate, suggesting that HCAs are not significant carbon sources in planta. Acetyl-bromide quantification of lignin demonstrated that R. solanacearum infections did not affect the gross quantity or distribution of stem lignin. However, the Δfcs mutant was significantly more susceptible to inhibition by two HCAs: caffeate and p-coumarate. Finally, plant colonization assays suggested that HCA degradation facilitates early stages of infection and root colonization. Together, these results indicated that ability to degrade HCAs contributes to bacterial wilt virulence by facilitating root entry and by protecting the pathogen from HCA toxicity.
Introduction
Plants produce thousands of phenolic compounds, which play roles in plant development and interactions with microbes (Mandal et al., 2010; Naoumkina et al., 2010). Among these are hydroxycinnamic acids (HCAs), which are monocyclic phenylpropanoid molecules. Roots exude HCAs and related phenolics to chelate metals, thereby facilitating uptake and transport of metals in the xylem sap (Ishimaru et al., 2011). In response to root pathogens, many plants release de novo synthesized HCAs into the rhizosphere, and grapevines infected with Xylella accumulate HCAs and HCA-conjugates in their xylem sap (Mandal and Mitra, 2008; Lanoue et al., 2010; Wallis and Chen, 2012). HCAs are broadly antimicrobial; they disrupt membrane integrity and decouple the respiratory proton gradient (Fitzgerald et al., 2004; Harris et al., 2010). Additionally, HCAs reinforce protective physical barriers in plants by cross-linking primary cell wall polysaccharides and by serving as precursors for the phenolic polymer lignin (Fry et al., 2000; Naoumkina et al., 2010; Campos et al., 2014).
Plants defend their vascular systems with phenolic-storing cells stationed along the xylem (Beckman, 2000). These phenolic storing cells decompartmentalize in response to infection and release phenolics into the xylem lumen, in a process similar to neutrophil degranulation in animal immunity. Exposing tomato roots to a xylem-dwelling fungal vascular wilt pathogen, Fusarium oxysporum f. sp. lycopersici, leads to increased accumulation of the HCAs ferulate and p-coumarate (Mandal and Mitra, 2008). Ultrastructure studies of xylem infected with the vascular pathogen Ralstonia solanacearum show phenomena consistent with phenolic release (Mueller and Beckman, 1984; Grimault et al., 1994; Rahman et al., 1999; Nakaho et al., 2000).
R. solanacearum causes bacterial wilt disease, which limits production of key crops like potato, banana, peanut, and tomato (Elphinstone, 2005). This soil-dwelling pathogen generally enters hosts through the roots and then colonizes the xylem elements throughout the plant. Extensive colonization of the xylem ultimately blocks water transport, leading to stunting and wilting. R. solanacearum strains form a large, heterogeneous species complex that collectively infects hundreds of different plant species(Peeters et al., 2013).
Several lines of evidence suggest that hydroxycinnamic acids are involved in tomato interactions with R. solanacearum. Quantitative resistance of tomato cultivars against R. solanacearum is correlated with early expression of phenylalanine ammonia lyase (PAL), which catalyzes the first step in phenylpropanoid biosynthesis (Vanitha et al., 2009). Transcriptomic analysis showed that multiple phenylpropanoid biosynthesis genes are upregulated in R. solanacearum-infected, resistant tomato plants compared to healthy plants (Ishihara et al., 2012) (Milling and Allen, unpublished). We previously found that drug efflux pumps protect R. solanacearum from the toxicity of many plant defense chemicals, including the HCA caffeate (Brown et al., 2007). More specifically, the genomes of many R. solanacearum strains encode an enzymatic pathway that is homologous to a Pseudomonas fluorescens pathway that breaks down the HCAs ferulate, p-coumarate, and caffeate to central carbon metabolites (Narbad and Gasson, 1998) (Fig. 1). These HCA degradation pathway genes are expressed by R. solanacearum cells growing in tomato xylem vessels at the onset of wilt symptoms (Salanoubat et al., 2002; Jacobs et al., 2012).
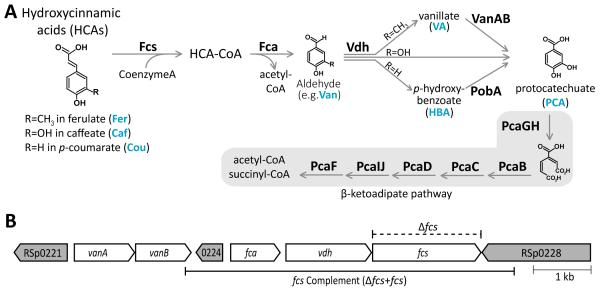
Fig. 1.
Hydroxycinnamic acid (HCA) degradation pathway and genes in R. solanacearum GMI1000. A, The HCA degradation pathway is shown with enzyme names in boldface. Fcs, Fca, and Vdh convert the HCAs ferulate, p-coumarate, and caffeate to the phenolic acids vanillate, p-hydroxybenzoate, and protocatechuate, respectively. VanAB and PobA convert vanillate and p-hydroxybenzoate to protocatechuate, which is further metabolized by the β-ketoadipate enzymes; B, A locus containing genes encoding multiple enzymes in the HCA degradation pathway. White arrows indicate ORFs encoding HCA degradation, and grey arrows indicate neighboring ORFS. RSp0221, RSp0224, and RSp0228 encode transcriptional regulators of the LysR family, MarR family, and Fis family, respectively. The dashed line above the genes indicates the region that was precisely excised to create the feruloyl-CoA synthetase deletion mutant (Δfcs). The solid line below the genes indicates the region used to genetically complement the Δfcs mutation in the Δfcs+fcs strain.
We explored the hypothesis that HCA degradation contributes to bacterial wilt disease. We found that HCA degradation is widely conserved in the R. solanacearum species complex. A feruloyl-CoA synthetase mutant (Δfcs) that cannot degrade HCAs had reduced virulence on tomato, delayed colonization of tomato roots, and increased susceptibility to the toxicity of the HCAs caffeate and p-coumarate.
Results
Organization of HCA degradation genes in R. solanacearum GMI1000
HCA degradation enzymes encoded by the genes fcs, fca, vdh, vanAB, and pobA convert the HCAs p-coumarate, caffeate, and ferulate to protocatechuate and acetyl-CoA (Fig. 1A). The β-ketoadipate enzymes encoded by the pca genes further metabolize protocatechuate to the central carbon metabolites succinyl-CoA and a second acetyl-CoA. In R. solanacearum strain GMI1000, the HCA degradation and β-ketoadipate genes are organized as five putative operons at three genomic loci (Fig. 1B): fca-vdh-fcs (RSp0225-0227), vanAB (RSp0222-0223), pobA (RSc02242), pcaGH (RSc1141-1142), and pcaIJFBDC (RSc2249-2255).
We used the Orthologous MAtrix (OMA) browser to investigate the conservation of genes for HCA and protocatechuate degradation across bacteria (Supplementary Table S2). OMA uses a strict algorithm to categorize orthologous proteins from complete publically available genome sequences (Altenhoff et al., 2011). The eukaryotic β-ketoadipate genes are not homologous to the bacterial genes, so we did not analyze eukaryotic genomes. OMA analysis indicated that HCA degradation is a rare trait among the 1,281 bacterial strains considered. Many plant pathogenic bacteria lacked HCA degradation genes, including the necrotroph Dickeya dadantii 3937, which uses feruloyl-esterases to cleave HCAs from cell wall polysaccharides (Hassan and Hugouvieux-Cotte-Pattat, 2011), and Xylella fastidiosa, which encounters HCAs in grapevine xylem (Wallis and Chen, 2012). Although the OMA database tends to yield false negatives, we gained insight on the prevalence and distribution of this pathway in bacteria because HCA degradation has been functionally characterized in several of the strains included in the OMA database (Parke and Ornston, 2003; Plaggenborg et al., 2003; Abdelkafi et al., 2006; Kim et al., 2008; Pérez-Pantoja et al., 2008; Romero-Silva et al., 2013; Campillo et al., 2014). For example, although OMA analysis indicated that Cupriavidus pinatubonensis JMP134 (formerly C. necator and Ralstonia eutropha) lacks 4/14 HCA degradation genes, this strain is known to degrade HCAs. Therefore, we hypothesized that the 33 strains containing more than 10/14 HCA degradation genes likely degrade HCAs. These strains are predominantly in genera known to spend part of their lifecycles in soil: Burkholderia, Brucella, and Pseudomonas. OMA analysis identified several plant-associated genera that appear to have functional β-ketoadipate pathways but lack the upstream HCA degrading enzymes: Xanthomonas campestris, Rhizobium spp., and Agrobacterium spp. (although Campilo et al. (2014) show that Agrobacterium strain C58 degrades HCAs).
