Summary
Metabolic adaptations are critical to the ability of bacterial pathogens to grow within host cells and are normally preceded by sensing of host-specific metabolic signals, which in turn can influence the pathogen's virulence state. Previously, we reported that the intracellular bacterial pathogen Listeria monocytogenes responds to low availability of branched-chain amino acids (BCAA) within mammalian cells by up-regulating both BCAA biosynthesis and virulence genes. The induction of virulence genes required the BCAA-responsive transcription regulator, CodY, but the molecular mechanism governing this mode of regulation was unclear. In this report, we demonstrate that CodY directly binds the coding sequence of the L. monocytogenes master virulence activator gene, prfA, 15 nt downstream of its start codon, and that this binding results in up-regulation of prfA transcription specifically under low concentrations of BCAA. Mutating this site abolished CodY binding and reduced prfA transcription in macrophages, and attenuated bacterial virulence in mice. Notably, the mutated binding site did not alter prfA transcription or PrfA activity under other conditions that are known to activate PrfA, such as during growth in the presence of glucose-1-phosphate. This study highlights the tight crosstalk between L. monocytogenes metabolism and virulence' while revealing novel features of CodY-mediated regulation.
Introduction
Microorganisms respond to different physical and chemical signals, such as temperature, salinity and nutrient availability, in order to adapt to diverse ecological niches. These signals mainly serve to regulate the metabolic status of the organism, but can also trigger induction of a niche-specific set of genes that further optimize the organism's growth. While observations of such phenomena in pathogenic bacteria were made years ago, the molecular mechanisms that link the sensing of environmental signals to regulation of specific genes during infection are just beginning to be deciphered. Within the host, pathogens sense nutritional and physical signals that lead to the induction of genes specifically expressed in the host (Abu Kwaik & Bumann, 2013, Brown et al., 2008). These genes promote the pathogen's ability to invade and manipulate the host, as well to adapt to the host metabolic environment (Fuchs et al., 2012, Bretl et al., 2011). For example, Legionella pneumophila senses changes in threonine concentrations within host cells and switches from the replicative to the transmissive phase (Sauer et al., 2005). Vibrio cholerae senses and responds to reduced oxygen tension by up-regulation of virulence genes via a thiol-based sensor domain located within its virulence transcription regulator (Liu et al., 2011). Other examples include the response of Salmonella typhimurium to high iron concentrations, leading to enhanced adherence to and invasion of enterocytes (Kortman et al., 2012), and induction by Listeria monocytogenes of virulence genes in response to glucose-1-phosphate, a sugar that is mainly available in mammalian cells (Ripio et al., 1997a). Though there are many examples of signaling-dependent gene expression in host-pathogen interactions, the mechanisms by which metabolic signals are sensed and transduced to regulatory cascades that co-regulate metabolism and virulence remain largely unknown.
L. monocytogenes, is a Gram-positive, foodborne intracellular pathogen and the causative agent of listeriosis in animals and humans (Swaminathan & Gerner-Smidt, 2007). L. monocytogenes invades host cells either passively via phagocytosis or actively by expressing surface proteins that induce bacterial internalization (Cossart, 2011). Once inside the cell, L. monocytogenes is initially contained in a vacuole from which it rapidly escapes by expressing the pore-forming cytolysin listeriolysin O (LLO), two additional phospholipases, PlcA and PlcB, and components of the Com system (Rabinovich et al., 2012, Mengaud et al., 1987, Portnoy et al., 1988, Geoffroy et al., 1991, Leimeister-Wachter et al., 1991, Goebel et al., 1988). In the host cell cytosol, L. monocytogenes replicates and gains mobility via polymerization of host actin filaments, enabling the bacteria to spread from cell to cell without being exposed to the extracellular environment (Tilney & Portnoy, 1989). Remarkably, most of the L. monocytogenes virulence factors are positively regulated by the master activator of virulence, PrfA (de las Heras et al., 2011). Regulation of the prfA gene is highly complex and multilayered. prfA is transcribed from three distinct promoters: two proximal promoters, P1 and P2, located upstream of the prfA translation initiation codon and one distal promoter, P3, located upstream of the plcA gene and from which expression results in a bicistronic plcA-prfA transcript (Freitag & Portnoy, 1994). Transcription from the P1 and P2 promoters requires the sigma factors SigA and SigA or SigB, respectively, while SigA-dependent transcription from P3 is dependent on PrfA itself (Rauch et al., 2005, Freitag & Portnoy, 1994, Lobel et al., 2012). In addition, prfA is negatively regulated by the trans-acting small RNAs, SreA and SreB, both of which are synthesized as S-adenosylmethionine-dependent riboswitches under rich nutritional conditions, and that block PrfA synthesis by pairing with the 5’ untranslated region (UTR) of prfA mRNA (Loh et al., 2009). Moreover, the prfA 5’-UTR contains a thermosensory structure that allows prfA mRNA translation at 37°C, but blocks translation at 30°C by sequestering the ribosome binding site (Johansson et al., 2002). Lastly, the 5’ coding region stabilizes prfA mRNA and thus increases the efficiency of its translation (Loh et al., 2012). In addition to its transcriptional regulation, PrfA is also regulated post-translationally. When grown in rich laboratory media L. monocytogenes expresses PrfA but it remains inactive (Renzoni et al., 1997). Upon invasion of host cells, however, PrfA becomes active, leading to a rapid activation of virulence genes (Scortti et al., 2007). Since PrfA is a member of the cyclic-AMP receptor protein (Crp) family of transcriptional regulators, of which many require the binding of a cofactor for full activity, it has been postulated that upon invasion into host cells a specific cofactor binds PrfA and activates it. While the identity of this putative cofactor remains unknown, mutations were identified within PrfA that render it constitutively active (PrfA* mutations) (Ripio et al., 1997a, Mineret al., 2008, Ripio et al., 1997b, Shetron-Rama et al., 2003, Vega et al., 2004, Wong & Freitag, 2004). Together these observations suggest that conformational changes in PrfA regulate its activity (Freitag et al., 2009).
As for metabolic signals, PrfA is known to be regulated by sugar availability. It was shown that sugars transported by the PEP-dependent phosphotransferase system (PTS), such as glucose and cellobiose, repress PrfA activity, while the presence of non-PTS host-derived sugars, such as glucose-1-phosphate and glycerol, trigger prfA transcription and PrfA activity. (Mertins et al., 2006, Joseph et al., 2008, Milenbachs et al., 1997). Notably, the activation of PrfA in rich media conditions was shown to require activated charcoal in the medium, though the reason for this effect has not been clearly determined (Ripio et al., 1996). Iron availability also seems to play a role in prfA regulation, though the mechanism is not clear (Conte et al., 1996, Böckmannet al., 1996). We have recently discovered that branched-chain amino acids (BCAA) [isoleucine, leucine and valine] also serve as a metabolic signal for induction of prfA and its downstream regulated genes (Lobel et al., 2012). Limiting BCAA availability activates virulence, a response that requires a sensor of BCAA, the transcription factor called CodY, a known global regulator of metabolic genes in low G+C Gram-positive bacteria (Sonenshein, 2005). CodY, which has been studied most thoroughly in Bacillus subtilis, responds to intracellular availability of BCAA (and GTP) via direct binding to these metabolites (Levdikov et al., 2006, Shivers & Sonenshein, 2004). Multiple reports have demonstrated that interaction with isoleucine allows CodY to bind to the promoters of hundreds of genes (Belitsky & Sonenshein, 2013, Preis et al., 2009, Shivers et al., 2006, Molle et al., 2003, Sonenshein, 2007). In its isoleucine-bound state, CodY acts as a repressor of many metabolic pathways, including the BCAA biosynthesis pathway (encoded in part by the ilv operon), and as a positive regulator of other genes (Shivers et al., 2006, Molle et al., 2003, Preis et al., 2009). Upon intracellular depletion of BCAA, CodY dissociates from these promoters thus altering their transcription (Sonenshein, 2007). Interestingly, some genus/species-specific differences in the functionality of this regulator were reported. Whereas the activity of CodY limits virulence of Staphylococcus aureus (Montgomery et al., 2012, Rivera et al., 2012) and Clostridium difficile (Dineen et al., 2010), CodY enhances the expression of virulence genes in L. monocytogenes, Bacillus anthracis and several other human bacterial pathogens, though the molecular mechanism that mediates this enhancement has not been deciphered (Kreth et al., 2011, Chateau et al., 2011, van Schaik et al., 2009, Hendriksen et al., 2008, Flores et al., 2013, Li et al., 2013, Lobel et al., 2012).
In this report, we demonstrate that CodY directly activates expression of the prfA gene by binding to a sequence located unexpectedly within the 5’ coding region. Using electrophoretic mobility shift (EMSA) and chromatin immuno-precipitation (ChIP) assays, we identified a CodY-binding site, located ~ 15-bp downstream of the prfA start codon, that is necessary for prfA expression under low BCAA concentrations. Mutating this site abolished CodY binding and resulted in lower transcription of prfA and PrfA-dependent virulence genes under low BCAA conditions. Moreover, both a ΔcodY mutant strain and a mutant altered in the prfA CodY-binding site were attenuated for virulence in both cultured macrophages and mice. Overall, this study defines a direct regulatory link between CodY and PrfA, and highlights the tight interaction between in vivo metabolism (i.e., during infection) and virulence in L. monocytogenes.
Results
CodY is required for the induction of virulence genes in response to low concentrations of BCAA, but not in response to glucose 1-phosphate
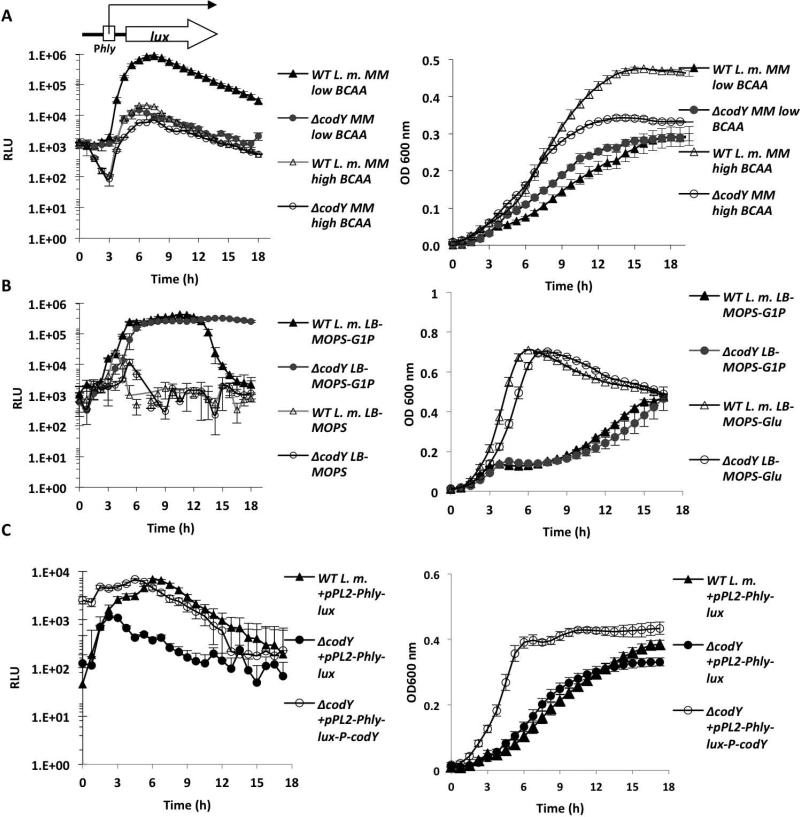
As mentioned above, two major metabolic signals are currently known to trigger robust expression of L. monocytogenes virulence genes and thus switch the bacteria to the virulent state: one is the availability of phosphorylated hexoses (e.g., glucose-1-phosphate), and the second is the low availability of BCAA. Both conditions are found in the cytosol of mammalian cells (Chico-Calero et al., 2002, Ripio et al., 1997a, Lobel et al., 2012) and our previous work demonstrated the involvement of CodY in the response to low BCAA (Lobel et al., 2012). To explore the possibility that the two metabolic signals use the same regulatory pathway, we first examined whether CodY is also required for the induction of virulence genes in bacteria growing in glucose-1-phosphate containing media. To this end, we used a lux reporter system to measure the expression of the hly gene (encoding LLO toxin) during growth in a defined minimal medium (MM) (Phan-Thanh & Gormon, 1997) containing relatively low or high concentrations of BCAA (~80 µM and ~800 μM, respectively, of each BCAA; named “low-BCAA MM” or “high-BCAA MM”) and in LB-MOPS activated charcoal medium supplemented with glucose-1-phosphate or glucose as a control (25 mM of each, “LB-MOPS-G1P” or “LB-MOPS-Glu”, respectively). To follow PrfA-dependent hly transcription, an integrative plasmid containing the lux operon under the control of the hly promoter (pPL2-Phlylux) (Bron et al., 2006, Lobel et al., 2012) was introduced by conjugation into the wild-type (WT) L. monocytogenes strain 10403S and its ΔcodY mutant derivative. Transconjugants were subjected to growth in the different media and parallel measurements of luminescence and optical density (OD600) were taken. As shown in Figure 1, hly was highly induced under low-BCAA MM and LB-MOPS-G1P conditions (~100-fold and ~1000-fold, respectively) and was not induced in the control media, high-BCAA MM and LB-MOPS-Glu. Notably, CodY was absolutely necessary for hly transcription during growth in low-BCAA MM, but was largely dispensable during growth in LB-MOPS-G1P medium (Figure 1 A-B). In the latter condition a biphasic luminescence profile was reproducibly observed. A ΔcodY complemented strain constitutively expressing codY from the integrative plasmid pPL2-Phlylux-P-codY exhibited WT levels of hly transcription under low-BCAA MM conditions, strengthening the premise that CodY indeed mediates hly induction in response to low availability of BCAA (Figure 1C). Together, the results indicate that the two metabolic signals (glucose 1-phosphate and BCAA) exploit distinct regulatory pathways that trigger hly transcription, and that CodY is primarily responsible for the induction under low concentrations of BCAA.
