SUMMARY
Telomere maintenance by telomerase is impaired in the stem cell disease dyskeratosis congenita and during human aging. Telomerase depends upon a complex pathway for enzyme assembly, localization in Cajal bodies and association with telomeres. Here, we identify the chaperonin CCT/TRiC as a critical regulator of telomerase trafficking, using a high content genome-wide siRNA screen in human cells for factors required for Cajal body-localization. We find that TRiC is required for folding the telomerase cofactor TCAB1, which controls trafficking of telomerase and small Cajal body RNAs (scaRNAs). Depletion of TRiC causes loss of TCAB1 protein, mislocalization of telomerase and scaRNAs to nucleoli, and failure of telomere elongation. DC patient-derived mutations in TCAB1 impair folding by TRiC, disrupting telomerase function and leading to severe disease. Our findings establish a critical role for TRiC-mediated protein folding in the telomerase pathway and link proteostasis, telomere maintenance and human disease.
INTRODUCTION
Telomeres are nucleoprotein structures that protect chromosome ends and serve as substrates for the enzyme telomerase (Palm and de Lange, 2008). Telomeres shorten progressively with cell division due to incomplete replication of the lagging DNA strand, and this shortening is offset by processive elongation of telomeres by telomerase (Pfeiffer and Lingner, 2013). Impaired maintenance of telomeres contributes to the pathogenesis of many disease states, including dyskeratosis congenita, pulmonary fibrosis, aplastic anemia, and liver cirrhosis (Armanios and Blackburn, 2012). Conversely, effective maintenance of telomeres by telomerase is thought to be important in progression of human cancers (Artandi and DePinho, 2010; Hahn et al., 1999; Horn et al., 2013; Huang et al., 2013; Killela et al., 2013). The telomerase ribonucleoprotein requires a complex series of biochemical steps to enable enzymatic function at telomeres. Efforts to develop telomerase therapeutics useful in diverse diseases require a more complete understanding of these steps and the molecules that govern them. Unbiased approaches to identify telomerase regulators in human cells have thus far been limited by difficulty in detecting telomerase components, precluding the development of genetic screens to interrogate this pathway.
Active telomerase enzyme is comprised of a catalytic core – the telomerase RNA component, TERC, and the telomerase reverse transcriptase, TERT – in addition to several additional proteins required for proper telomerase function. The biogenesis of functional telomerase is mediated by a series of maturation, assembly and trafficking steps that take place within the nucleus as well as within Cajal bodies – subnuclear structures devoted to RNA modification and assembly (Egan and Collins, 2012). Human TERC shares sequence motifs in common with small Cajal body RNAs (scaRNAs), which act as guides for post-transcriptional modification of splicing RNAs within Cajal bodies (Darzacq et al., 2002; Jady et al., 2004; Zhu et al., 2004). The processes regulating telomerase assembly overlap considerably with biogenesis pathways for scaRNAs and related small nucleolar RNAs (snoRNAs), all of which share an H/ACA sequence recognized by the dyskerin core complex – dyskerin, NHP2 and NOP10. TERC stability is dependent on the dyskerin core complex, and on the assembly factors NAF1, Shq1 and pontin/reptin (Egan and Collins, 2012). Incorporation of TERT protein into an RNP containing TERC and the dyskerin core complex yields a telomerase enzyme that is stable and catalytically active (Cohen et al., 2007; Mitchell et al., 1999).
After these initial stages of assembly, telomerase localizes to Cajal bodies by association with TCAB1, which recognizes the CAB box sequence common to TERC and scaRNAs (Cristofari et al., 2007; Tycowski et al., 2009; Venteicher et al., 2009). Telomerase is specifically recruited to telomeres through an interaction between TERT and the OB-fold of the telomere binding protein TPP1 (Abreu et al., 2010; Nandakumar et al., 2012; Sexton et al., 2012; Zhong et al., 2012), and this step requires TCAB1 (Stern et al., 2012; Zhong et al., 2012). TCAB1 is required for trafficking of telomerase to Cajal bodies and for telomere maintenance (Venteicher et al., 2009). This requirement is highlighted by the existence of TCAB1 mutations in patients with dyskeratosis congenita (DC), a stem cell disease caused by a failure of telomere maintenance (Zhong et al., 2011). DC is characterized by an epidermal triad (oral leukoplakia, nail dystrophy and skin pigmentation abnormalities), bone marrow failure, pulmonary fibrosis, and increased cancer (Armanios and Blackburn, 2012). Mutations in DC target each of the known components of telomerase and interfere with many steps in the telomerase pathway, including assembly, trafficking, recruitment to telomeres and catalytic activity (Batista and Artandi, 2013). TCAB1 mutations occur in an autosomal recessive form of DC, in which single amino acid substitutions in TCAB1 protein cause a marked reduction in protein accumulation, and disruption of telomerase localization from Cajal bodies to nucleoli (Zhong et al., 2011). Mislocalization of telomerase to nucleoli causes telomere shortening and DC, however, the mechanism by which these mutations disrupt TCAB1 function is not understood.