The hydroxycinnamic acid (HCA) degradation pathway is broadly conserved in the R. solanacearum species complex
Because hydroxycinnamic acids are common plant metabolites, the ability to degrade these metabolites could benefit R. solanacearum. To explore the genetic conservation of the HCA degradation pathway in the large and heterogeneous R. solanacearum species complex, we searched for homologs of HCA degradation genes in the genomes of 23 available R. solanacearum strains (Fig. 2A). Only two strains lacked multiple HCA degradation genes: Phylotype IIA strain K60 and Phylotype IV Blood Disease Bacterium (BDB) strain R229. We identified homologs of each HCA degradation gene in the remaining 21 strains (91%), but in eight of these strains, one or more genes were located on a contig border or were annotated as putative pseudogenes.
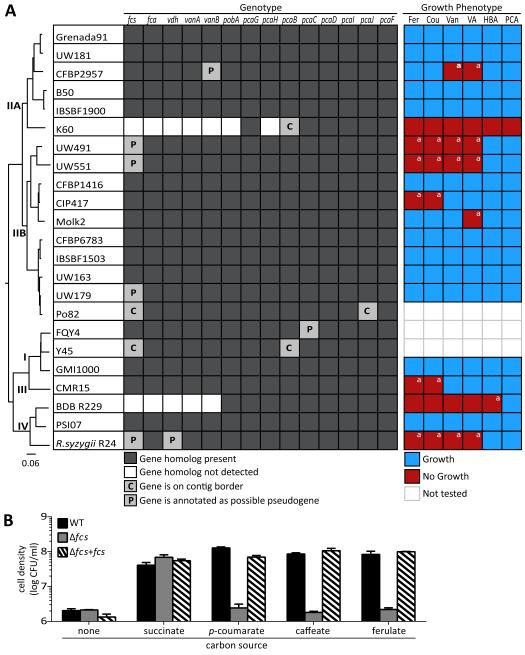
Fig. 2.
HCA degradation is widely conserved in the R. solanacearum species complex. A, Genetic and functional conservation of HCA degradation. A whole genome comparison phylogenetic tree is shown on the left. Presence of HCA degradation genes and growth of R. solanacearum strains on ferulate (Fer), p-coumarate (Cou), vanillin (Van), vanillate (VA), p-hydroxybenzoate (HBA), and protocatechuate (PCA) are indicated. aGrowth phenotype differs from genotype prediction; B, fcs encodes a functional feruloyl-CoA synthetase in strain GMI1000. Strains were grown in minimal media supplemented with 0.2 mM succinate, p-coumarate, caffeate, ferulate, or no carbon (−) for 72 hr. Bars represent the mean of 3 biological replicates and error bars indicate standard error of the mean.
The number of potential pseudogenes and genes lying on contig borders made it difficult to predict the HCA degradation ability of a third of the sequenced strains, so we functionally characterized the HCA degradation ability of all available strains. We could not analyze strains Po82, FQY-4, and Y45 since the authors of these published genomes would not share their strains (Li et al., 2011; Xu et al., 2011; Cao et al., 2013). Each strain was tested for its ability to grow on the HCAs ferulate (Fer) and p-coumarate (Cou) as well as on the pathway intermediates vanillin (Van), vanillate (VA), p-hydroxybenzoate (HBA), and protocatechuate (PCA) (see Fig. 1A). Most strains grew on all tested compounds, except where genomic data indicated an incomplete pathway (Fig 2A). For example, because it lacks fcs, fca, vdh, vanB, pobA, and pcaH, strain K60 did not grow on any tested substrate. Similarly, BDB strain R229, lacking fcs, fca, vdh, and vanB, grew only on protocatechuate.
In several cases, bioinformatic data did not accurately predict biological function. Surprisingly, although strains CFBP2957 and Molk2 grew on ferulate, they did not grow on the ferulate degradation intermediates vanillin (in the case of CFPB2957) or vanillin and vanillate (in the case of Molk2). It is possible that higher sensitivity to toxicity of vanillin and vanillate prevented these strains from growing. Several strains with a putatively pseudogenized fcs displayed contrasting growth phenotypes. While strain UW179 grew on all compounds, the phylotype II sequevar 1 (Race 3 biovar 2) strains UW491 and UW551 unexpectedly did not grow on ferulate, p-coumarate, vanillin, or vanillate. Further, strains CIP417 and CMR15 grew on intermediate metabolites but not ferulate or p-coumarate even though these strains apparently possess complete sets of HCA degradation genes. Taken together, these analyses indicate that most R. solanacearum strains can degrade at least some HCAs; the unexpected positive and negative results for growth on HCAs also highlight the importance of functional experiments to confirm genomic analyses.
To determine whether HCA degradation contributes to bacterial wilt disease, we created an fcs deletion mutant in the background of phylotype I strain GMI1000. Hereafter, strain GMI1000 is referred to as wildtype or WT and the GMI1000 deletion mutant lacking the feruloyl-CoA synthetase open reading frame is referred to as the Δfcs mutant. While WT used HCAs and intermediate phenolics as a carbon source (Fig. 2), the Δfcs mutant did not grow on the HCAs p-coumarate, caffeate, or ferulate, as predicted (Fig. 2B). Additionally, the mutant grew as well as wildtype on all pathway intermediates (data not shown). This result confirmed the bioinformatic annotation of this gene, and also confirmed the deletion of the fcs gene. Genetic complementation of the mutant with the cloned fcs operon under control of the native promoter restored its growth on HCAs (Fig. 2B).
HCA degradation contributes to R. solanacearum virulence on tomato
We used a naturalistic soil soak virulence assay to measure the contribution of HCA degradation to R. solanacearum virulence on tomato. Bacterial suspensions were poured into the soil of individually potted unwounded tomato plants, and symptom development was measured over time.
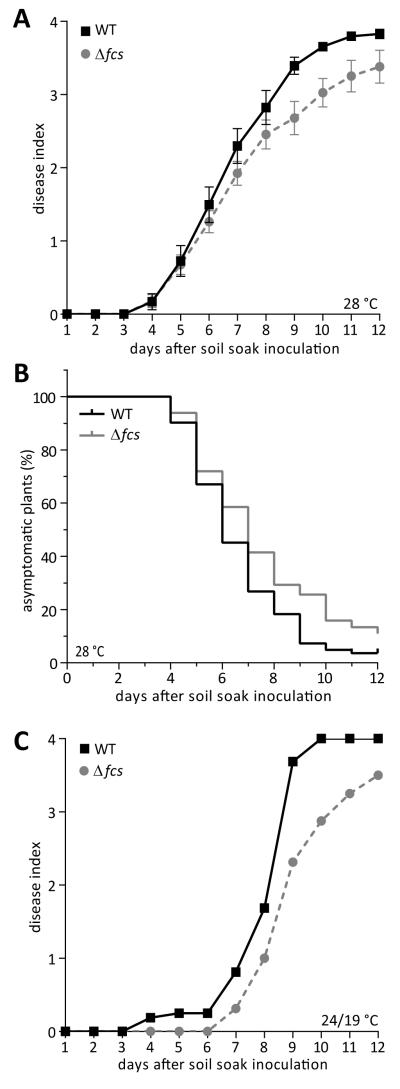
The Δfcs mutant displayed a modest but significant reduction in virulence on plants grown at the tropical temperature 28°C (Fig. 3A; repeated measure ANOVA; P=0.0123). The reduced virulence of the Δfcs mutant did not result from an altered rate of symptom progression. Once they became symptomatic, plants inoculated with either strain progressed to end-stage disease at the same rate (average time was 1.2 and 1.1 days for WT and Δfcs inoculated plants, respectively, between the first symptoms and the highest disease index rating; P=0.402, unpaired t-test). To determine whether the virulence defect was due to a delay in symptom onset, we used survival analysis. Although survival analysis was originally developed to analyze patient outcome data in clinical trials, this statistical tool can analyze any discrete biological events in a time course. This analysis revealed that symptom onset was earlier in WT-inoculated plants than in Δfcs-inoculated plants (Fig. 3B; log-rank Mantel-Cox test; P = 0.0118). The median time until symptom onset was 6 days after WT inoculation and 7 days after Δfcs mutant inoculation. This result suggested that the virulence defect of the Δfcs mutant affects an early stage of the infection process before symptom onset.
Fig. 3.
HCA degradation is required for full virulence of R. solanacearum. A, Disease progress of WT and Δfcs mutant strains on susceptible tomato plants grown at 28 °C. Twenty-one-day-old unwounded plants (cv. Bonny Best) grown at constant 28 °C were inoculated by pouring a bacterial suspension into the soil of each pot. Symptoms were rated using a 0 to 4 disease index scale. Each point represents the mean disease index of a total of 82 plants per strain, in 6 biological replicates. Bars indicate standard error of the mean. Disease progress of the Δfcs mutant was significantly slower than that of wild-type (P=0.0123, two-way repeated measures ANOVA); B, Survival analysis of the above dataset showing the rate of symptom onset after inoculations with WT and the Δfcs mutant; C, Disease progress of strains on tomato plants grown in a 24 °C day /19 °C night cycle (1 biological replicate with N=16 plants per strain).