Figure 1. Induction of hly gene under low BCAA conditions is CodY-dependent, while its induction by glucose-1-phosphate is CodY-independent.
A. Luminescence (left panels) and optical density (right panels) measurements of WT L. monocytogenes and ΔcodY bacteria harboring the pPL2-Phlylux plasmid indicating Phly promoter activity during growth in low-BCAA MM and high-BCAA MM. B. Luminescence (RLU) and optical density measurements of WT L. monocytogenes and ΔcodY bacteria harboring the pPL2-Phlylux plasmid during growth in LB-MOPS-G1P and LB-MOPS-Glu media. C. Complementation of ΔcodY strain. A copy of the codY gene under the regulation of the SPAC/lacOid promoter (taken from pLIV2 plasmid) was introduced into the ΔcodY strain on the integrative plasmid pPL2 together with the hly-lux reporter system (pPL2-Phlylux-P-codY). Both ΔcodY and its complemented strain were grown in low-BCAA MM and the Phly promoter activity was measured. Results are averages of at least 4 independent experiments, representing 3 biological replicates in each experiment. Error bars represent standard deviation.
CodY preferentially binds the regulatory region of the prfA gene under low concentrations of BCAA
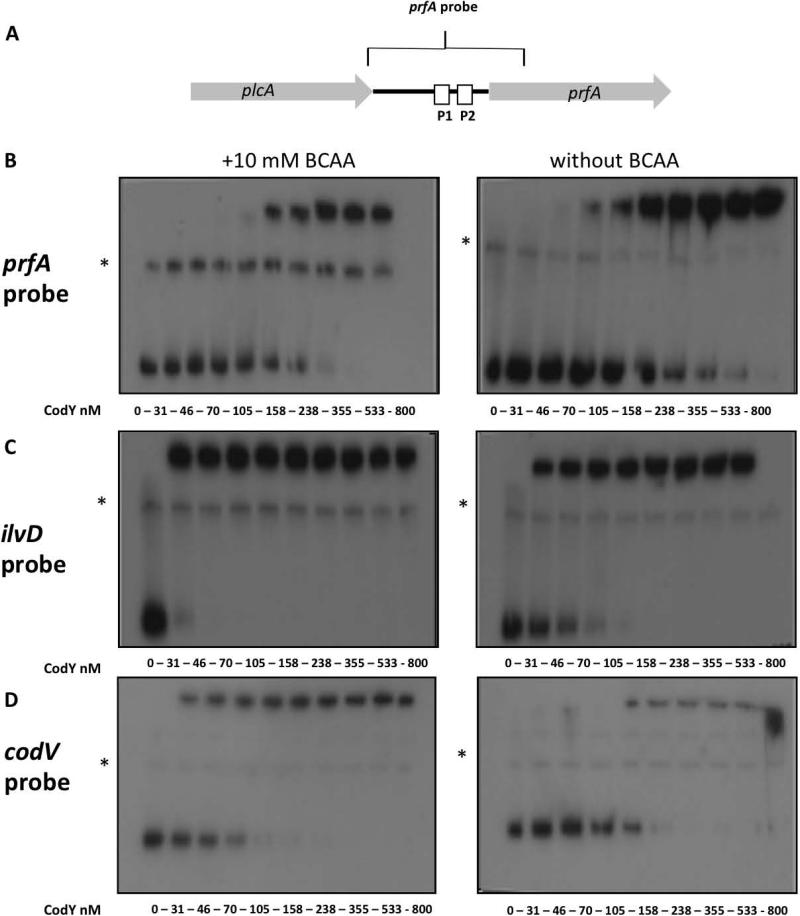
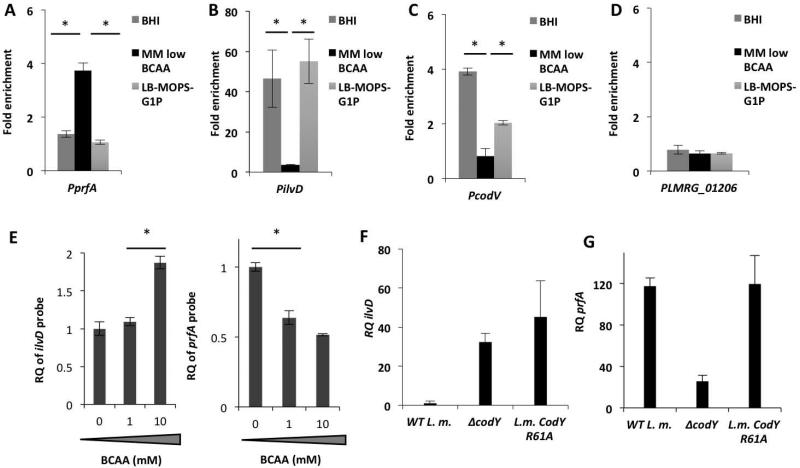
Since we have previously shown that induction of the hly promoter under conditions of limiting BCAA is not only CodY-dependent but also PrfA-dependent, and that prfA transcription is reduced in a ΔcodY mutant, we reasoned that the regulatory effect of CodY on hly and additional virulence genes is mediated via PrfA itself. We have demonstrated that CodY specifically activates the transcription of prfA from its two proximal promoters P1 and P2, though we did not determine whether this regulation occurs directly or indirectly via another factor (Lobel et al., 2012). To investigate if CodY directly regulates prfA by binding to its regulatory region, we performed electrophoretic mobility shift assays (EMSA), in which increasing amounts of purified recombinant 6His-tagged CodY (CodY-His6) were incubated with a prfA promoter probe containing the complete plcA-prfA intergenic region plus 100-bp of the prfA coding sequence (Figure 2A). For CodY purification, a codY gene with six histidine codons appended to the 3’ end was cloned in the pET28 vector and expressed as described in the Materials and Methods section. The EMSA assay was performed under two conditions, with and without BCAA (10 mM of each), and included two control DNA probes: one containing the ilvD promoter region of the ilv operon known to be directly regulated by CodY (Molle et al., 2003) and the second containing the promoter of the codVWXY operon (encoding the codY gene), as in Lactococus lactis the codY gene was shown to be auto-regulated (den Hengst et al., 2005). In line with our expectations, the EMSA assays demonstrated that CodY binds to all three probes, albeit with different affinities. CodY binding to the prfA probe exhibited a KD of 236 ± 49 nM, whereas its binding to the ilvD and codV probes exhibited a much lower KD of 3.1 ± 1.5 nM and 51.74 ± 1.68 nM, respectively, under conditions containing 10 mM of BCAA (Figure 2 B-D) (average KD values were calculated based on 3 regression analyses of 3 independent EMSA gel experiments, Figure S1). As expected, a significant decrease in CodY binding to the ilvD and the codV control probes was observed in the absence of BCAA (KD = 48 ± 20.7 nM and 96 ± 11.4 nM, respectively), though CodY binding to the prfA probe under these conditions remained largely unchanged (KD = 232 ± 66 nM) (Figure 2 and S1). We also performed EMSA assays with specific unlabeled competitor probes to verify the specificity of CodY binding (Figure S2). While these in vitro experiments have demonstrated binding of CodY to the regulatory region of the prfA gene (though with lower affinity in comparison to the control probes), they did not demonstrate a role for BCAA in this binding as insinuated by the in vivo experiments. To better examine this question we set up to assess CodY ability to bind the prfA regulatory region under in vivo conditions, i.e., during bacterial growth in rich or low BCAA-containing media. For this purpose we performed a chromatin immunoprecipitation assay in combination with real-time, quantitative PCR (ChIP-RT-qPCR) to analyze the binding of His-tagged CodY to regulatory DNA sequences in vivo (Waldminghaus & Skarstad, 2010). First, a derivative of strain 10403S was constructed that has a sixhistidine tag at the 3’-end of the chromosomal codY gene (L.m. codY-6his strain). The functionality of the His-tagged CodY was similar to that of the native CodY, as shown by its ability to support bacterial growth in brain heart infusion (BHI) medium and to repress ilvD transcription in this rich medium (Figure S3). The L.m. codY-6his strain was then grown in the two rich media, BHI and LB-MOPS-G1P, and in low-BCAA MM. During mid-exponential growth phase, bacteria were treated with formaldehyde to covalently crosslink proteins to DNA. Bacteria were then lysed, CodY-His6 was immunoprecipitated, and CodY-bound DNA was isolated and quantitated by RT-qPCR using specific primers for the regulatory regions of prfA, ilvD, codV and LMRG_01206 as a negative control. Using this method, the association of CodY with prfA, ilvD, codV and LMRG_01206 promoter regions was evaluated, this time under relevant in vivo conditions. Notably, we found an enriched association of CodY with the prfA regulatory region during growth in low-BCAA MM in comparison to BHI or LB-MOPS-G1P (both contain excess amounts of BCAA), whereas the ilvD and codV promoter-containing regions were more highly associated with CodY during growth in the rich media than in low-BCAA MM (Figure 3 A-D). Notably, the results demonstrate the opposite effect of BCAA on CodY binding to the different promoters and support the hypothesis that, under conditions of limiting BCAA in vivo, CodY binds the prfA regulatory region to enhance its expression. Intrigued by the observation that the EMSA experiment failed to detect the effect of BCAA on CodY binding to the prfA probe, we hypothesized that this could be due in part to the lack of natural competitor binding sites within this in vitro system. In vivo CodY binds multiple promoters/binding sites with different affinities and it is most likely that the dynamics of CodY binding to weak binding sites, such as the one of prfA, is greatly influenced by the competition between the different binding sites in each given condition (Belitsky & Sonenshein, 2013). To address this question we developed a different in vitro system that is based on a CodY pull-down competition-binding assay. In this set up, both ilvD and prfA probes were incubated with purified His-tagged CodY and competed for its binding under different BCAA concentrations. Briefly, purified CodY-His6 was incubated with the prfA probe and the ilvD probe under increasing concentrations of BCAA (0, 1 and 10 mM). Following incubation, CodY was precipitated using cobalt Talon beads and the amounts of prfA and ilvD probes bound to CodY were quantitated by RT-qPCR using specific primers. The results of this experiment indicated an opposite trend of CodY binding to the different probes. While binding of CodY to the ilvD probe was enhanced when BCAA concentrations were increased, the prfA probe competed more effectively for CodY binding when BCAA concentrations were reduced (Figure 3E). These in vitro results are in accordance with the in vivo data showing that CodY binds prfA to a greater extent when BCAA are limited and support the premise that gene regulation by CodY is influenced by its competitive binding to multiple sites with different affinities under varying BCAA concentrations (Belitsky & Sonenshein, 2013). In line with this model, we next examined whether reducing the affinity of CodY to BCAA (by mutating its BCAA binding site) will favor binding of CodY to PprfA in comparison to PilvD. Since the affinity of CodY to its different binding sites greatly depends on BCAA concentration, such an experiment could further demonstrate the differential effect of BCAA on CodY binding. To this end, we selectively mutated the BCAA-binding site within CodY, and assessed whether such a mutant of CodY could still activate prfA. The BCAA-binding site of CodY was characterized structurally in B. subtilis and was shown to be highly conserved (Levdikov et al., 2006). Moreover, mutations made within this site reduced to varying extents the ability of CodY to respond to isoleucine and thus to repress metabolic genes (Villapakkam et al., 2009). Here we chose to change arginine-61 to alanine (R61A), as this residue is part of the BCAA-binding pocket and was shown to contribute to the isoleucine response (Levdikov et al., 2006, Villapakkam et al., 2009). The CodY-R61A strain together with WT L.m. and ΔcodY bacteria were subjected to growth in the rich medium BHI and in low-BCAA MM and the transcription levels of the ilvD and prfA genes were analyzed using RT-qPCR. The CodY-R61A bacteria grown in BHI accumulated ilvD transcripts at a high level' similar to that seen in ΔcodY bacteria, whereas WT bacteria exhibited much lower expression of this gene (Figure 3F). This result suggested that the CodY-R61A mutant had lost BCAA-dependent repressing activity to a significant extent. On the contrary, a different picture was seen in low-BCAA MM where the CodY-R61A mutant was still able to activate the prfA gene' similarly to WT bacteria, whereas the ΔcodY mutant was not (Figure 3G). These results imply that in high BCAA conditions CodY is preferentially bound to some binding sites (e.g., ilvD) and when BCAA levels drop CodY is still able to bind to other sites (e.g., prfA). These observations accord with the data showing that CodY's affinity to the ilvD probe is 80-fold higher than to prfA probe when BCAA are present, but only 5-fold higher when BCAA are absent (Figure 2). Altogether, these experiments indicated a direct in vitro and in vivo association between CodY and the regulatory region of the prfA gene under conditions that are consistent with those that support CodY-dependent activation of prfA transcription (Figure 1 and (Lobel et al., 2012)).