Since HCA degradation genes were highly expressed when R. solanacearum infected plants at cool temperatures (Meng, Jacobs and Allen, unpublished), we also quantified the virulence of the Δfcs mutant in a growth chamber at 24°C day and 19°C night (Fig 3C). Under these cooler conditions, the Δfcs mutant also displayed a virulence defect.
HCA degradation is not required for R. solanacearum growth in plant-associated environments
We hypothesized that HCA degradation contributes to R. solanacearum virulence by providing the bacterium with a carbon source in the competitive and nutrient-limited niches in and around plants. Plant roots exude HCAs into the rhizosphere, and HCAs comprise up to 10% of the water-soluble carbon in soil (Smolander et al., 2005). Thus, the ability to use HCAs as a carbon source could provide bacteria with a competitive edge in the soil. We asked whether HCA degradation increased growth of R. solanacearum in water-soluble potting soil extract and in root exudate from sterile tomato seedlings. The Δfcs mutant grew as well as its wild type parent in both substrates (Fig. 4A-B). However, sterile tomato seedlings may produce less HCAs than mature plants with a diverse microbiome since pathogens induce production and release of HCAs into the rhizosphere (Neumann and Römheld, 2007). HCAs chelate and transport metals in the xylem sap, and concentrations of HCA conjugates increase in grapevines infected with Xylella (Ishimaru et al., 2011; Wallis and Chen, 2012). Therefore we asked whether HCA degradation provides a growth benefit to R. solanacearum in xylem sap. Xylem sap was harvested by detopping healthy tomato plants and allowing root pressure to exude the sap. The sap was filter sterilized and used as a growth substrate for WT, Δfcs, and Δfcs+fcs (complemented) bacteria. HCA degradation ability did not affect growth of any strain in healthy xylem sap (Fig. 4C). Because R. solanacearum infections induce expression of tomato phenolic biosynthesis genes (Ishihara et al., 2012; Mitra, Milling, and Allen unpublished), we hypothesized that xylem sap from infected plants contains higher HCA concentrations that would benefit growth of the WT strain. However, we detected no differences in growth between WT and Δfcs when they were grown in sap harvested from plants infected with WT or the Δfcs mutant (data not shown). Because the Δfcs mutant grew normally on these substrates, we infer that HCAs were not significant sources of carbon or present in sufficient concentrations to inhibit bacterial growth.
Fig. 4.
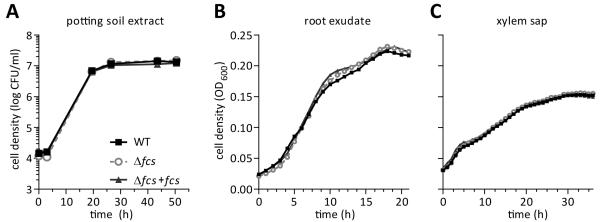
Hydroxycinnamic acid degradation does not enhance R. solanacearum growth in plant associated environments. A-C, ex vivo bacterial growth in: A, water extract of potting soil; B, tomato root exudate; and C, tomato xylem sap harvested from stems of un-inoculated, healthy plants. Graphs show the mean of 3 replicates.
HCA degradation contributes to colonization of tomato roots
Studies of R. solanacearum growth in potting soil extract, root exudate from sterile seedlings, and xylem sap cannot reflect the complex process of invading and multiplying in hosts. Therefore, we transitioned to whole-plant assays. To measure the overall fitness of the Δfcs mutant in a naturalistic infection, we used a competition assay where plants were co-inoculated with a 1:1 suspension of WT and Δfcs mutant bacteria using the soil-soaking method. At the first sign of wilt symptoms, we quantified the population size of each strain in the midstem (Fig. 5C). With a median competitive index (CI) of 0.46, the Δfcs mutant was significantly less fit than WT bacteria (Wilcoxon signed rank test; P<0.0001). The WT strain outcompeted the Δfcs mutant by 2.2-fold. This assay requires strains to compete during several stages of the R. solanacearum infection cycle: survival in bulk soil and colonization of host rhizosphere, roots, and stems. The cumulative effects of these competitive interactions are assessed by comparing population sizes of the two strains in tomato stems. The observed reduced competitive fitness of the Δfcs mutant indicates that HCA degradation contributes to at least one stage of the R. solanacearum infection cycle.
Fig. 5.
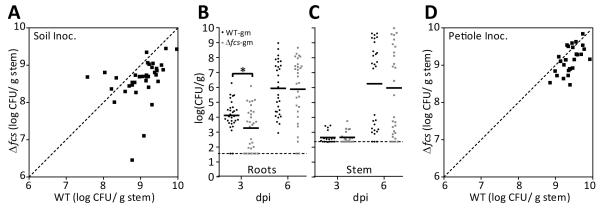
HCA degradation contributes to root entry and competitive fitness following soil soak inoculation of tomato. A-B, Plants grown at 28 °C were soil-soak inoculated with suspensions of WT or Δfcs bacteria. At 3- and 6-days post inoculation (dpi); A, 300 mg root tissue; or B, 100 mg of midstem tissue were harvested, ground, and dilution plated to determine cell density of R. solanacearum (N=30 plants for root colonization at 3 and 6 dpi; N=20 for stem colonization at 3 dpi and N=30 at 6 dpi); Solid lines represent the median population sizes and the dashed lines represent the limit of detection. WT-gm colonized roots better than Δfcs-gm at 3 days after inoculation (P<0.0094; t-test); C, Competitive fitness of WT vs. Δfcs bacteria following soil-soak inoculation. Tomato plants were co-inoculated with mixtures of reciprocally-marked WT and Δfcs strains. At the first stage of disease (less than 25% leaves wilted), midstem tissue was harvested, ground, and dilution plated. Population size of each strain was normalized by initial inoculum. Median competitive index (CI) of the Δfcs mutant was 0.46 (P<0.0001, Wilcoxon Signed Rank Test; N=13 plants per co-inoculation, 26 total); D, Competition of Δfcs and WT bacteria in tomato stem following direct stem inoculation. Tomato plants were co-inoculated via a cut leaf petiole with 4,000 CFU in a 1:1 suspension of reciprocally-marked WT and Δfcs strains. Midstem tissue was harvested at the first sign of symptoms, ground, and dilution plated. Population size of each strain was normalized by initial inoculum. Median CI of the Δfcs mutant was 0.71 (P=0.225, Wilcoxon Signed Rank Test; N=14 plants per co-inoculation, 28 total).
To more narrowly investigate the role of HCA degradation in early stages of infection, we individually soil-soak inoculated tomato plants with gentamicin-marked WT and Δfcs bacteria and quantified population sizes of the strains in surface-sterilized roots (Fig. 5A). At 3 dpi, the population sizes of the Δfcs mutant were lower than those of WT bacteria in roots (P<0.0094, t-test), but by 6 dpi, the population size of the mutant caught up to wildtype levels. These results indicate that HCA degradation contributed significantly to R. solanacearum’s ability to enter and/or grow within the root.
To investigate the role of HCA degradation when the bacterium is in tomato stems, we used a two-pronged approach. First we quantified population sizes of the WT and Δfcs strains in the midstem stem after individual soil-soak inoculations (Fig 5B). At 3 dpi, few stems were colonized with detectable levels of bacteria. At 6 dpi, the average stem population size of WT bacteria was slightly, but insignificantly, higher than that of the Δfcs mutant. We next used an in planta competition assay that can reveal subtle colonization defects that are missed in individual colonization assays (Yao and Allen, 2006; Macho et al., 2010). For this assay, 2,000 CFU of reciprocally-marked WT and Δfcs bacteria were co-inoculated into tomato plants via a cut petiole. At symptom onset, the midstem population size of each strain was determined by grinding stem tissue and dilution plating. The population size of the WT strain was slightly larger than that of the Δfcs mutant in this in-stem competition assay, but the two strains were not significantly different. Overall, colonization assays indicated that HCA degradation may improve stem colonization and showed that HCA degradation contributed significantly to the bacterium’s ability to colonize roots.
R. solanacearum HCA degradation does not detectably affect quantity or distribution of lignin in susceptible tomato stems
As phenylpropanoids, HCAs are precursors to many plant physical defenses, such as diferulate cross-links in primary cell walls and lignin in secondary cell walls. Plant hosts often respond to pathogens by increasing biosynthesis and deposition of phenylpropanoids (Dixon and Paiva, 1995). Previous studies found that bacterial wilt-resistant tomato plants express phenylpropanoid biosynthesis genes early during R. solanacearum infections (Vanitha et al., 2009). We hypothesized that HCA degradation by the pathogen may reduce or prevent lignin formation by decreasing the pool of lignin precursors (HCAs). To test this prediction, we used the acetyl bromide assay to measure gross lignin amounts in whole stems from healthy tomato plants and from plants infected with WT or Δfcs bacteria. The lignin content in stems of the wilt-susceptible Bonny Best cultivar did not increase in response to infection with either R. solanacearum strain (Fig. 6A). Analysis of whole stems could overlook variation in lignin distribution between conditions, but a histopathological analysis of stem cross sections using the lignin-specific stain phloroglucinol revealed that lignin was similarly distributed in all samples, mainly around the xylem vessels in the vascular bundles (Fig. 6B-D). This result suggested that R. solanacearum’s HCA degradation pathway does not detectably alter the amount or the distribution of host lignin, at least under the conditions tested.
Fig. 6.
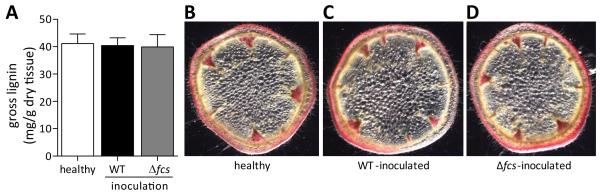
HCA degradation by R. solanacearum did not affect total lignin quantity or distribution in tomato stems. A, Mean gross lignin content in tomato stems at 6 days post soil-soak inoculation. Whole stems of healthy (mock-inoculated) or infected tomato plants (n=6 per condition) were dried, ground, and analyzed by the acetyl bromide lignin quantification assay using wood pulp inulin as a standard. Error bars indicate standard error of the mean. Similar results were obtained at 3 and 9 days after inoculation; B-D, Phloroglucinol HCl-stained cross-sections of stems from representative healthy or symptomatic infected plants. Pink precipitate indicates lignin.
These experiments do not determine if HCA degradation affects the amount of diferulate cross-links in the primary cell wall. Several plant pathogens with large repertoires of cell-wall degrading enzymes use feruloyl esterases to cleave ferulate from cell wall sugars (DiGuistini et al., 2011; Hassan and Hugouvieux-Cotte-Pattat, 2011; Balcerzak et al., 2012). It is possible that R. solanacearum encounters diferulate bridges when entering roots or when degrading pit membranes between xylem vessels. Therefore, we tested for feruloyl esterase activity by growing WT strain GMI1000 on an HCA-glucoside analog, ethyl-ferulate. The strain could not use ethyl-ferulate as a sole carbon source (data not shown), which indicates a lack of feruloyl esterase activity. To test whether HCA degradation helps R. solanacearum pass through pit membranes and spread in tomato stem, wildtype and Δfcs mutant strains were directly inoculated into the xylem of four week old tomato plants via a cut petiole. At 6-10 days after inoculation, bacterial population sizes were quantified by grinding and dilution plating stem tissue harvested at the point of inoculation and distal sites (3 and 6 cm above the point of inoculation). There were no strain-to-strain differences in population sizes in the distal stem (data not shown). These results suggest that HCA degradation does not measurably contribute to R. solanacearum spread in tomato stems.
HCA degradation protects R. solanacearum from caffeate and p-coumarate toxicity
HCAs are broadly toxic to microbes. They can directly disrupt membrane integrity, and they are converted to reactive quinones under oxidative conditions, such as after an ROS burst during infection of a eukaryotic host (Li and Steffens, 2002; Fitzgerald et al., 2004). We hypothesized that R. solanacearum uses its HCA degradation pathway to detoxify these potentially lethal chemicals. Using a minimum inhibitory concentration (MIC) growth assay, we compared growth of WT and Δfcs bacteria in the presence of increasing concentrations of the three HCAs: p-coumarate, caffeate, and ferulate. For WT, the MICs of the p-coumarate and caffeate were 1,500 μM, and the MIC of ferulate was 3,000 μM (Fig. 7). The growth of the Δfcs mutant was dramatically reduced relative to growth of the WT strain at sub-MIC concentrations of both p-coumarate and caffeate with the Δfcs mutant showing a significant growth defect in as little as 23 μM of p-coumarate and caffeate. The Δfcs mutant suffered near-complete growth inhibition at 375 μM p-coumarate even though WT bacteria were unaffected by this concentration (Fig. 7A and B). In contrast, there was no difference in growth between WT and Δfcs bacteria at any ferulate concentration (Fig. 7C). Adding a wild-type copy of the fcs operon to the mutant restored full wild-type levels of p-coumarate and caffeate tolerance to the complemented strain. Together, these results suggested that HCA degradation protects R. solanacearum from toxicity of caffeate and p-coumarate but not ferulate.
Fig. 7.
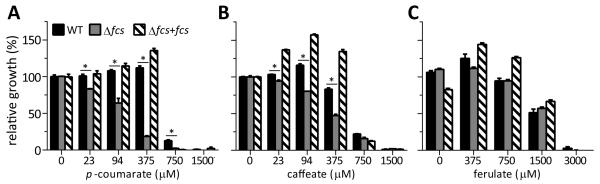
HCA degradation protected R. solanacearum from HCA toxicity. Bacterial growth in succinate minimal medium supplemented with increasing concentrations of: A, p-coumarate (p-Cou); B, caffeate (Caf); or C, ferulate (Fer). Culture optical density was measured by a plate reader 48 hr after inoculation with 105 CFU/ml of bacteria. Growth of each strain was calculated relative to that of wild-type bacteria growing without HCAs. Error bars indicate standard error of the mean. The WT strain was less inhibited than the Δfcs mutant by p-coumarate and caffeate (t-test; P< 0.005).
Discussion
Plant-associated bacteria experience a complex cocktail of secondary metabolites produced by their eukaryotic hosts. Some of these compounds may provide nutrition, while many are inhibitory or toxic. We found that the ability to degrade a group of such compounds, the HCAs, is a quantitative virulence factor for R. solanacearum. More specific analyses revealed that HCA degradation contributes to bacterial wilt pathogenesis, possibly by protecting the pathogen from inhibition by toxic HCAs during root colonization.
Several lines of evidence support our model that HCA degradation contributes to R. solanacearum fitness at early stages of disease. First, we found that the HCA degradation and β-ketoadipate pathways are found predominantly in soil-inhabiting bacteria. In contrast, we did not detect conservation of HCA degradation genes in the xylem-colonizing bacteria Clavibacter michiganensis, Xylella fastidiosa, Dickeya dadantii, or Erwinia amylovora. Second, the virulence defect of the Δfcs mutant includes a delay in the first appearance of symptoms. The delay in symptom onset likely results from this mutant’s slower colonization of host roots. Surprisingly, the delay in root colonization by the Δfcs mutant did not result in smaller eventual population sizes in the stems of infected plants. This suggests that once the pathogen gains entry to xylem vessels, a strain unable to degrade HCAs can grow to the same final density as its wild-type parent. Thus, HCA degradation appears to be most useful to R. solanacearum in host roots and rhizospheres.
Our in vitro inhibition assay demonstrates that the mutant is more susceptible to toxicity of certain HCAs. Although the HCA concentrations required for growth inhibition in vitro are 10 to 100-fold higher than concentrations measured in planta, concentrations of these compounds in the xylem of roots and stems may be locally high where phenolics are released by sentinel phenolic-storing-cells (Beckman, 2000; Alvarez et al., 2008; Mandal and Mitra, 2008; Wallis and Chen, 2012). Additionally, the metabolic state of R. solanacearum cells affected their susceptibility to HCAs; HCAs were more inhibitory when R. solanacearum was grown in glucose minimal media than when grown in succinate minimal media (data not shown). Moreover, it takes less HCA to inhibit microbial growth when HCAs are present in mixtures than when only one HCA is present (Harris et al., 2010). R. solanacearum cells likely encounter mixtures of HCAs and other antimicrobial compounds when infecting plants. It is therefore possible that the effective inhibitory concentrations of caffeate and p-coumarate are lower in the complex chemical environment of an infected plant than in a single-chemical in vitro MIC assay.
Surprisingly, HCA degradation ability did not affect the toxicity of ferulate. R. solanacearum may have an fcs-independent pathway that specifically degrades ferulate, although this seems unlikely because the Δfcs mutant could not grow on ferulate. Alternatively, R. solanacearum could have a drug efflux pump that is highly effective at removing ferulate but less active on caffeate and p-coumarate. The drug efflux pumps encoded by dinF and acrA are very important for strain K60’s virulence on tomato (Brown et al., 2007). The acrA mutant had heightened susceptibility to caffeate toxicity, but other HCAs were not tested. Strain K60 lacks the HCA degradation pathway, so it would be interesting to determine if drug efflux pumps also protect other R. solanacearum strains from toxicity of caffeate and other HCAs.