Figure 2. CodY binds the regulatory regions of prfA gene, ilv operon and the cod operon.
A. Schematic representation of the full-length prfA probe (367-bp) encoding prfA intergenic regulatory promoter region and the first 100 nucleotides of prfA coding sequence. B. Electrophoretic mobility shift assay (EMSA) of CodY binding to prfA probe with (left panel) and without (right panel) 10 mM of BCAA (isoleucine, leucine and valine). C. EMSA analysis of CodY binding to the ilvD probe with and without 10 mM of BCAA. D. EMSA analysis of CodY binding to the codV probe with and without 10 mM of BCAA. Primers used for amplification of DNA probes are described in Table S2. Results are representative of at least 3 independent biological repeats. Non-specific bands are marked with asterisks (*).
Figure 3. CodY preferentially binds the regulatory region of the prfA gene under limiting concentrations of BCAA.
In vivo ChIP-RT-qPCR analysis of 6His-tagged CodY binding to regulatory region of A. prfA gene (PprfA), B. ilvD promoter (PilvD), C. codV promoter (PcodV) and D. LMRG_01206 promoter (PLMRG_01206) as a negative control, during bacterial growth in BHI, low-BCAA MM and LB-MOPS G1P media. Fold enrichment of CodY association with each one of the tested sequences was normalized to the control sequences of bglA and rpoD and to its no-ChIP control. Results are average of 4 independent biological repeats. Error bars represent standard deviation (SD), * represents P value < 0.05. E. In vitro CodY pull-down competition binding assay. Both ilvD and full-length prfA probes were mixed to compete for 6His-tagged CodY binding at increasing concentrations of BCAA. Following incubation CodY was precipitated using cobalt beads and the amounts of prfA and ilvD probes bound to CodY were quantitated by RT-qPCR using specific primers. F. RT-qPCR analysis of ilvD transcription levels in WT L.m., ΔcodY and L.m. CodY R61A bacteria grown to mid-log in BHI rich medium. G. RT-qPCR analysis of prfA transcription levels in WT L. m., ΔcodY and L.m. CodY R61A bacteria grown to mid-log in low-BCAA MM. The RQ values are normalized to ilvD or prfA expression in WT bacteria grown in BHI. Results are average of 3 independent biological repeats. Error bars represent 95% confidence interval, * represents P value < 0.05.
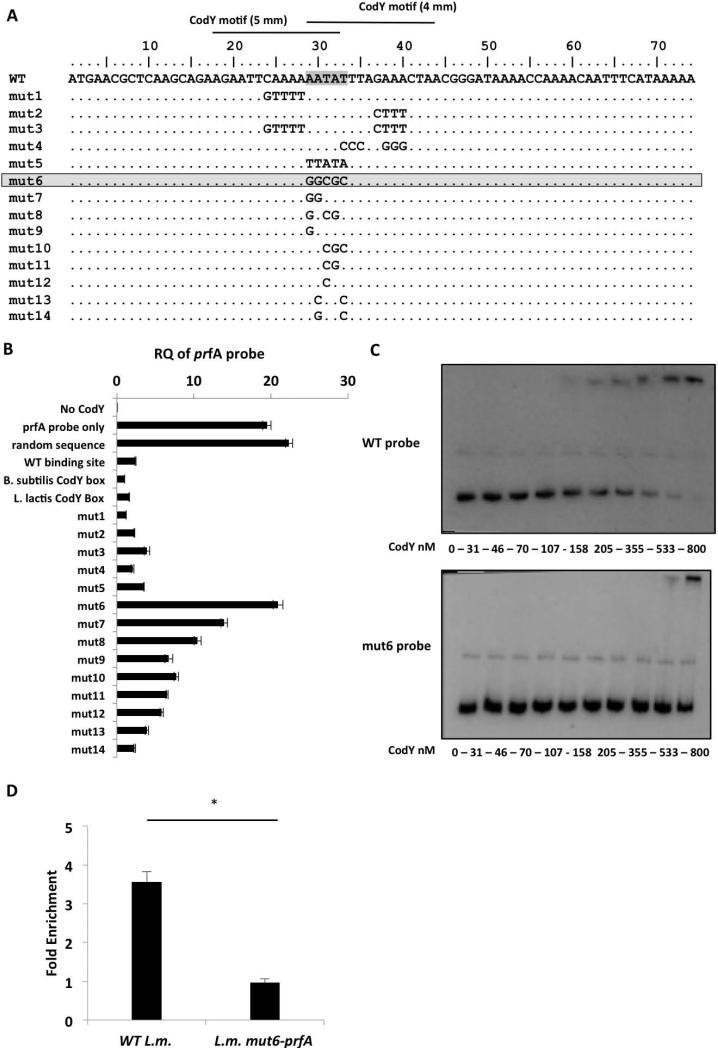
CodY binds a core sequence of AATAT within the coding region of the prfA gene
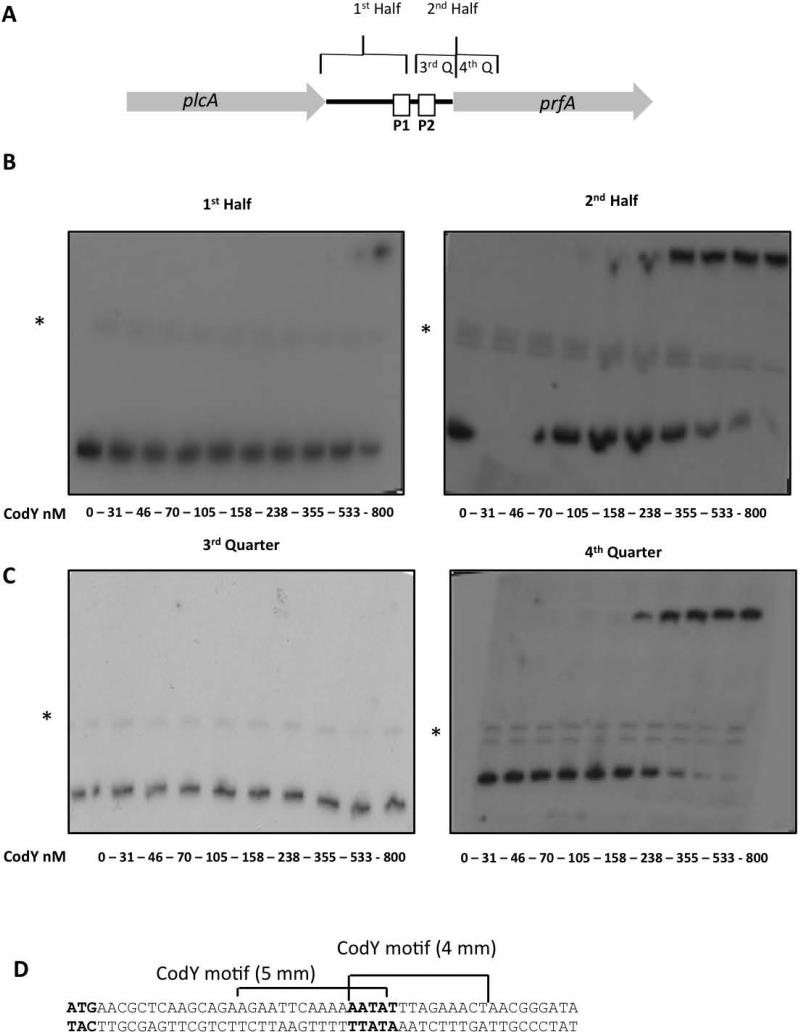
Next, we aimed to determine the exact location at which CodY binds the prfA regulatory region (as designated in the prfA probe, Figure 2A). To this end, a set of new probes was synthesized to include each of the two halves and four quarters of the full-length prfA probe (Figure 4A) and subjected to further EMSA analysis. As shown in Figure 4, CodY demonstrated specific binding to the downstream half of the full-length prfA probe (KD = 216 ± 25 nM, Figure 4B and Figure S1), and primarily to its 3’ end, as indicated by the exclusive binding of CodY to the fourth quarter (KD = 229 ±14.5, Figure 4C and Figure S1). Interestingly, the fourth quarter probe includes the first 100-bp of the prfA coding sequence. These results suggested that CodY might have an intragenic binding site within the prfA coding region and not, as usually found with transcription activators, upstream of the transcription start point. Interestingly, close examination of this 100-bp sequence revealed that it contains two overlapping 15-bp motifs similar to the CodY box consensus in both L. lactis and B. subtilis (each with 4- and 5-bp mismatches) (Belitsky & Sonenshein, 2013, den Hengst et al., 2005), that were located 15-45 bp downstream of the prfA gene ATG start codon (Figure 4D).
Figure 4. Mapping CodY binding site to the 5’ -coding sequence of prfA gene using EMSA.
A. Schematic representation of the prfA probe and its two halves and four quarters segments. B. EMSA analysis of CodY binding to the two halves of the prfA probe. C. EMSA analysis of CodY binding to the 3rd and 4th quarters of prfA probe. The results are representative of at least 3 independent biological repeats. Experiments were performed without BCAA. Non-specific bands are marked with asterisks (*). D. Sequence of the putative CodY boxes identified in the 4th quarter of the prfA probe.