Our virulence studies used a single strain, GMI1000, but the capacity to degrade HCAs is genetically well conserved across the R. solanacearum species complex. Although bioinformatic analysis accurately predicted the ability of strains to grow on various HCA compounds 83% of the time, several strains did not grow on all predicted HCA carbon sources. These disparities demonstrate that predictions based on genomic analysis require functional validation, especially to confirm enzyme substrate specificity(Airola et al., 2014). The multi-strain screen for HCA degradation ability revealed several surprising results. Although CFBP2957 and Molk2 grew on ferulate, they did not grow on ferulate degradation intermediates. It is possible that higher sensitivity to toxicity of vanillin and vanillate prevented these strains from growing. Alternatively, vanillin and vanillate may not induce expression of vdh and vanAB genes as these compounds appear to do in most R. solanacearum strains. Pseudogenization of the fcs gene in the Race 3 biovar 2 strains (UW491 and UW551) appears to prevent this strain from growing on most metabolites upstream of protocatechuate. This is in contrast to strain GMI1000 where deletion of fcs did not affect growth on any compounds besides HCAs.
Although R. solanacearum expresses its HCA degradation genes in stem xylem vessels during tomato pathogenesis, these genes are only expressed at moderate levels (Jacobs et al., 2012). It is not surprising that HCA degradation genes were not identified in our previous IVET screen for root exudate induced genes because that study used R. solanacearum strain K60, which has lost HCA degradation ability (Colburn-Clifford and Allen, 2010). Transcriptional analysis could be used to compare expression of HCA degradation genes at different stages in the R. solanacearum life cycle, particularly in the rhizosphere. Phenolics in root exudate are chemoattractants for many rhizosphere bacteria, including Agrobacterium tumefaciens (Mandal et al., 2010). Chemotaxis allows R. solanacearum to locate host plants, but it remains to be determined whether root-exuded phenolics serve as chemoattractants (Yao and Allen, 2006).
Plant phenolics influence expression of virulence genes in many plant mutualists and pathogens. Ferulate and other phenolics induce expression of Agrobacterium vir genes, and flavonoids induce Rhizobium nod genes, both of which are required for association with plants (Bhattacharya et al., 2010). Expression of the Dickeya dadantii type III secretion genes is induced by the phenolics o-coumarate and trans-cinnamate and repressed by p-coumarate (Li et al 2009 and Yang et al 2008). We cannot rule out the possibility that the virulence defect of the Δfcs mutant is due to HCA-mediated repression of the pathogen’s type III secretion system, although such repression cannot be complete because the Δfcs mutant still triggers a hypersensitive response in the non-host tobacco (data not shown).
In response to pathogen attack, plants can reinforce cell walls with lignin, which is an HCA polymer. Although we didn’t identify an obvious difference in stem lignin in susceptible tomato after R. solanacearum infections, we cannot rule out subtle but biologically important differences that would be undetectable in our gross analyses. Moreover, this trait may play an important role in resistant tomato cultivars. A previous study observed increased lignification in response to R. solanacearum infections in the quantitatively wilt-resistant tomato cv. LS-89, but not in susceptible tomato cv. Ponderosa (Ishihara et al., 2012). Histopathological studies of resistant and susceptible tomato found that R. solanacearum colonized fewer xylem vessels in wilt-resistant varieties (Grimault et al., 1994; Rahman et al., 1999). It is therefore possible that physical barriers to pathogen spread may be a component of tomato resistance to bacterial wilt.
Taken together, our results indicate that R. solanacearum’s ability to enzymatically disarm HCAs contributes to the success of this widespread pathogen. Pathogens have adopted multiple strategies to evade plant defense compounds. Pseudomonas syringae uses type III effectors to manipulate plant phenylpropanoids (Truman et al., 2006). Many plant pathogens protect themselves with drug efflux pumps, while others enzymatically degrade the plant defense compounds pisatin, tomatine, and HCAs (Tegtmeier and VanEtten, 1982; Tegos et al., 2002; Brown et al., 2007; Seipke and Loria, 2008; Michielse et al., 2012). Our results support a general model that root-infecting pathogens encounter toxic concentrations of HCAs, and that degradation of these defenses is important for pathogenic success.
Materials and Methods
Cultures and stock solutions
The bacterial strains and plasmids used in this study are described in Table 1. E. coli was grown in Luria-Bertani (LB) medium at 37°C. R. solanacearum was grown in CPG broth or TZC plates at 28 °C (Kelman, 1954). When appropriate, the antibiotics gentamicin (15 mg/L), kanamycin (25 mg/L), and ampicillin (50 m/L) were added. Boucher’s minimal medium (BMM) buffered with 10 mM MES (2-(N-morpholino) ethanesulfonic acid) pH 5.5 or 7.0 was used as a minimal medium (Boucher et al., 1985). For sampling from the soil or roots, R. solanacearum was plated on modified SMSA semi-selective medium (10 g/L peptone, 5 ml/L glycerol, 1 g/L casamino acids, 2.5 mg/L crystal violet, 2.5 mg/L TZC [tetrazolium chloride], 13 mg/L bacitracin, 0.3 mg/L penicillin, 2.5 mg/L chloramphenicol, 25 mg/L cycloheximide) (Engelbrecht, 1994). Stock solutions of phenolic compounds were prepared in DMSO. Chemicals were from Sigma-Aldrich, Fisher Scientific, or Difco Laboratories.
Table 1. Strains and plasmids used in this study.
| Strain or Plasmida | Relevant characteristicsb | Source or Reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | F−
mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697galU galK rpsL (Strr) endA1 nupG |
Life Technologies |
| R. solanacearum | ||
| GMI1000 (WT) | Wildtype, phylotype I | (Boucher et al., 1985) |
| GMI1000-Gm | GMI1000 transformed with pRCG-GWY, Gmr |
This study |
| GMI1000-Km | GMI1000 transformed with pRCK-GWY, Kanr |
This study |
| Δ fcs | GMI1000 with unmarked, precise deletion of the feruloyl-CoA synthetase (fcs) ORF |
This study |
| Δfcs-Gm | Δfcs transformed with pRCG-GWY, Gmr | This study |
| Δfcs-Km | Δfcs transformed with pRCK-GWY, Kanr | This study |
| Δ fcs+fcs | Complemented Δfcs with Tn7FcsComp integrated into chromosome at the selectively neutral att site, Gmr |
This study |
| Plasmids | ||
| pRCG-GWY | Vector that integrates downstream of glmS on the GMI1000 chromosome, Gmr |
(Monteiro et al., 2012) |
| pRCK-GWY | Vector that integrates downstream of glmS on the GMI1000 chromosome, Kanr |
(Monteiro et al., 2012) |
| pCR-blunt | Cloning vector | Life Technologies |
| pUFR80 | pUFR80, Sucs (sacB), Kanr | (Castañeda et al., 2005) |
| pUC18T- miniTn7T-Gm |
Vector that integrates into selectively neutral att site on R. solanacearum chromosome Gmr, Ampr |
(Choi et al., 2005) |
| pTNSl | Helper plasmid for pUC18T-miniTn7T-Gm encoding the site-specific TnsABCD Tn7 transposase, Ampr |
(Choi et al., 2005) |
| pUFR80-fcsKO | pUFR80 + fcs markerless deletion construct inserted into sacI/xbaI sites in MCS, Sucs (sacB), Kanr |
This study |
| pMiniTn7FcsComp |
fcs operon (fca-vdh-fcs) with native promoter cloned into hindIII/speI sites in pUC18T-miniTn7T-Gm, Gmr, Ampr |
This study |
A plate assay was used to detect HCA degradation ability in 20 R. solanacearum isolates. BMM MES pH 7.0 plates were supplemented with 1 mM succinate, ferulate, p-coumarate, vanillin, vanillate, p-hydroxybenzoate, or 5 mM protocatechuate. Compound concentration was chosen empirically as there was a trade-off between compound toxicity at high concentrations and minimal bacterial growth at low concentrations. To assess growth, 2 μl of a dense overnight culture of each strain was spotted onto the plates. After incubation at 28 °C for three to five days, growth of the strains on each substrate was assessed relative to growth on BMM plates without supplemented carbon. In several cases, plate growth phenotypes were indeterminate or contradicted predictions from genomic data. So, growth was further tested by culturing these strains for 48 hr in liquid BMM with the relevant carbon source and quantifying cell density by dilution plating.
Plant growth conditions
Wilt-susceptible tomato plants (cv. Bonny Best) were grown in Sunshine Redimix professional growing mix at 28 °C in a climate controlled growth chamber with a 12 hr day/ 12 hr night cycle. To test virulence during cool conditions, plants were grown in a climate controlled chamber with a 24 day/19 °C night cycle. Plants were watered with Hoagland solution.