To delineate the physiological relevance of the identified putative CodY-binding site(s) within the prfA gene we first performed a mutational analysis of this region to identify nucleotides critical for CodY binding. A series of double-stranded probes (~50 or 70-bp) containing the putative CodY-binding sites bearing mutations in different nucleotides, as well as the WT putative binding sites and CodY-binding sites found in B. subtilis and L. lactis, were synthesized and assessed for CodY binding under in vitro conditions (Figure 5A and Table S2). CodY binding to the different probes was evaluated using the same competition-binding assay described above, this time using the full-length prfA probe (Figure 2A) and each of the competitor probes. Purified CodY-His6 was incubated with the full-length prfA probe and with each of the short mutated probes in a ratio of 1:10, respectively. Following incubation, CodY was precipitated and the amount of the full-length prfA probe bound to CodY was quantitated by RT-qPCR using primers specific only to this prfA probe (Table S2). In this assay, mutated probes that preserved CodY binding effectively competed with the full-length prfA probe and thus reduced its amount bound to CodY, whereas probes that failed to compete did not alter CodY binding to the prfA probe. First, the binding of CodY to the full-length prfA probe was validated in comparison to a probe containing a random sequence, and, as expected, no competition was observed (the level of the full-length prfA probe bound to CodY was similar to that of the control sample containing the prfA probe alone) (Figure 5B). Conversely, probes containing the consensus L. lactis CodY-binding motif (den Hengst et al., 2005) or a high affinity B. subtilis CodY-binding motif (Belitsky & Sonenshein, 2013) competed efficiently for CodY binding. This latter result was also observed when a probe containing the L. monocytogenes WT prfA putative CodY-binding sites was used (Figure 5B). Next, the ability of the mutated probes to compete with the full-length prfA probe for CodY binding was measured. Notably, we found a 5-nt sequence (AATAT, overlapping the two putative CodY box motifs) to be the most critical for CodY binding, since changing this sequence to GGCGC (as in the probe mut6) completely abolished the ability to compete for CodY binding (Figure 5 A-B). Specific mutations made within these 5 nucleotides only partially affected CodY binding (probes mut7 to mut13), suggesting that together these 5-nt represent a critical sequence that is necessary for the interaction with CodY (Figure 5B). Interestingly, increasing the G-C content of another sequence downstream of the AATAT site (mut4 probe), which overlapped one of the CodY boxes, had no effect on CodY binding, suggesting that CodY might bind upstream of this putative site. Next, an EMSA analysis comparing CodY binding to the WT and to the mut6 binding sites (WT probe and mut6 probe, respectively, in the frame of the 2nd half of the prfA probe) confirmed that CodY binds to the mut6 probe with a much lower affinity, demonstrating a KD of > 800 nM in comparison to a KD of 216 ± 25.5 nM with the WT probe (Figure 5C, and Figure S1). Furthermore, performing EMSA assays using the mut6 prfA probe as a competitor to the WT prfA probe showed that the mut6 probe does not compete with the WT prfA probe for CodY binding (Figure S2). To further examine the importance of the AATAT sequence, CodY binding in vivo was assessed after the mut6 mutation was introduced into the chromosomal prfA gene by allelic exchange (L.m. mut6-prfA mutant). The mutation was introduced into the background of the L.m. codY-6his strain to allow ChIP analysis. The ability of CodY to bind the WT or the mutated binding site sequence within the chromosomal prfA gene was evaluated in vivo during bacterial growth in low-BCAA MM using ChIP-RT-qPCR analysis as described above. The results of this experiment clearly demonstrated that the AATAT sequence is important for CodY binding in vivo, as the L.m. mut6-prfA mutant DNA was not enriched in the CodY-bound fraction whereas the WT sequence was enriched 3.5-fold compared to the control genes (Figure 5D). Taken together, these results establish the existence of a CodY-binding site(s) within the 5’-end of the prfA gene and define the AATAT nucleotides as critical for CodY binding in vitro and in vivo.
Figure 5. Mutational analysis of the CodY-binding region identifies an AATAT sequence that is critical for CodY binding.
A. Representation of the 74-bp probes synthesized to contain different mutations within the putative CodY-binding region (mut-1 to -14). Critical nucleotides for CodY binding are highlighted. B. A CodY pull-down competition binding assay between the 367-bp prfA probe and each one of the 74-bp competitor probes. The assay is based on a DNA pull down assay using a 6His-tagged CodY protein. Analysis of CodY binding to the prfA probe was done using RT-qPCR with primers specific to the full-length prfA probe. Controls include a sample without CodY (No-CodY), a sample without a competitor (prfA probe only), a sample with random sequence probe as competitor and probes containing the WT prfA CodY-binding site, B. subtilis or L. lactis CodY-boxes as competitors. Results are average of 3 independent biological repeats. Error bars represent standard error of the mean. C. EMSA analysis of CodY binding to prfA 2nd half probe (WT probe) and to prfA 2nd half probe containing the mut6 mutation (mut6 probe). Results are representative of 3 independent biological repeats. D. ChIP-RT-qPCR analysis of CodY binding to WT and L.m. mut6-prfA prfA gene during growth in low-BCAA MM. Results are average of at least 3 independent biological repeats. Error bars represent standard deviation (SD), * represents P value < 0.05.
CodY positively regulates PrfA via binding to its identified site within the prfA-coding region
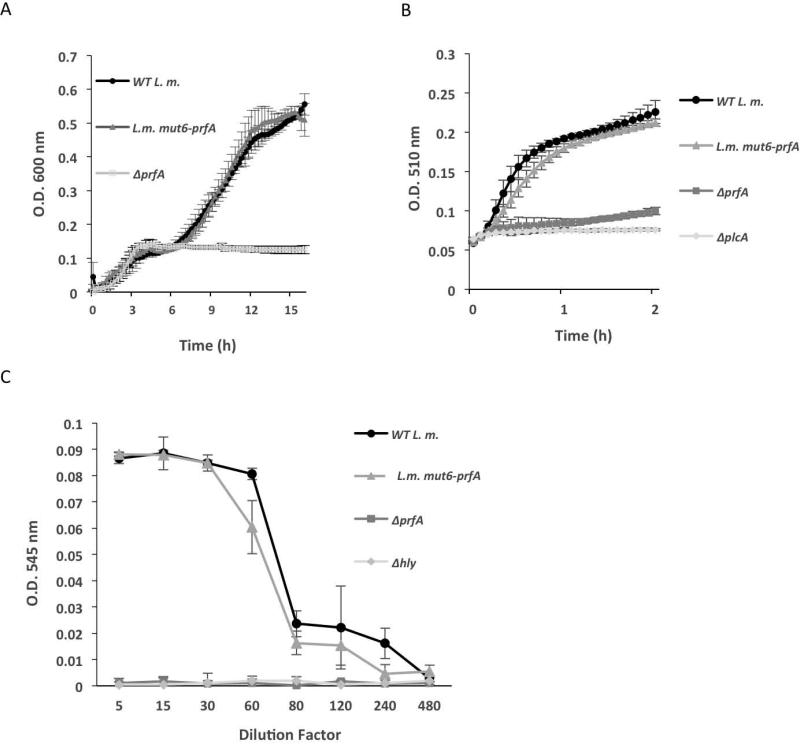
Having identified the sequence critical for CodY to bind to the prfA gene, we investigated the biological importance of this interaction in the regulation of L. monocytogenes virulence. Ideally, we wanted to employ the L.m. mut6-prfA mutant for this purpose. However, as this mutation results in two amino acid substitutions in the PrfA protein (K10R and Y11R), we first questioned whether these mutations impair PrfA structure and activity. To this end, we compared the growth of WT and L.m. mut6-prfA strains in LB-MOPS-G1P medium, which is known to require an active PrfA protein and is independent of CodY (Figure 1B). Of note, PrfA is required to activate the expression of the glucose-1-phosphate transporter, UhpT, which facilitates the uptake and utilization of this sugar (Chico-Calero et al., 2002). As shown in Figure 6A, both strains grew similarly in the LB-MOPS-G1P medium, whereas a ΔprfA mutant was unable to grow. Since during growth in this medium L. monocytogenes expresses and secretes several virulence factors in a PrfA-dependent manner, we next analyzed and compared the activity of two such factors, LLO and PlcA, in the supernatants of WT and L.m. mut6-prfA cultures. The activity of LLO was measured by a red blood cell hemolysis assay, whereas the activity of PlcA was measured by a PI-PLC specific assay, with ΔprfA, Δhly and ΔplcA mutants used as control strains (Rabinovich et al., 2012). The WT and L.m. mut6-prfA strains exhibited similar levels of LLO and PlcA activities in their supernatants, suggesting that both strains express comparable levels of active PrfA proteins in LB-MOPS-G1P medium (Figure 6 B-C). Overall, the results indicate that the prfA-mut6 mutation does not interfere with PrfA protein activity. Another support for this premise came from a 3D-structural comparative analysis between the solved PrfA structure and the predicted structure of the mut6-PrfA protein (based on homology modeling), which demonstrated no change in the structure of the two proteins, including critical domains, such as the DNA binding and dimerization domains (Figure S4).
Figure 6. mut6-prfA is not impaired in PrfA activity.
A. Growth of WT L. m., L.m. mut6-prfA and ΔprfA bacteria in LB-MOPS-G1P medium. B. PlcA activity in the supernatants of WT L.m., L.m. mut6-prfA, ΔprfA and ΔplcA mutants measured using a PI-PLC phospholipase activity assay. Bacteria were grown in LB-MOPS-G1P medium and supernatants were harvested at mid-exponential growth. Results are average of 3 independent biological repeats. Error bars represent standard error of the mean. C. Analysis of LLO activity in the supernatants of WT L.m., L.m. mut6-prfA, ΔprfA and Δhly mutants using hemolysis assay in red blood cells. Bacteria were grown in LB-MOPS-G1P medium and supernatants were harvested at mid-exponential growth.
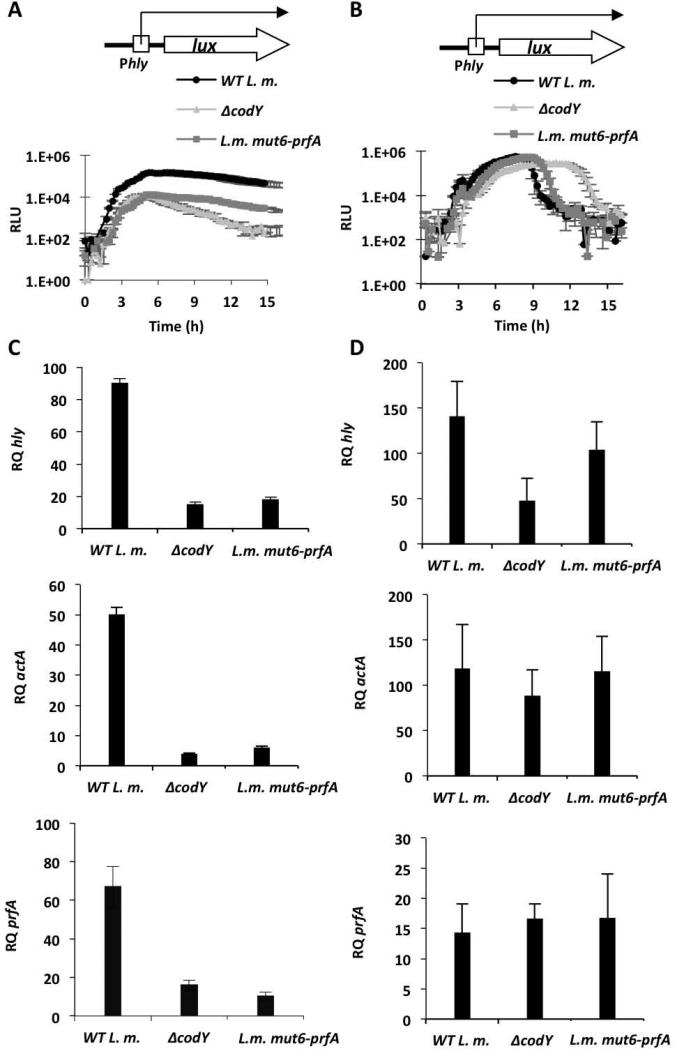
Next, the effect of the mutated CodY-binding site on the regulation of prfA and other virulence genes was tested. The pPL2-Phlylux reporter plasmid was introduced into the WT, ΔcodY and L.m. mut6-prfA strains to monitor hly transcription during growth in low-BCAA MM and LB-MOPS-G1P media. As shown in Figure 7, CodY and the CodY-binding site were both necessary for the induction of hly under conditions of limiting concentrations of BCAA, whereas they were largely dispensable during growth in LB-MOPS-G1P (Figure 7 A-B). The biphasic/delayed luminescence pattern observed with the ΔcodY mutant during growth in LB-MOPS-G1P (also Figure 1B) was not evident in the L.m. mut6-prfA mutant, suggesting that a codY mutation causes this phenotype by a mechanism independent of the AATAT sequence (Figure 7B). Further analysis of the transcription levels of hly, actA and, most importantly, prfA during bacterial growth confirmed that both CodY and its binding site in prfA are necessary to activate the transcription of prfA and the virulence genes in low BCAA medium, but not during growth in LB-MOPS-G1P medium (Figure 7 C-D). Based on these data we conclude that CodY specifically regulates the transcription of prfA and its downstream virulence genes via direct binding to the identified site located within the prfA gene.
Figure 7. CodY and its prfA-intragenic binding site are important for prfA activation at low concentrations of BCAA.