Genomic analysis of R. solanacearum species complex and identification of HCA degradation gene homologs
The phylogenetic tree was designed around a matrix of genomic distances obtained using the Maximum Unique Match index (MUMi) algorithm (Deloger et al., 2009). MUMi values were computed from pairwise genome comparisons made with MUMMer 3.0 (Kurtz et al., 2004). The distances were then clustered together into a tree using the neighbor-joining method (Saitou and Nei, 1987).
We identified homologs of the GMI1000 and UW551 HCA degradation pathway using the OMA algorithm with translated coding sequences from the genomes of the other R. solanacearum isolates (Altenhoff et al., 2011).
Strain construction
The Δfcs strain was created using a sacB suicide vector designed to precisely excise the fcs ORF. Briefly, we amplified ~1 kb regions directly upstream (fcsKOupF: 5′-CTCGACGATGCGGACCTG-3′; fcsKOupR: 5′-GACAGCGACCTCGCATCAG-3′) and downstream of the fcs ORF (fcsKOdwnF: 5′-ctcatgcgaggtcgctgtcGAGTGTTGAGCGGGGCC-3′; fcsKOdwnR: 5′-GGAAGGCGAATTCGAGCG-3′) by PCR, fused the fragments by splice by overlap extension PCR (SOE-PCR) (Heckman and Pease, 2007), and blunt-end ligated them into the pCR-blunt subcloning vector (Life Technologies). This knockout construct was transferred by restriction digestion and ligation to the sacB vector pUFR80 to create pUFR80-fcsKO (Castañeda et al., 2005). R. solanacearum strain GMI1000 (WT) was transformed with pUFR80-fcsKO by electroporation and plated on kanamycin media to select for merodiploids that were sucrose sensitive and KanR. A clone was then counter-selected on CPG+5% sucrose to select for excision of the sacB-containing vector backbone. This process either restored the wildtype genotype or yielded a markerless deletion of the fcs ORF. Colony PCR using fcsKOupF and fcsKOdwnR primers was used to screen for loss of the fcs ORF.
A miniTn7 vector was used to complement the Δfcs strain (Choi et al., 2005). The miniTn7 transposon integrates into the selectively neutral att site downstream of glmS. We amplified the 5.6 kb putative operon encompassing 500 bp upstream of fca, the fca ORF, the vdh ORF and the fcs ORF using primers fcsoperonF (5′-TGCACCAGGACCAATACCTC-3′) and fcsoperonR (5′-CTCAACGTGTTCCCCATCCA-3′). The resulting PCR product was subcloned into pCR-blunt and transferred by restriction enzyme digestion and ligation into pUC18t-miniTn7-Gm to create pTn7fcsComp. The Δfcs R. solanacearum strain was transformed with pMiniTn7fcsComp and the helper vector pTNS1 encoding the transposase TnsABC+D and plated on gentamicin media. Complementation was confirmed by restoration of the ability to grow on HCAs.
To create antibiotic marked strains for colonization and competition assays, GMI1000 and the Δfcs strain were transformed with the chromosomal insertion vectors pRCG-GWY or pRCK-GWY carrying gentamicin or kanamycin cassettes, respectively (Monteiro et al., 2012).
Virulence assay
To assess virulence following soil soak inoculation, 17- to 21-day old plants with unwounded roots were inoculated by pouring bacterial suspensions into the soil to a final concentration of 1 × 108 CFU/g soil (Tans-Kersten et al., 1998). Symptoms on each plant were rated daily using a disease index scale of 0-4 corresponding to wilt severity: 0, asymptomatic plants; 1, less than 25%; 2, less than 50%; 3, less than 75%; and 4, up to 100% leaves wilted.
Colonization and competition assays
To assess colonization ability of individual strains, plants were soil soak inoculated with either WT-Gm or Δfcs-Gm. At 3 and 6 days after inoculation, bacterial populations were determined in surface-sterilized root and midstem stem tissue. To surface sterilize the roots, soil was removed by gentle shaking and washing. Then roots were swirled in a 10% bleach solution for 15 sec and rinsed three times in successive water baths to remove remaining bleach. Roots were sectioned into evenly distributed slices totaling 0.3 g. From the same plants, a 0.1 g midstem stem slice was sampled. Tissue was ground in water with 0.28 mm metal beads using a homogenizer (MoBio). Stem grinding required 2 cycles, and root grinding required 3 cycles of 2200 rpm for 1.5 min with a 4 min rest between cycles. Homogenized root and stem tissue were dilution plated onto SMSA and CPG with gentamicin, respectively.
A competition assay was used to investigate subtler differences in stem colonization ability (Yao and Allen, 2006). For soil soak inoculations, plants were inoculated with a 1:1 mixture of antibiotic-marked WT:Δfcs bacteria totaling 1 × 108 CFU/g soil. Marker swapping was used to ensure that competitive fitness differences were not caused by the antibiotic resistance marker: thus a set of plants were inoculated with a WT-Gm+ Δfcs-Km mixture and another set were inoculated with a WT-Km+ Δfcs-Gm mixture. Antibiotic markers did not significantly impact the fitness of either strain (Fig. S1). At the first sign of wilt symptoms (disease index = 1), plants were harvested and population sizes of each strain in the stem were determined by grinding and dilution plating on selective media. Population size was normalized to initial inoculum of each strain. Then, competitive index (CI) was calculated by dividing the normalized Δfcs population size by the normalized WT population size from the same plant. For cut-petiole inoculations, bacteria were directly introduced into the stem by placing a 2 μl drop of bacterial suspension onto a freshly cut petiole. Each plant was inoculated with 4,000 cells in a 1:1 mixture of marked WT and Δfcs bacteria.
Growth in xylem sap, root exudate, and potting soil extract
Xylem sap was collected from healthy and soil soak inoculated plants displaying the first signs of symptoms, as previously described (Jacobs et al., 2012). Plants were detopped with a sharp blade, and sap was allowed to pool on the stump by root pressure. The first drop was discarded to avoid contamination by cell debris, and the stump was rinsed with water and blotted dry. Sap was only collected for 30 minutes to avoid damage-response-induced changes in sap composition. Samples were flash-frozen and kept at −80°C until use. Growth of strains on 0.2 uM filter-sterilized xylem sap, root exudate was measured in a plate reader (Bio-Tek). Overnight cultures were washed and adjusted to OD600nm of 1.0. In a half-area 96-well plate (Corning), 45 μl of each growth substrate was combined with 5 μl of bacterial suspensions. Optical density was measured hourly until growth plateaued. Each experiment was repeated twice.
Root exudate was collected as described (Yao and Allen, 2006). Briefly, seeds were sterilized and germinated on 1% water agar plates in the dark for 3 days. Sterile roots were transferred into a 50 ml conical tube containing 5 ml of BMM with 10 mM MES pH 7.0. Tubes were incubated in the dark for 24 hr, and root exudate was used immediately as previous studies reported loss of potency with time.
To collect water-soluble potting soil extract, 1 g of potting soil was suspended in 50 ml of distilled water in a 50 ml conical tube and incubated horizontally with shaking for 2 hr at room temperature (Smolander et al., 2005). Filtered potting soil extract was used immediately. Dense overnight cultures were washed, and 0.5 μl of the cell suspension was inoculated into 5 ml of potting soil extract. Cell density was determined periodically by dilution plating.
Lignin quantification and visualization
Twenty-one day-old tomato plants were left healthy or inoculated by pouring WT or Δfcs bacteria into the soil to a final concentration of 1 × 108 CFU/g soil. Total stem was harvested 3, 6, and 9 days after inoculation and desiccated, yielding approximately 30 mg dry weight/plant. Total lignin was quantified by the spectroscopic acetyl bromide assay (Fukushima and Hatfield, 2004). Wood pulp inulin was used as a lignin standard. Lignin in cross-sections of tomato stems was stained by phloroglucinol:HCl (Nakano and Meshitsuka, 1992).
Growth inhibition assay
A growth inhibition assay modified from the standard minimum inhibitory concentration (MIC) assay was used to test the toxicity of HCAs (Brown et al., 2007). BMM MES pH 5.5 +10 mM succinate supplemented with 23-3000 μM of ferulate, caffeate, or p-coumarate or with no inhibiting compound was inoculated with bacterial strains to 1 × 105 CFU/ml final concentration. After incubating strains at 28 °C with shaking, cell density was measured by optical density in a Bio-Tek plate reader. Growth of strains in each condition was calculated relative to growth of the WT strain without inhibitory HCAs.
Supplementary Material
e-Xtra Fig. S1. Kanamycin and gentamicin resistance markers did not affect competitive fitness of strains. A, Soil soak competition data of WT and Δfcs bacteria from Fig. 5; B, Petiole competition data of WT and Δfcs bacteria from Fig. 5 Results from the WT-kan:Δfcs-gm inoculation are shown in filled squares and results from the WT-gm:Δfcs-kan inoculation are shown in open circles.
Acknowledgements
The authors gratefully acknowledge Paul Weimer for help with lignin analysis, Mehdi Kabbage for help with microscopy, and Frederique van Gijsegem, Devanshi Khokhani, and Philippe Prior for valuable discussions. We thank Brianna Fochs for technical help. This research was supported by the USDA-ARS Floral and Nursery Crops Research Initiative and the UW-Madison College of Agricultural and Life Sciences. TML was supported by NIH National Research Service Award T32 GM07215 and by a Agriculture and Food Research Initiative Competitive Grant from the USDA National Institute of Food and Agriculture.
Footnotes
Author Contributions
T. Lowe designed most experiments and analyzed data. F. Ailloud constructed the whole-genome phylogenetic tree and analyzed the genetic conservation of the HCA degradation pathway. C. Allen advised T. Lowe in experimental design and analysis. All authors participated in drafting the manuscript and approved of the final version for submission.
Literature Cited
- Abdelkafi S, Sayadi S, Gam A, Ben Z, Casalot L, Labat M. Bioconversion of ferulic acid to vanillic acid by Halomonas elongata isolated from table-olive fermentation. FEMS Microbiol. Lett. 2006;262:115–120. doi: 10.1111/j.1574-6968.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- Airola MV, Tumolo JM, Snider J, Hannun YA. Identification and biochemical characterization of an acid sphingomyelinase-like protein from the bacterial plant pathogen Ralstonia solanacearum that hydrolyzes ATP to AMP but not sphingomyelin to ceramide. PLoS ONE. 2014;9:e105830. doi: 10.1371/journal.pone.0105830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez S, Marsh EL, Schroeder SG, Schachtman DP. Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ. 2008;31:325–340. doi: 10.1111/j.1365-3040.2007.01770.x. [DOI] [PubMed] [Google Scholar]
- Balcerzak M, Harris LJ, Subramaniam R, Ouellet T. The feruloyl esterase gene family of Fusarium graminearum is differentially regulated by aromatic compounds and hosts. Fungal Biol. 2012;116:478–488. doi: 10.1016/j.funbio.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Beckman CH. Phenolic-storing cells: keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Path. 2000;57:101–110. [Google Scholar]
- Bhattacharya A, Sood P, Citovsky V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010;11:705–719. doi: 10.1111/j.1364-3703.2010.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher C, Barberis P, Trigalet A, Demery D. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 1985;131:2449–2457. [Google Scholar]
- Brown DG, Swanson JK, Allen C. Two host-induced Ralstonia solanacearum genes, acrA and dinF, encode multidrug efflux pumps and contribute to bacterial wilt virulence. Appl. Environ. Microbiol. 2007;73:2777–2786. doi: 10.1128/AEM.00984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillo T, Renoud S, Kerzaon I, Vial L, Baude J, Gaillard V, Bellvert F, Chamignon C, Comte G, Nesme X. Analysis of hydroxycinnamic acid degradation in Agrobacterium fabrum reveals a coenzyme A-dependent, beta-oxidative deacetylation pathway. Appl. Env. Microbiol. 2014;80:3341–3349. doi: 10.1128/AEM.00475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos L, Lisón P, López-Gresa MP, Rodrigo I, Zacarés L, Conejero V, Bellés JM. Transgenic tomato plants overexpressing tyramine N-hydroxycinnamoyltransferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae. Mol. Plant-Microbe Interact. 2014;27:1159–1169. doi: 10.1094/MPMI-04-14-0104-R. [DOI] [PubMed] [Google Scholar]
- Cao Y, Tian B, Liu Y, Cai L, Wang H, Lu N, Wang M, Shang S, Luo Z, Shi J. Genome sequencing of Ralstonia solanacearum FQY_4, isolated from a bacterial wilt nursery used for breeding crop resistance. Genome Announc. 2013;1:e00125–00113. doi: 10.1128/genomeA.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda A, Reddy JD, El-Yacoubi B, Gabriel DW. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant-Microbe Interact. 2005;18:1306–1317. doi: 10.1094/MPMI-18-1306. [DOI] [PubMed] [Google Scholar]
- Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Colburn-Clifford J, Allen C. A cbb3-type cytochrome C oxidase contributes to Ralstonia solanacearum R3bv2 growth in microaerobic environments and to bacterial wilt disease development in tomato. Mol. Plant-Microbe Interact. 2010;23:1042–1052. doi: 10.1094/MPMI-23-8-1042. [DOI] [PubMed] [Google Scholar]
- Deloger M, El Karoui M, Petit MA. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J. Bacteriol. 2009;191:91–99. doi: 10.1128/JB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGuistini S, Wang Y, Liao NY, Taylor G, Tanguay P, Feau N, Henrissat B, Chan SK, Hesse-Orce U, Alamouti SM. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2504–2509. doi: 10.1073/pnas.1011289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphinstone JG. The current bacterial wilt situation: A global overview. In: Allen C, Prior P, Hayward AC, editors. Bacterial Wilt: The Disease and the Ralstonia solanacearum Species Complex. American Phytopathological Society Press; St Paul: 2005. pp. 9–28. [Google Scholar]
- Engelbrecht M. Modification of a semi-selective medium for the isolation and quantification of Pseudomonas solanacearum. Bacterial Wilt Newsletter. 1994:3–5. [Google Scholar]
- Fitzgerald D, Stratford M, Gasson M, Ueckert J, Bos A, Narbad A. Mode of antimicrobial action of vanillin against Escherichia coli, Lactobacillus plantarum and Listeria innocua. J. Appl. Microbiol. 2004;97:104–113. doi: 10.1111/j.1365-2672.2004.02275.x. [DOI] [PubMed] [Google Scholar]
- Fry SC, Willis SC, Paterson AE. Intraprotoplasmic and wall-localised formation of arabinoxylan-bound diferulates and larger ferulate coupling-products in maize cell-suspension cultures. Planta. 2000;211:679–692. doi: 10.1007/s004250000330. [DOI] [PubMed] [Google Scholar]
- Fukushima RS, Hatfield RD. Comparison of the acetyl bromide spectrophotometric method with other analytical lignin methods for determining lignin concentration in forage samples. J. Agric. Food Chem. 2004;52:3713–3720. doi: 10.1021/jf035497l. [DOI] [PubMed] [Google Scholar]
- Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, Duan YP, Flores-Cruz Z, Huang Q, Clifford JM, Presting G, González ET, Reddy J, Elphinstone J, Swanson J, Yao J, Mulholland V, Liu L, Farmerie W, Patnaikuni M, Balogh B, Norman D, Alvarez A, Castillo JA, Jones J, Saddler G, Walunas T, Zhukov A, Mikhailova N. Identification of open reading frames unique to a select agent: Ralstonia solanacearum Race 3 biovar 2. Mol. Plant-Microbe Interact. 2006;19:69–79. doi: 10.1094/MPMI-19-0069. [DOI] [PubMed] [Google Scholar]
- Grimault V, Gelie B, Lemattre M, Prior P, Schmit J. Comparative histology of resistant and susceptible tomato cultivars infected by Pseudomonas solanacearum. Physiol. Mol. Plant Pathol. 1994;44:105–123. [Google Scholar]
- Harris V, Jiranek V, Ford CM, Grbin PR. Inhibitory effect of hydroxycinnamic acids on Dekkera spp. Appl. Microbiol. Biotechnol. 2010;86:721–729. doi: 10.1007/s00253-009-2352-6. [DOI] [PubMed] [Google Scholar]
- Hassan S, Hugouvieux-Cotte-Pattat N. Identification of two feruloyl esterases in Dickeya dadantii 3937 and induction of the major feruloyl esterase and of pectate lyases by ferulic acid. J. Bacteriol. 2011;193:963–970. doi: 10.1128/JB.01239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Mitsuhara I, Takahashi H, Nakaho K. Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato. PLoS ONE. 2012;7:e46763. doi: 10.1371/journal.pone.0046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Kakei Y, Shimo H, Bashir K, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK. A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J. Biol. Chem. 2011;286:24649–24655. doi: 10.1074/jbc.M111.221168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JM, Babujee L, Meng F, Milling A, Allen C. The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio. 2012;3:e00114–00112. doi: 10.1128/mBio.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman A. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in tetrazolium medium. Phytopathol. 1954;44:693–695. [Google Scholar]
- Kim D, Kim SW, Choi KY, Lee JS, Kim E. Molecular cloning and functional characterization of the genes encoding benzoate and p-hydroxybenzoate degradation by the halophilic Chromohalobacter sp. strain HS-2. FEMS Microbiol. Lett. 2008;280:235–241. doi: 10.1111/j.1574-6968.2008.01067.x. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoue A, Burlat V, Henkes GJ, Koch I, Schurr U, Röse US. De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol. 2010;185:577–588. doi: 10.1111/j.1469-8137.2009.03066.x. [DOI] [PubMed] [Google Scholar]