A. Luminescence of WT L. m., ΔcodY and L.m. mut6-prfA bacteria harboring the integrative reporter plasmid pPL2-Phlylux during growth in low-BCAA MM. B. Luminescence of WT L. m., ΔcodY and L.m. mut6-prfA bacteria harboring the integrative reporter plasmid pPL2-Phlylux during growth in LB-MOPS-G1P medium. Error bars represent standard deviation. C. RT-qPCR analysis of hly, actA and prfA genes in WT L. m., ΔcodY and L.m. mut6-prfA bacteria grown in low-BCAA MM. D. RT-qPCR analysis of hly, actA and prfA genes in WT L. m., ΔcodY and L.m. mut6-prfA bacteria grown in LB-MOPS-G1P medium. The results represent 3 independent biological repeats. Transcription levels are represented as relative quantity (RQ), relative to the transcription levels during growth in BHI. Error bars represent 95% confidence interval.
Both CodY and its binding site within the prfA gene are important to promote virulence in macrophage cells and in mice
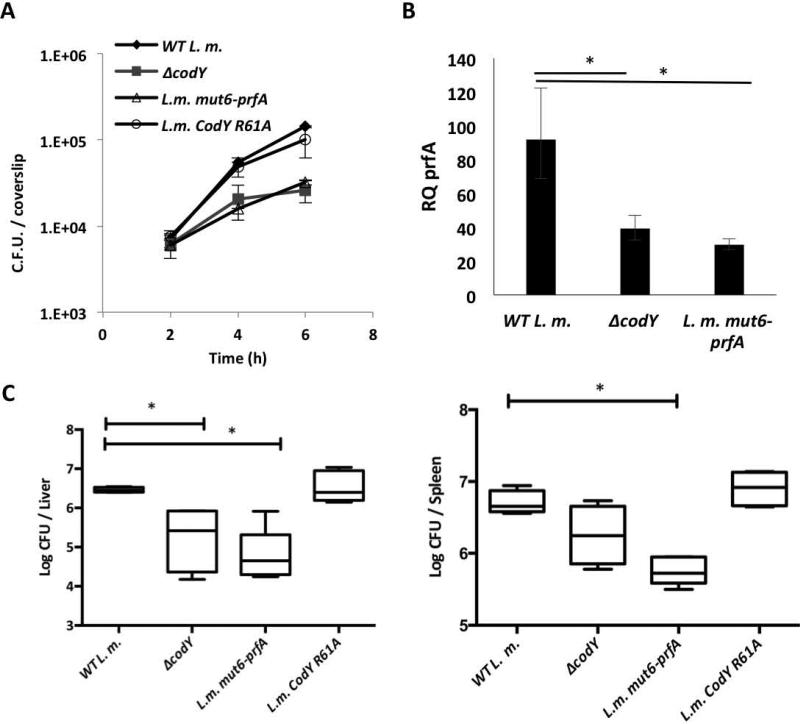
Lastly, the roles of CodY and its binding site were evaluated during infection of macrophage cells and mice. First, the intracellular growth ability of the WT, ΔcodY, L.m. mut6-prfA and L.m. CodY R61A strains was monitored in bone marrow-derived (BMD) macrophages. Notably, both ΔcodY and L.m. mut6-prfA mutants exhibited a similar intracellular growth defect in comparison to WT bacteria and to bacteria harboring the codY R61A mutation (Figure 8A). These results further indicated that while both CodY and its DNA binding site are important for intracellular growth, efficient binding of BCAA may not be required. Transcription analysis of prfA gene during intracellular growth of ΔcodY and L.m. mut6-prfA mutants confirmed that both CodY and its binding site play a role in its activation during infection (Figure 8B). Next, the fitness of WT, ΔcodY and L.m. mut6-prfA mutant strains was evaluated in young C57BL/6 female mice. Mice were injected intravenously with 4×104 of WT, ΔcodY, L.m. CodY R61A or L.m. mut6-prfA bacteria (5 mice in each group) and bacterial counts in the spleens and livers of the infected mice were analyzed at 72 hours post-infection. As shown in Figure 8C, both ΔcodY and L.m. mut6-prfA mutants colonized the livers and spleens of the infected mice to a lesser extent than did WT and L.m, CodY R61A bacteria, exhibiting 5-10 fold decreases in colony-forming units (CFUs) recovered from both organs. These results clearly indicate that both CodY and its binding site within the prfA gene contribute to L. monocytogenes virulence.
Figure 8. CodY and its prfA-intragenic binding site play a major role in L. monocytogenes virulence.
A. Intracellular growth of WT L. m., ΔcodY, L.m. mut6-prfA and L.m. CodY R61A bacteria in BMD macrophages. Results are representative of 3 independent biological repeats. Error bars represent the standard error of the mean. B. RT-qPCR analysis of prfA during intracellular growth of WT L. m., ΔcodY and L.m. mut6-prfA bacteria in bone marrow derived (BMD) macrophages. Results are average of 3 independent biological repeats. Transcription levels are represented as relative quantity (RQ), relative to levels in WT L.m bacteria grown to mid-log in BHI rich medium. Error bars represent 95% confidence interval, * represents P value < 0.05. C. Intravenous infection of C57BL/6 mice with WT L. m., ΔcodY, L.m. mut6-prfA and L. m. CodY R61A bacteria. Bacterial colony forming units (CFUs) were numerated at 72 h.p.i. from livers and spleens taken from 10 infected mice for each strain. The results are mean of 2 independent experiments in which 5 mice were infected in each group. Mann-Whitney tests were performed for statistical significance (* P value < 0.05).
Discussion
Bacterial pathogens have co-evolved physical and metabolic adaptations in order to invade and grow successfully within mammalian cells, but how and at what level these seemingly different adaptations are intertwined has remained elusive. This report, together with our previous studies, reveals an interesting case in which host cell metabolism plays an important role in activation of bacterial virulence. We have previously shown that upon L. monocytogenes invasion of macrophage cells, certain pathways (e.g., those for biosynthesis of purines, histidine, arginine and BCAA) are activated in the bacteria in order to cope with the low availability of nutrients within the mammalian cell's cytosol (Lobel et al., 2012). Among these pathways we found that the BCAA biosynthesis appears to be the main metabolic pathway interlinked with L. monocytogenes virulence. Low concentrations of BCAA were found to trigger enhanced induction of PrfA and the virulence genes it controls in vitro, suggesting that L. monocytogenes might recognize BCAA deficiency as a signature of the mammalian nutritional environment (Lobel et al., 2012).
In the present study we discovered a direct link between the BCAA-sensing regulator CodY and the expression of PrfA. We found that CodY binds within the 5’ end of the prfA coding sequence in vitro and that binding in bacterial cells occurs preferentially under conditions of limiting concentrations of BCAA. A five-bp sequence within the CodY-binding site proved to be necessary for CodY-dependent activation of prfA transcription, implying that binding of CodY to that site is important for virulence gene expression in vitro and in vivo, and for colonization of liver and spleen. Furthermore, we found that activation of prfA transcription by CodY is less dependent on BCAA binding than is CodY-dependent repression of ilvD. In summary, these data demonstrate that CodY plays a direct role in prfA activation and affects virulence specifically when BCAA are in limiting amounts. As discussed in more detail below, this is the first report indicating that CodY activity as a regulatory protein correlates with a drop in BCAA concentration in vivo and that CodY can activate transcription by binding within a coding sequence.
While in most cases analyzed to date CodY has been shown to function as a repressor whose activity is increased when BCAA availability is high, there are several examples of positive regulation by CodY, again when BCAA are available (Shivers & Sonenshein, 2004). Many transcription factors (the classic example is AraC) are known to function as both activators and repressors (van Hijum et al., 2009) and, in some cases, to demonstrate both activities at the same target gene when the regulator is in its effector-bound or effector-unbound states (Schleif, 2010). Other regulatory proteins bind to different targets in the effector-bound (holo) (Wunsche et al.) and – unbound (apo) states (Balderas-Martinez et al., 2013). For example, Lrp (a regulatory protein in Gram-negative bacteria that controls both metabolic and virulence genes) and the Fur regulators have been shown to be active in both their holo and apo forms (Carpenter et al., 2009, Butcher et al., 2012, Baek et al., 2011, Newman & Lin, 1995, van der Woude et al., 1995, Cho et al., 2008). In fact, Lrp was shown to repress genes in its holo form, while activating others in the apo form (Chen et al., 2005). Such examples raise the possibility that CodY may also have more complex modes of action than presently known and may be more versatile than anticipated. Since binding of BCAA to CodY induces a significant conformational change that appears to separate the C-terminal winged helix-turn-helix motifs of the dimeric protein (Levdikov et al., 2006, Levdikov et al., 2009), thereby facilitating binding to DNA, we did not expect that limiting the availability of BCAA would enable CodY to bind DNA. Nevertheless, the observation that a CodY protein that is mutated within its BCAA-binding site loses its ability to repress ilvD but retains ability to activate prfA raises the possibility that CodY might possess an additional functional conformation when BCAA are limited.
A second unexpected conclusion of this study is that CodY activates prfA transcription by binding to a site within the coding region. Gene activation typically occurs via binding of transcription factors to sequences upstream of the RNA polymerase binding site, enabling the factors to interact directly with RNA polymerase without blocking the progress of the transcription machinery (van Hijum et al., 2009). Nevertheless, intragenic binding sites can also play a role in activation of transcription, as opposed to a more conventional role in repression (Munson & Scott, 2000, Qi & Hulett, 1998, Munson et al., 2001, Mitra et al., 2005, Ouyang et al., 2011, Liu et al., 1998, Gal-Mor et al., 2011, Shi et al., 2004, Feng et al., 2003). For example, B. subtilis PhoP, which regulates the response to phosphate starvation, activates genes by binding to sites internal to their coding sequences thereby enhancing full promoter activity (Liu et al., 1998). In E. coli, the AraC-family Rns virulence activator promotes its own transcription by binding to two intragenic sites (Munson & Scott, 2000). Binding to these internal sites leads to a stronger interaction of RNA polymerase with the promoter region and the formation of an open transcription initiation complex. ChIP-Seq analyses of other bacterial global regulators have demonstrated that the binding of transcription activators is not restricted to upstream sequences but can involve binding within coding regions (Wang et al., 2013, Park et al., 2013, Butcher et al., 2012, Butcher et al., 2011, Cho et al., 2008, Munson & Scott, 2000, Martin & Rosner, 2001). Interestingly, a recent genome-wide analysis of CodY-binding sites in B. subtilis revealed that 47% of the total sites identified were within coding regions. Some of these sites have been shown to mediate negative regulation (Belitsky & Sonenshein, 2013); though others may be involved in gene activation. It is important to note, however, that although the studies reported here support the idea that binding of CodY within a coding sequence can lead to gene activation, the exact molecular mechanism underlying this positive regulation remains to be delineated.
In this regard, it is interesting that the 5’-UTR of prfA mRNA is highly dynamic, responding to different metabolic and environmental cues. The RNA thermosensing structure was shown to overlap with a region that interacts with two non-coding RNAs that are themselves SAM-responsive riboswitches (Loh et al., 2009). One could imagine that CodY binding to a site just downstream of the 5’-UTR region might slow down the RNA polymerase leading to stabilization of upstream RNA structures (of the 5’-transcribed UTR) that facilitate continuation of transcription rather than termination. In such a case, the relatively weak binding of CodY to the intragenic site may still allow the RNA polymerase to proceed with elongation of transcription. In that way, CodY binding to prfA might be strong enough to permit interaction with RNA polymerase or the 5’-UTR, but not so strong as to interfere with transcription elongation. Notably, similar mechanisms were proposed for PhoP and other regulators and were suggested to represent a new mode of regulation by activators (Liu et al., 1998). Alternative mechanisms are equally possible, such as those involving DNA-looping or cooperative interaction with additional regulators, as already shown for CodY and CcpA (Wunsche et al., 2012, Shivers et al., 2006), and/or adjacent binding sites (van Hijum et al., 2009).
The ability of CodY to bind to the relatively weak prfA site under conditions of limiting BCAA raises important mechanistic questions. Binding of CodY to many B. subtilis sites is highly cooperative (Belitsky & Sonenshein, 2013). If the same is true of L. monocytogenes CodY binding sites, but the prfA site demonstrates non-cooperative binding, the prfA site would compete well with other sites at low BCAA levels, i.e., when the concentration of ligand-bound CodY is below the threshold for cooperativity. At concentrations above the threshold, binding to prfA would be relatively poor, i.e., cooperative binding sites would compete much more effectively for CodY, explaining why prfA expression would be activated by CodY only at limiting BCAA concentrations. Our data demonstrating that the prfA site competes better for CodY binding when BCAA are limited are in line with this premise.
Overall the data presented here support a model whereby L. monocytogenes exploits a simple yet elegant coupling mechanism to sense a host-specific metabolic cue and induce virulence genes. Namely, a global regulator that functions predominantly as a repressor (when bound to BCAA) is recruited upon a shift in conditions (i.e., isoleucine limitation) to function as a prfA activator. Using this mechanism, L. monocytogenes can respond to a drop in BCAA availability (as happens upon invasion of host cells) by releasing repression of many metabolic pathways, including BCAA biosynthesis, that facilitate growth and survival. In addition, ligand-limited CodY appears to have the ability to activate transcription of prfA, leading to induction of the major Listeria virulence genes. Interestingly, if Listeria had evolved to use CodY in what seems to be the conventional way (i.e., as a BCAA-dependent repressor of prfA), invasion of the host would still lead to PrfA synthesis when BCAA are limiting, but strong repression by CodY in high BCAA could interfere with other, CodY-independent mechanisms of prfA regulation, such as in response to G-1-P. Importantly, this study highlights the regulation of PrfA by BCAA and CodY, though it represents only one signaling pathway out of many that have been shown to affect PrfA expression. How these distinct pathways are integrated into the overall scheme of PrfA regulation in vivo is not completely clear and will no doubt be the topic of future studies.
Experimental Procedures
Ethics Statement
Experimental protocols were approved by the Tel Aviv university Animal Care and Use Committee (L-09-008) according to the Israel Welfare Law (1994) and the National Research Council guide (Guide for the Care and Use of Laboratory Animals 2010).
Bacterial strains and growth media
L. monocytogenes 10403S was used as the wild type strain (WT) and served as the parental strain to generate allelic exchange mutant strains (Table S1). E. coli XL-1 Blue (Stratagene®) was used for vector propagation and E. coli strain SM-10 was used for transfer of plasmids to L. monocytogenes by conjugation. L. monocytogenes strains were grown in brain heart infusion (BHI, Merck®) rich medium, or in LB-MOPS medium that was pre-incubated with 0.2% activated charcoal and supplemented with either 25 mM of glucose (Glu) or glucose-1-phosphate (G1P) (Ripio et al., 1997a) or in minimal defined medium (MM) (Phan-Thanh & Gormon, 1997) at 37°C with agitation. MM contains 100 μg ml−1 of each of the BCAAs, which are 760 μM for leucine and isoleucine and 850 μM for valine. For growth under limiting concentrations of nutrients, MM was freshly made with 10-fold less of the BCAA [isoleucine, leucine and valine] (10 μg ml−1 for each amino acid, which is 76 μM for leucine and isoleucine and 85 μM for valine.) and termed low-BCAA MM. Lists of bacterial strains and plasmids used in this study are provided in Table S1.
In vitro growth of L. monocytogenes in different laboratory media
Bacteria from overnight cultures grown in BHI medium were adjusted to O.D.600 0.03 in fresh BHI medium or low/high -BCAA MM (10 μg and 100 μg of each BCAA per ml, respectively) or LB-MOPS-G1P or Glu (25mM) activated charcoal medium and grown in a Synergy HT Biotek® plate reader at 37°C for 24 h. O.D. measurements were made every 15 min. For luminescence assays, L. monocytogenes strains harboring the Phly-luciferase reporter system (pPL2-Phlylux) were used and luminescence measurements, presented as relative luminescence units (RLU, relative to the blank medium luminescence), were taken every 15 min.
Generation of L.m. codY-6his, L.m. mut6-prfA and L.m. CodY R61A strains
Upstream and downstream regions of the codY and prfA genes were amplified using Phusion DNA polymerase (NEB®) and specific primers containing the 6His-tag or the mut6 mutation. The PCR products were cloned in pKSV7oriT vector (Smith & Youngman, 1992) (A list of all primers used in this study is provided in Table S2). Cloned plasmids were sequenced and conjugated to L. monocytogenes using E. coli SM-10 strain (Simon R, 1983). L. monocytogenes conjugants were then grown at 41°C for two days on BHI with chloramphenicol to promote plasmid integration into the bacterial chromosome by homologous recombination. For plasmid curing, bacteria were passaged several times in fresh BHI without chloramphenicol at 30°C to allow plasmid excision via the generation of an allelic exchange. Bacteria were then seeded on BHI plates and chloramphenicol sensitive colonies picked for validation of allelic exchange by PCR, enzyme restriction analysis and sequencing.
Purification of CodY-His6
L. monocytogenes CodY-6His was expressed in E. coli strain BL-21 from the pET28 expression plasmid. 10 ml of overnight bacterial culture were diluted in 0.5 L of LB medium supplemented with 30 μg ml−1 of kanamycin. Bacteria were grown till O.D.600 0.3, and then induced with 1 mM IPTG for 4 h. The bacteria were then harvested by centrifugation (4000 rpm, 10 min), washed in 50 ml of cold Buffer A (0.3M NaCl, 50 mM NaH2PO4, pH 8) and resuspended in 15 ml of buffer A with 10 mM imidazole and 1 mM PMSF. Bacteria were lysed by an Ultra high-pressure homogenizer (Stansted Fluid Power®) at 12000 psi. Cell debris was removed by centrifugation at 16,000 g for 20 min and the lysate was incubated with 1ml of Ni-NTA beads (Sigma ®) for 1h at 4°C with tilting. The Ni-NTA beads were then loaded on a column and washed with 10 ml wash Buffer A supplemented with 10 mM imidazole. The protein was eluted by 250 mM imidazole in Buffer A and dialyzed against 100 ml of Buffer A. Protein concentration was determined using a Nanodrop 1000 (Thermo®) spectrophotometer. A small sample was separated on SDS-PAGE gel followed by Commassie staining to test for the purity of the protein. For DNA footprinting analysis a slightly different protocol for CodY purification was used. L. monocytogenes CodY was purified as described previously for B. subtilis CodY (Belitsky & Sonenshein, 2008) using pET24b plasmid (pET24b-codY) (Bennett et al., 2007) and codY expression was induced using 0.2 mM IPTG.
Electrophoretic Mobility Shift Assay
Purified CodY-6His was incubated with 5 ng of target DNA in Binding Buffer (20 mM Tris-Cl pH 8, 50 mM KCl, 2 mM MgCl2, 0.5 mM EDTA, 1mM DTT, 0.05% NP-40, 5% glycerol, 25 μg ml−1 salmon sperm DNA) for 15 min at room temperature. The samples were then loaded onto a pre-run 8% native acrylamide gel and separated for 1.5 h at 200 V. The DNA was transferred to a positively charged nylon membrane (Pall®) by overnight electrophoresis (15V) in TBE buffer. Following DNA strand separation in 0.4 M NaOH, crosslinking was performed by UV exposure. The membrane was probed with a radiolabeled DNA generated using NEBlot® kit (New England BioLabs) for 16 h, and then washed 3 times with SSC X 2 + 0.2% SDS, SSC X 0.2 + 0.2% SDS and SSC X 2. Finally the membrane was visualized by a radioactive sensitive phosphor-screen (Sigma®), emitting light on a light sensitive film (FUJI®). EMSA assay used for competition experiments was performed as detailed above, with the exception of labeling the DNA probes with DIG using Roche DIG Gel Shift kit®, adding 10X of competitor unlabeled DNA probe. Detection of labeled DNA was done using Roche DIG detection kit®. For calculations of average KD values, the fraction of the free DNA probe at each CodY concentration was quantitated by densitometric analysis using ImageJ software for each EMSA gel (Schneider et al., 2012). Regression analysis was performed for each gel and the Kd value was calculated. Average Kd values were based on 2-3 regression analyses of each probe. To demonstrate the reproducibility of the EMSA gels, averaged quantifications of 2-3 biological repeats of each probe were fit using exponential least-squares regression analysis (shown in figure S2). The Kd values were determined as the concentration in which 50% of the DNA probes were unbound as deduced from the regression analysis. List of primers used for the amplification of target DNA sequences/probes is found in Table S2.
ChIP RT-qPCR analysis
The chromatin immunoprecipitation (ChIP) protocol was adapted from ref (Waldminghaus & Skarstad, 2010). Briefly, L. monocytogenes codY-6His strain was grown in BHI or low-BCAA MM to O.D.600 0.3 or in LB-MOPS-G1P media to O.D.600 0.6. Then 1.5% of formaldehyde was added for 20 min at room temperature, followed by quenching with 0.5 M glycine for 5 min. The cells were harvested by centrifugation, and washed twice with cold TBS. At this point, the cells were frozen in liquid N2 and kept in -80°C. Following thawing, the cells were resuspended in 200 μl of resuspension buffer (20% sucrose, 10 mM TRIS pH 8, 50 mM NaCl, 10 mM EDTA with freshly added 10 mg ml−1 lysozyme and 2 U of Mutanolysin, Sigma). The samples were incubated at 37°C for 30 min and then 800 μl of IP buffer (50 mM HEPES-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X 100, 0.1 % sodium deoxycholate and 0.1 % SDS) and 1 mM of PMSF were added. The samples were then sonicated (6 rounds of 30 sec) and cell debris was removed by centrifugation at 14,000 g for 10 min at 4°C. The lysate was then transferred to a new 1.7 ml tube and 20 μl of 50% slurry protein A/G (Sigma) and 5 μl of anti-6His antibody (ABCAM® ab-18184) were added and incubated overnight with slow rotation at 4°C. Beads were collected by centrifugation (1 min, 3500 g) and the supernatant, which contains unbound DNA, was transferred to a new 1.7 ml tube to serve as control DNA for further analysis. The beads were washed twice with 500 μl cold IP buffer, once with 500 μl of cold high salt IP buffer (0.5 M NaCl), once with 500 μl cold wash buffer (10mM Tris pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate) and once with 500 μl of cold TE pH 8 buffer. Finally the beads were resuspended in 100 μl of elution buffer (50 mM Tris pH 8, 10 mM EDTA and 1% SDS) and incubated at 65°C for 10 min. Beads were removed by 1 min centrifugation. 100 μl of ChIP DNA or control DNA were supplemented with 80 μl of TE buffer with 2.5 μl RNase (8 mg ml−1, Fermentas®) and incubated at 42°C for 1.5 h. DNA was released by incubation with 20 μl of proteinase K (Fermentas®) for 2 h at 42°C, following 16 h at 65°C. DNA was purified using QIAGEN MinElute kit® and eluted in 10 μl water. RT-qPCR analysis was performed on 1–10 ng of ChIP and control DNA using a Step One Plus Real Time PCR system (Applied Biosystems®). DNA levels were normalized to two house keeping genes: bglA and rpoD using Step One™ software. The enrichment value of each ChIP sample was calculated based on its cognate DNA control sample.
In vitro CodY pull-down competition binding assay
The assay is based on a DNA pull down protocol that was modified from (Dineen et al., 2010). For the screen of CodY binding site mutants (figure 6)' 300 nM purified CodY-His6 were incubated with 7nM of prfA probe in 250 μl pull down buffer (20 mM TRIS pH 8, 50 mM sodium glutamate, 5% glycerol, 10mM MgCl2, 0.05% NP-40, 25 μg ml−1 E. coli tRNA) with or without ~400nM of competitor DNA probe for 25 min at room temperature. For the prfA and ilvD probes competition assay (figure 5D), 200 nM purified CodY-His6 were incubated with 30 nM ilvD probe and 300 nM prfA probe in 250 μl pull down buffer supplemented with 0, 1 or 10 mM of BCAA for 25 min at room temperature. Following incubation, 50 μl of TALON® cobalt affinity beads (pre-washed with the same buffer) were added for 20 min with an occasional tilting. Beads were washed 6 times in pull down buffer (if BCAA were present in the binding reaction, the same concentration was included in these washes), centrifuged at 1000 g for 2 min at each wash and finally resuspended in 100 μl of 10 mM TRIS, pH 8 buffer. DNA was released by boiling for 10 min followed by incubation with 1 μl of Proteinase K (Fermentas®) for 1 h at 65°C. Proteinase K was inactivated by boiling for 10 min and beads were removed by centrifugation. The resulting supernatant was diluted 1:10 in DNase free water and analyzed by RT-qPCR analysis using a Step One Plus Real Time PCR system (Applied Biosystems®) with primers specific to the indicated probes.