- Li L, Steffens JC. Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta. 2002;215:239–247. doi: 10.1007/s00425-002-0750-4. [DOI] [PubMed] [Google Scholar]
- Li Z, Wu S, Bai X, Liu Y, Lu J, Liu Y, Xiao B, Lu X, Fan L. Genome sequence of the tobacco bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 2011;193:6088–6089. doi: 10.1128/JB.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Guidot A, Barberis P, Beuzón CR, Genin S. A competitive index assay identifies several Ralstonia solanacearum Type III effector mutant strains with reduced fitness in host plants. Mol. Plant-Microbe Interact. 2010;23:1197–1205. doi: 10.1094/MPMI-23-9-1197. [DOI] [PubMed] [Google Scholar]
- Mandal S, Mitra A. Accumulation of cell wall-bound phenolic metabolites and their upliftment in hairy root cultures of tomato (Lycopersicon esculentum Mill.) Biotechnol. Lett. 2008;30:1253–1258. doi: 10.1007/s10529-008-9666-9. [DOI] [PubMed] [Google Scholar]
- Mandal SM, Chakraborty D, Dey S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 2010;5:359–368. doi: 10.4161/psb.5.4.10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse CB, Reijnen L, Olivain C, Alabouvette C, Rep M. Degradation of aromatic compounds through the β-ketoadipate pathway is required for pathogenicity of the tomato wilt pathogen Fusarium oxysporum f. sp. lycopersici. Mol. Plant Pathol. 2012;13:1089–1100. doi: 10.1111/j.1364-3703.2012.00818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro F, Solé M, Dijk I.v., Valls M. A chromosomal insertion toolbox for promoter probing, mutant complementation, and pathogenicity studies in Ralstonia solanacearum. Mol. Plant-Microbe Interact. 2012;25:557–568. doi: 10.1094/MPMI-07-11-0201. [DOI] [PubMed] [Google Scholar]
- Mueller W, Beckman C. Ultrastructure of the cell wall of vessel contact cells in the xylem of tomato stems. Ann. Botany. 1984;53:107–114. [Google Scholar]
- Nakaho K, Hibino H, Miyagawa H. Possible mechanisms limiting movement of Ralstonia solanacearum in resistant tomato tissues. J. Phytopathol. 2000;148:181–190. [Google Scholar]
- Nakano J, Meshitsuka G. The detection of lignin. In: Lin SY, Dence CW, editors. Methods Lignin Chemistry. Springer-Verlag; Berlin: 1992. pp. 23–32. [Google Scholar]
- Naoumkina MA, Zhao Q, GALLEGO-GIRALDO L, Dai X, Zhao PX, Dixon RA. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010;11:829–846. doi: 10.1111/j.1364-3703.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbad A, Gasson MJ. Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology. 1998;144:1397–1405. doi: 10.1099/00221287-144-5-1397. [DOI] [PubMed] [Google Scholar]
- Neumann G, Römheld V. The release of root exudates as affected by the plant physiological status. In: Pinton R, Veranini Z, Nannipieri P, editors. The Rhizosphere Biochemistry and Organic Substances at the Soil-Plant Interface. CRC Press; New York: 2007. pp. 23–72. [Google Scholar]
- Parke D, Ornston LN. Hydroxycinnamate (hca) catabolic genes from Acinetobacter sp. strain ADP1 are repressed by HcaR and are induced by hydroxycinnamoyl-coenzyme A thioesters. Appl. Environ. Microbiol. 2003;69:5398–5409. doi: 10.1128/AEM.69.9.5398-5409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Guidot A, Vailleau F, Valls M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 2013;14:651–662. doi: 10.1111/mpp.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pantoja D, De la Iglesia R, Pieper DH, González B. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol. Rev. 2008;32:736–794. doi: 10.1111/j.1574-6976.2008.00122.x. [DOI] [PubMed] [Google Scholar]
- Plaggenborg R, Overhage J, Steinbüchel A, Priefert H. Functional analyses of genes involved in the metabolism of ferulic acid in Pseudomonas putida KT2440. Appl. Microbiol. Biotechnol. 2003;61:528–535. doi: 10.1007/s00253-003-1260-4. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Abdullah H, Vanhaeke M. Histopathology of susceptible and resistant Capsicum annuum cultivars infected with Ralstonia solanacearum. J. Phytopathol. 1999;147:129–140. [Google Scholar]
- Remenant B, Babujee L, Lajus A, Médigue C, Prior P, Allen C. Sequencing of K60, type strain of the major plant pathogen Ralstonia solanacearum. J. Bacteriol. 2012;194:2742–2743. doi: 10.1128/JB.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenant B, Cambiaire J-CD, Cellier G, Barbe V, Medigue C, Jacobs JM, Fegan M, Allen C, Prior P. Phylotype IV strains of Ralstonia solanacearum, R. syzygii and the Blood Disease Bacterium form a single genomic species despite their divergent life-styles. PLoS ONE. 2011;6:e24356. doi: 10.1371/journal.pone.0024356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenant B, Coupat-Goutaland B, Guidot A, Cellier G, Wicker E, Allen C, Fegan M, Pruvost O, Elbaz M, Calteau A, Salvignol G, Mornico D, Mangenot S, Barbe V, Médigue C, Prior P. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics. 2010;11:379. doi: 10.1186/1471-2164-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Silva MJ, Méndez V, Agullo L, Seeger M. Genomic and functional analyses of the gentisate and protocatechuate ring-cleavage pathways and related 3-hydroxybenzoate and 4-hydroxybenzoate peripheral pathways in Burkholderia xenovorans LB400. PloS ONE. 2013;8:e56038. doi: 10.1371/journal.pone.0056038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, Chandler M, Choisne N, Claudel-Renard C, Cunnac S, Demange N, Gaspin C, Lavie M, Moisan A, Robert C, Saurin W, Schiex T, Siguier P, Thebault P, Whalen M, Wincker P, Levy M, Weissenbach J, Boucher CA. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- Seipke RF, Loria R. Streptomyces scabies 87-22 possesses a functional tomatinase. J. Bacteriol. 2008;190:7684–7692. doi: 10.1128/JB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolander A, Loponen J, Suominen K, Kitunen V. Organic matter characteristics and C and N transformations in the humus layer under two tree species, Betula pendula and Picea abies. Soil Biol. Biochem. 2005;37:1309–1318. [Google Scholar]
- Tans-Kersten J, Guan Y, Allen C. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 1998;64:4918–4923. doi: 10.1128/aem.64.12.4918-4923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegos G, Stermitz FR, Lomovskaya O, Lewis K. Multidrug pump inhibitors uncover remarkable activity of plant antimicrobials. Antimicrobial Agents and Chemotherapy. 2002;46:3133–3141. doi: 10.1128/AAC.46.10.3133-3141.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeier K, VanEtten H. The role of pisatin tolerance and degradation in the virulence of Nectria haematococca on peas: a genetic analysis. Phytopathology. 1982;72:608–612. [Google Scholar]
- Truman W, Zabala MT, Grant M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. 2006;46:14–33. doi: 10.1111/j.1365-313X.2006.02672.x. [DOI] [PubMed] [Google Scholar]
- Vanitha SC, Niranjana SR, Umesha S. Role of phenylalanine ammonia lyase and polyphenol oxidase in host resistance to bacterial wilt of tomato. J. Phytopathol. 2009;157:552–557. [Google Scholar]
- Wallis CM, Chen J. Grapevine phenolic compounds in xylem sap and tissues are significantly altered during infection by Xylella fastidiosa. Phytopathology. 2012;102:816–826. doi: 10.1094/PHYTO-04-12-0074-R. [DOI] [PubMed] [Google Scholar]
- Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Guilbaud C, Fegan M, Prior P. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 2007;73:6790–6801. doi: 10.1128/AEM.00841-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zheng H.-j., Liu L, Pan Z.-c., Prior P, Tang B, Xu J.-s., Zhang H, Tian Q, Zhang L.-q. The complete genome sequence of plant pathogen Ralstonia solanacearum strain Po82. J. Bacteriol. 2011;193:4261–4262. doi: 10.1128/JB.05384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Allen C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 2006;188:3697–3708. doi: 10.1128/JB.188.10.3697-3708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
e-Xtra Fig. S1. Kanamycin and gentamicin resistance markers did not affect competitive fitness of strains. A, Soil soak competition data of WT and Δfcs bacteria from Fig. 5; B, Petiole competition data of WT and Δfcs bacteria from Fig. 5 Results from the WT-kan:Δfcs-gm inoculation are shown in filled squares and results from the WT-gm:Δfcs-kan inoculation are shown in open circles.