Analysis of LLO and PlcA activity
L. monocytogenes bacteria were grown in LB-MOPS-G1P medium at 37°C overnight (12 h), and supernatants were separated by centrifugation. The hemolytic activity assay was performed as described previously (Glomski et al., 2002): bacterial supernatants were treated with 5 mM dithiothreitol (DTT), serially diluted in PBS, and incubated with 0.5% sheep red blood cell suspension (NovaMed®); hemolysis was measured by following the change in absorbance at 540 nm. The PI-PLC activity assay was adapted from Geoffroy et al. (Geoffroy et al., 1991): 1 ml of sodium-cholate (58 mM), CaCl2 (10 mM), and 0.036 g phosphatidyl-inositol (P6636; Sigma®) were mixed with 7 ml NaCl (0.15M). One hundred μl of the assay solution was then mixed with 100 μl of bacterial supernatants and incubated in a plate reader at 37°C for 10 h, following turbidity assessment at 510 nm.
Bacterial RNA purification
RNA was harvested from bacteria grown to mid-log phase (O.D.600 = 0.35) in low-BCAA MM. Precultures were grown in MM overnight prior to the experiments. In LB-MOPS-G1P experiments, RNA was harvested from bacteria at post log-phase (O.D.600 ~ 0.7). RNA was extracted by a standard phenol-chloroform extraction protocol including a DNase I treatment. RNA from bacteria growing inside macrophages at 2 hours post infection was harvested as described previously (Rabinovich et al., 2012). Briefly, bacteria were harvested by filtration and the filters were frozen rapidly in liquid nitrogen. Later, bacteria were released from filters by washing and bacterial RNA was isolated using phenol-chloroform extraction. Bacterial RNA was further amplified using MessageAmpTM II Bacteria Prokaryotic RNA Kit (Ambion®).
Real time quantitative PCR analysis of gene expression
One microgram (1μg) of RNA was reverse transcribed to cDNA using the QScript reverse transcription kit (Roche®). RT-qPCR was performed on 16 ng of cDNA using SYBER Green (Roche®) in a Step-one Plus real time PCR system (Applied Biosystems®). The transcription level of each gene of interest was normalized to that of a reference gene: bglA in the intracellular experiments and rpoD mRNA in the low-BCAA MM and LB-MOPS-G-1-P medium experiments. Statistical analysis was performed using the StepOne™ V2.3 software. RT-qPCR primers are described in Table S2.
Intracellular growth of L. monocytogenes
Bone marrow-derived macrophages (BMDM) used for infection experiments were isolated from 6-8 week old female C57/BL6 mice (Harlan laboratories) as described previously (Celada et al., 1984). BMDM were cultured in DMEM-based media supplemented with 20% fetal bovine serum, sodium pyruvate (1 mM), L-glutamine (2 mM), β-Mercaptoethanol (0.05 mM), and M-CSF (L929-conditioned medium). Approximately 8×106 L. monocytogenes bacteria were used to infect 2×106 macrophage cells seeded in a 60 mm Petri dish, resulting in 1-2 bacteria per cell. Thirty minutes after infection, macrophage monolayers were washed three times with PBS and fresh medium added. At 1-hour post infection (h.p.i.) gentamicin (50 μg ml−1) was added to limit bacterial extracellular growth. Intracellular growth was evaluated as follows. Macrophages were seeded on 13 glass cover slips in a 60 mm plate. At each time point three cover slips were removed and transferred to 2 ml of sterile water, which released intracellular bacteria. Then serial dilutions of this 2 ml were plated on BHI plates and colony-forming units (CFUs) counted the next day.
In vivo mice infections
L. monocytogenes bacteria were grown in BHI medium at 30°C overnight. Bacterial cultures were washed twice in Ringer's lactate solution and counted (~ 2×109 bacteria per ml). C57BL/6 (6-8 weeks old) female mice (Harlan Laboratories, Ltd, Israel) were infected via tail vein injections with 4×104 bacteria in 200 μl of PBS. Animals were observed daily for any signs of illnesses and were euthanized 72 hours post-infection. Spleens and livers were harvested and homogenized in 0.2% saponin, and the numbers of viable bacteria in each organ were determined by plating serial dilutions of homogenates onto BHI agar plates. The experiment involved 5 mice in each group and was repeated twice, yielding similar results.
Supplementary Material
Figure S1. Densitometry analysis of EMSA gels of prfA, ilvD and codV probes in the presence or absence of 10 mM BCAA. Averaged regression analyses for each probe. Analysis based on densitometry measurements of the free DNA probe at each CodY concentration using ImageJ software (Schneider et al., 2012). The lower left panel shows the analysis for ilvD probe in the presence of 10 mM BCAA at 4-32 nM concentrations of CodY: the representative EMSA analysis (right) and the quantification analysis (left). Quantifications of 2-3 biological repeats were fitted via exponential least-squares regression analysis. The average KD values depicted in the manuscript are based on 2-3 regression analyses made for each probe and are not derived from the averaged graphs presented here. Error bars represent standard error of the mean.
Figure S2. Specific competition EMSA assays of labeled and unlabeled prfA, ilvD and codV probes. Electrophoretic mobility shift assay (EMSA) of CodY binding to the prfA (A), ilvD probe (B) and codV (C) probes with and without specific unlabeled competitors. For prfA probe competition, the prfA-mut6 probe was also used as a specific competitor. The EMSAs were performed without BCAA and the concentration of CodY used for each probe is equivalent to the Kd50 of CodY for each probe (Figure S1). Primers used for amplification of DNA probes are described in Table S2. The results represent two independent biological repeats (N=2).
Figure S3. The 6His-CodY protein is fully active. A. Growth analysis of WT, ΔcodY and L.m. CodY-6his strains in BHI medium. Results are average of 3 biological repeats (N=3). Error bars represent standard deviation. B. RT-qPCR analysis of ilvD transcription in WT, ΔcodY and L.m. CodY-6his strains grown in BHI medium. Results are average of 3 biological repeats (N=3). Error bars represent 95% confidence interval.
Figure S4. Predicted structure of mut6-PrfA protein. The dimer structure of mut6-PrfA based on the solved WT PrfA structure obtained from the Protein Database Bank (PDB, accession number 2BEO) was predicted using the SWISS-MODEL homology modeling server (http://swissmodel.expasy.org/). The mut6-PrfA dimer structure (yellow) was aligned to the WT PrfA structure (blue) using the PyMol software. Based on this analysis the Root Mean Square deviation between the alpha carbons of the structures is 0.222 Å.
Acknowledgments
We thank Sivan Friedman for her help with experiments. We thank Ran Nir-Paz for his help with mice infection experiments. We are grateful to Colin Hill for giving us the pPL2-Phlylux plasmid. This work was supported by the Israel Science Foundation and the ERA-net pathogenomics (funded by MOH Israel) grants to AAH and in part by a research grant (R01GM042219) from the US National Institute of General Medical Sciences to ALS. LL is funded by the Friends of Tel Aviv University in Argentina and the Constantiner Institute for Molecular Genetics.
References
- Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: ‘Nutritional virulence’ as an emerging paradigm. Cellular Microbiology. 2013;15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- Baek CH, Kang HY, Roland KL, Curtiss R., 3rd Lrp acts as both a positive and negative regulator for type 1 fimbriae production in Salmonella enterica serovar Typhimurium. PloS one. 2011;6:e26896. doi: 10.1371/journal.pone.0026896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderas-Martinez YI, Savageau M, Salgado H, Perez-Rueda E, Morett E, Collado-Vides J. Transcription factors in Escherichia coli prefer the holo conformation. PloS one. 2013;8:e65723. doi: 10.1371/journal.pone.0065723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. Journal of bacteriology. 2008;190:1224–1236. doi: 10.1128/JB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63:1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- Böckmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Molecular microbiology. 1996;22:643–653. doi: 10.1046/j.1365-2958.1996.d01-1722.x. [DOI] [PubMed] [Google Scholar]
- Bretl DJ, Demetriadou C, Zahrt TC. Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiology and molecular biology reviews : MMBR. 2011;75:566–582. doi: 10.1128/MMBR.05004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron PA, Monk IR, Corr SC, Hill C, Gahan CG. Novel luciferase reporter system for in vitro and organ-specific monitoring of differential gene expression in Listeria monocytogenes. Appl Environ Microbiol. 2006;72:2876–2884. doi: 10.1128/AEM.72.4.2876-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Palmer KL, Whiteley M. Revisiting the host as a growth medium. Nature Reviews Microbiology. 2008;6:657–666. doi: 10.1038/nrmicro1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher BG, Bronstein PA, Myers CR, Stodghill PV, Bolton JJ, Markel EJ, Filiatrault MJ, Swingle B, Gaballa A, Helmann JD, Schneider DJ, Cartinhour SW. Characterization of the Fur regulon in Pseudomonas syringae pv. tomato DC3000. Journal of bacteriology. 2011;193:4598–4611. doi: 10.1128/JB.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BM, Whitmire JM, Merrell DS. This is not your mother's repressor: the complex role of fur in pathogenesis. Infection and immunity. 2009;77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celada A, Gray PW, Rinderknecht E, Schreiber RD. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. The Journal of experimental medicine. 1984;160:55–74. doi: 10.1084/jem.160.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateau A, van Schaik W, Six A, Aucher W, Fouet A. CodY regulation is required for full virulence and heme iron acquisition in Bacillus anthracis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:4445–4456. doi: 10.1096/fj.11-188912. [DOI] [PubMed] [Google Scholar]
- Chen S, Iannolo M, Calvo JM. Cooperative binding of the leucineresponsive regulatory protein (Lrp) to DNA. Journal of molecular biology. 2005;345:251–264. doi: 10.1016/j.jmb.2004.10.047. [DOI] [PubMed] [Google Scholar]
- Chico-Calero I, Suárez M, González-Zorn B, Scortti M, Slaghuis J, Goebel W, Vázquez-Boland JA, Consortium ELG. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MP, Longhi C, Polidoro M, Petrone G, Buonfiglio V, Di Santo S, Papi E, Seganti L, Visca P, Valenti P. Iron availability affects entry of Listeria monocytogenes into the enterocytelike cell line Caco-2. Infection and immunity. 1996;64:3925–3929. doi: 10.1128/iai.64.9.3925-3929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. Regulation of Listeria virulence: PrfA master and commander. Current Opinion in Microbiology. 2011;14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- den Hengst CD, van Hijum SAFT, Geurts JMW, Nauta A, Kok J, Kuipers OP. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. The Journal of biological chemistry. 2005;280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- Dineen SS, McBride SM, Sonenshein AL. Integration of Metabolism and Virulence by Clostridium difficile CodY. Journal of bacteriology. 2010;192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Oropeza R, Kenney LJ. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Molecular microbiology. 2003;48:1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x. [DOI] [PubMed] [Google Scholar]
- Flores AR, Olsen RJ, Wunsche A, Kumaraswami M, Shelburne SA, 3rd, Carroll RK, Musser JM. Natural variation in the promoter of the gene encoding the mga regulator alters host-pathogen interactions in group a streptococcus carrier strains. Infection and immunity. 2013;81:4128–4138. doi: 10.1128/IAI.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Port GC, Miner MD. Listeria monocytogenes -from saprophyte to intracellular pathogen. Nature reviews. Microbiology. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Portnoy DA. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Molecular microbiology. 1994;12:845–853. doi: 10.1111/j.1365-2958.1994.tb01070.x. [DOI] [PubMed] [Google Scholar]
- Fuchs TM, Eisenreich W, Heesemann J, Goebel W. Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra-and intracellular habitats. FEMS microbiology reviews. 2012;36:435–462. doi: 10.1111/j.1574-6976.2011.00301.x. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Elhadad D, Deng W, Rahav G, Finlay BB. The Salmonella enterica PhoP directly activates the horizontally acquired SPI-2 gene sseL and is functionally different from a S. bongori ortholog. PloS one. 2011;6:e20024. doi: 10.1371/journal.pone.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy C, Raveneau J, Beretti JL, Lecroisey A, Vazquez-Boland JA, Alouf JE, Berche P. Purification and characterization of an extracellular 29-kilodalton phospholipase C from Listeria monocytogenes. Infection and immunity. 1991;59:2382–2388. doi: 10.1128/iai.59.7.2382-2388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glomski IJ, Gedde MM, Tsang AW, Swanson JA, Portnoy DA. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. The Journal of cell biology. 2002;156:1029–1038. doi: 10.1083/jcb.200201081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W, Kathariou S, Kuhn M, Sokolovic Z, Kreft J, Kohler S, Funke D, Chakraborty T, Leimeister-Wachter M. Hemolysin from Listeria--biochemistry, genetics and function in pathogenesis. Infection 16 Suppl. 1988;2:S149–156. doi: 10.1007/BF01639739. [DOI] [PubMed] [Google Scholar]
- Hendriksen WT, Bootsma HJ, Estevao S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PWM. CodY of Streptococcus pneumoniae: Link between Nutritional Gene Regulation and Colonization. Journal of bacteriology. 2008;190:590–601. doi: 10.1128/JB.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Joseph B, Mertins S, Stoll R, Schar J, Umesha KR, Luo Q, Muller-Altrock S, Goebel W. Glycerol Metabolism and PrfA Activity in Listeria monocytogenes. Journal of bacteriology. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortman GA, Boleij A, Swinkels DW, Tjalsma H. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PloS one. 2012;7:e29968. doi: 10.1371/journal.pone.0029968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Chen Z, Ferretti J, Malke H. Counteractive Balancing of Transcriptome Expression Involving CodY and CovRS in Streptococcus pyogenes. Journal of bacteriology. 2011;193:4153–4165. doi: 10.1128/JB.00061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister-Wachter M, Domann E, Chakraborty T. Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is coordinately expressed with listeriolysin in Listeria monocytogenes. Molecular microbiology. 1991;5:361–366. doi: 10.1111/j.1365-2958.1991.tb02117.x. [DOI] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Colledge VL, Lebedev AA, Williamson DC, Sonenshein AL, Wilkinson AJ. Structural Rearrangement Accompanying Ligand Binding in the GAF Domain of CodY from Bacillus subtilis. Journal of molecular biology. 2009;390:1007–1018. doi: 10.1016/j.jmb.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. The structure of CodY, a GTP-and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. The Journal of biological chemistry. 2006;281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- Li J, Ma M, Sarker MR, McClane BA. CodY Is a Global Regulator of Virulence-Associated Properties for Clostridium perfringens Type D Strain CN3718. mBio. 2013;4 doi: 10.1128/mBio.00770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Qi Y, Hulett FM. Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Molecular microbiology. 1998;28:119–130. doi: 10.1046/j.1365-2958.1998.00779.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yang M, Peterfreund GL, Tsou AM, Selamoglu N, Daldal F, Zhong Z, Kan B, Zhu J. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Listeria monocytogenes Virulence. PLoS Genetics. 2012;8:e1002887. doi: 10.1371/journal.pgen.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-Acting Riboswitch Controls Expression of the Virulence Regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Loh E, Memarpour F, Vaitkevicius K, Kallipolitis BH, Johansson J, Sonden B. An unstructured 5'-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Research. 2012;40:1818–1827. doi: 10.1093/nar/gkr850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. The AraC transcriptional activators. Curr Opin Microbiol. 2001;4:132–137. doi: 10.1016/s1369-5274(00)00178-8. [DOI] [PubMed] [Google Scholar]
- Mengaud J, Chenevert J, Geoffroy C, Gaillard JL, Cossart P. Identification of the structural gene encoding the SH-activated hemolysin of Listeria monocytogenes: listeriolysin O is homologous to streptolysin O and pneumolysin. Infection and immunity. 1987;55:3225–3227. doi: 10.1128/iai.55.12.3225-3227.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins S, Joseph B, Goetz M, Ecke R, Seidel G, Sprehe M, Hillen W, Goebel W, Muller-Altrock S. Interference of Components of the Phosphoenolpyruvate Phosphotransferase System with the Central Virulence Gene Regulator PrfA of Listeria monocytogenes. Journal of bacteriology. 2006;189:473–490. doi: 10.1128/JB.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenbachs AA, Brown DP, Moors M, Youngman P. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Molecular microbiology. 1997;23:1075–1085. doi: 10.1046/j.1365-2958.1997.2711634.x. [DOI] [PubMed] [Google Scholar]
- Miner MD, Port GC, Bouwer HG, Chang JC, Freitag NE. A novel prfA mutation that promotes Listeria monocytogenes cytosol entry but reduces bacterial spread and cytotoxicity. Microbial pathogenesis. 2008;45:273–281. doi: 10.1016/j.micpath.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Das HK, Dixit A. Identification of a positive transcription regulatory element within the coding region of the nifLA operon in Azotobacter vinelandii. Applied and Environmental Microbiology. 2005;71:3716–3724. doi: 10.1128/AEM.71.7.3716-3724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. Journal of bacteriology. 2003;185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. CodY deletion enhances in vivo virulence of communityassociated methicillin-resistant Staphylococcus aureus clone USA300. Infection and immunity. 2012;80:2382–2389. doi: 10.1128/IAI.06172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson GP, Holcomb LG, Scott JR. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infection and immunity. 2001;69:186–193. doi: 10.1128/IAI.69.1.186-193.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson GP, Scott JR. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Molecular microbiology. 2000;36:1391–1402. doi: 10.1046/j.1365-2958.2000.01957.x. [DOI] [PubMed] [Google Scholar]
- Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annual Review of Microbiology. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Deka RK, Norgard MV. BosR (BB0647) controls the RpoNRpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS pathogens. 2011;7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally. PLoS Genet. 2013;9:e1003839. doi: 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan-Thanh L, Gormon T. A chemically defined minimal medium for the optimal culture of Listeria. International journal of food microbiology. 1997;35:91–95. doi: 10.1016/s0168-1605(96)01205-6. [DOI] [PubMed] [Google Scholar]
- Portnoy DA, Jacks PS, Hinrichs DJ. Role of hemolysin for the intracellular growth of Listeria monocytogenes. The Journal of experimental medicine. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis H, Eckart RA, Gudipati RK, Heidrich N, Brantl S. CodY activates transcription of a small RNA in Bacillus subtilis. Journal of bacteriology. 2009;191:5446–5457. doi: 10.1128/JB.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Hulett FM. PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Molecular microbiology. 1998;28:1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012;150:792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- Rauch M, Luo Q, Muller-Altrock S, Goebel W. SigB-Dependent In Vitro Transcription of prfA and Some Newly Identified Genes of Listeria monocytogenes Whose Expression Is Affected by PrfA In Vivo. Journal of bacteriology. 2005;187:800–804. doi: 10.1128/JB.187.2.800-804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzoni A, Klarsfeld A, Dramsi S, Cossart P. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infection and immunity. 1997;65:1515–1518. doi: 10.1128/iai.65.4.1515-1518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripio MT, Brehm K, Lara M, Suarez M, Vazquez-Boland JA. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. Journal of bacteriology. 1997a;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripio MT, Dominguez-Bernal G, Lara M, Suarez M, Vazquez-Boland JA. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. Journal of bacteriology. 1997b;179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripio MT, Dominguez-Bernal G, Suarez M, Brehm K, Berche P, Vazquez-Boland JA. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Research in microbiology. 1996;147:371–384. doi: 10.1016/0923-2508(96)84712-7. [DOI] [PubMed] [Google Scholar]
- Rivera FE, Miller HK, Kolar SL, Stevens SM, Jr., Shaw LN. The impact of CodY on virulence determinant production in communityassociated methicillin-resistant Staphylococcus aureus. Proteomics. 2012;12:263–268. doi: 10.1002/pmic.201100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Bachman MA, Swanson MS. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS microbiology reviews. 2010;34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortti M, Monzo HJ, Lacharme-Lora L, Lewis DA, Vazquez-Boland JA. The PrfA virulence regulon. Microbes and infection / Institut Pasteur. 2007;9:1196–1207. doi: 10.1016/j.micinf.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shetron-Rama LM, Mueller K, Bravo JM, Bouwer HG, Way SS, Freitag NE. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Molecular microbiology. 2003;48:1537–1551. doi: 10.1046/j.1365-2958.2003.03534.x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Latifi T, Cromie MJ, Groisman EA. Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J Biol Chem. 2004;279:38618–38625. doi: 10.1074/jbc.M406149200. [DOI] [PubMed] [Google Scholar]
- Shivers RP, Dineen SS, Sonenshein AL. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Molecular microbiology. 2006;62:811–822. doi: 10.1111/j.1365-2958.2006.05410.x. [DOI] [PubMed] [Google Scholar]
- Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Molecular microbiology. 2004;53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- Simon R UP, Pühler A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/technology. 1983;1:784–791. [Google Scholar]
- Smith K, Youngman P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nature reviews. Microbiology. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes and infection / Institut Pasteur. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. The Journal of cell biology. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Kaltenbach LS, Low DA. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Molecular microbiology. 1995;17:303–312. doi: 10.1111/j.1365-2958.1995.mmi_17020303.x. [DOI] [PubMed] [Google Scholar]
- van Hijum SAFT, Medema MH, Kuipers OP. Mechanisms and Evolution of Control Logic in Prokaryotic Transcriptional Regulation. Microbiology and Molecular Biology Reviews. 2009;73:481–509. doi: 10.1128/MMBR.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W, Chateau A, Dillies MA, Coppee JY, Sonenshein AL, Fouet A. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infection and immunity. 2009;77:4437–4445. doi: 10.1128/IAI.00716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Y, Rauch M, Banfield MJ, Ermolaeva S, Scortti M, Goebel W, Vazquez-Boland JA. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Molecular microbiology. 2004;52:1553–1565. doi: 10.1111/j.1365-2958.2004.04052.x. [DOI] [PubMed] [Google Scholar]
- Villapakkam AC, Handke LD, Belitsky BR, Levdikov VM, Wilkinson AJ, Sonenshein AL. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. Journal of bacteriology. 2009;191:6865–6876. doi: 10.1128/JB.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Skarstad K. ChIP on Chip: surprising results are often artifacts. BMC genomics. 2010;11:414. doi: 10.1186/1471-2164-11-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu B, Wang Q, Wang L. Genome-wide analysis of the salmonella Fis regulon and its regulatory mechanism on pathogenicity islands. PloS one. 2013;8:e64688. doi: 10.1371/journal.pone.0064688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Freitag NE. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. Journal of bacteriology. 2004;186:6265–6276. doi: 10.1128/JB.186.18.6265-6276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunsche A, Hammer E, Bartholomae M, Volker U, Burkovski A, Seidel G, Hillen W. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. The FEBS journal. 2012;279:2201–2214. doi: 10.1111/j.1742-4658.2012.08604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Densitometry analysis of EMSA gels of prfA, ilvD and codV probes in the presence or absence of 10 mM BCAA. Averaged regression analyses for each probe. Analysis based on densitometry measurements of the free DNA probe at each CodY concentration using ImageJ software (Schneider et al., 2012). The lower left panel shows the analysis for ilvD probe in the presence of 10 mM BCAA at 4-32 nM concentrations of CodY: the representative EMSA analysis (right) and the quantification analysis (left). Quantifications of 2-3 biological repeats were fitted via exponential least-squares regression analysis. The average KD values depicted in the manuscript are based on 2-3 regression analyses made for each probe and are not derived from the averaged graphs presented here. Error bars represent standard error of the mean.
Figure S2. Specific competition EMSA assays of labeled and unlabeled prfA, ilvD and codV probes. Electrophoretic mobility shift assay (EMSA) of CodY binding to the prfA (A), ilvD probe (B) and codV (C) probes with and without specific unlabeled competitors. For prfA probe competition, the prfA-mut6 probe was also used as a specific competitor. The EMSAs were performed without BCAA and the concentration of CodY used for each probe is equivalent to the Kd50 of CodY for each probe (Figure S1). Primers used for amplification of DNA probes are described in Table S2. The results represent two independent biological repeats (N=2).
Figure S3. The 6His-CodY protein is fully active. A. Growth analysis of WT, ΔcodY and L.m. CodY-6his strains in BHI medium. Results are average of 3 biological repeats (N=3). Error bars represent standard deviation. B. RT-qPCR analysis of ilvD transcription in WT, ΔcodY and L.m. CodY-6his strains grown in BHI medium. Results are average of 3 biological repeats (N=3). Error bars represent 95% confidence interval.
Figure S4. Predicted structure of mut6-PrfA protein. The dimer structure of mut6-PrfA based on the solved WT PrfA structure obtained from the Protein Database Bank (PDB, accession number 2BEO) was predicted using the SWISS-MODEL homology modeling server (http://swissmodel.expasy.org/). The mut6-PrfA dimer structure (yellow) was aligned to the WT PrfA structure (blue) using the PyMol software. Based on this analysis the Root Mean Square deviation between the alpha carbons of the structures is 0.222 Å.