Nascent polypeptides must navigate a complex thermodynamic and kinetic landscape to acquire and maintain their native, functional form. Coordinating this process across all the proteins in the cell requires a highly organized proteostasis network that regulates protein biogenesis, conformational maintenance, and degradation (Kim et al., 2013; Wolff et al., 2014). For many proteins, proper folding and function requires stabilization of folding intermediates, a process performed by molecular chaperones, which derive from one of several classes including HSP70, HSP90, and chaperonins. Responsible for the vast majority of chaperone-mediated folding within the cell, the HSP70 and HSP90 systems tend to act as monomers or homodimers to bind sections of nascent polypeptides, stabilizing regions of hydrophobicity until the polypeptides can properly fold. In contrast, chaperonins form barrel-like cages in which individual polypeptides are encapsulated, allowing folding to proceed isolated from the general cellular environment (Horwich et al., 2007).
As a general rule, fast-folding proteins are stabilized by the HSP70 and HSP90 systems, whereas difficult-to-fold proteins are transferred to the chaperonin system. TRiC (TCP-1 Ring Complex) is a large Group II chaperonin complex containing eight homologous subunits arranged in two stacked, octameric rings. TRiC was originally identified based on its essential role in folding the cytoskeletal proteins actin and tubulin (Frydman et al., 1992; Gao et al., 1992). Approaches to identify novel TRiC substrates suggest that its substrates are more numerous, although only a restricted set of proteins out of the thousands that flux through the proteostasis network depend on TRiC for their biogenesis (Yam et al., 2008). The primary function of TRiC is to participate in the folding of newly-synthesized polypeptides, as opposed to the refolding of stress-denatured proteins; consequently, unlike other arms of the proteostasis network, TRiC expression is linked to protein synthesis and is not induced by stress (Albanèse et al., 2006). TRiC has been shown to inhibit polyglutamine aggregation, and thus may have neuroprotective properties regarding the development of age-related misfolding diseases such as Huntington's disease (Behrends et al., 2006; Kitamura et al., 2006; Tam et al., 2006).
In this study, we deploy a genetic screen to identify new regulators of telomerase trafficking using a genome-wide, high content, RNA FISH-based siRNA screen in human cells. The screen identified subunits of the chaperonin TRiC and we demonstrate an essential role for TRiC in telomerase function and trafficking of telomerase and scaRNAs to Cajal bodies. These effects are due to the essential role of TRiC in folding TCAB1, and we find that DC patient mutations in TCAB1 disrupt this critical step in TCAB1 biogenesis, providing a molecular explanation for these cases of human disease. These findings reveal important connections linking proteostasis pathways to telomerase function and telomere disease states.
RESULTS
TRiC is a recurrent hit in a genetic screen for regulators of telomerase trafficking
The application of a genetic screen interrogating the telomerase trafficking pathway has potential to identify new regulators of telomerase. However, low expression of the telomerase catalytic core, TERT and TERC, has precluded the development of such assays. To overcome these limitations, we designed and performed a genome-wide, high content siRNA screen for genes required for localization of TERC and TCAB1 in Cajal bodies. To capture a sufficient population of cells, we acquired all images at 200x magnification. Although endogenous TERC foci were readily detectable at higher magnifications (i.e. 630x), 200x magnification was insufficient to resolve individual foci (Figure S1A); instead, we generated clonal HeLa S3 cells stably overexpressing TERC at levels approximately nine-fold higher than endogenous TERC (Figure S1B). Overexpressed TERC was readily detectable in foci at 200x (Figure S1A, S1B), and retained its dependence on dyskerin and TCAB1 for localization to Cajal bodies, validating the utility of this approach (Figure S1C).
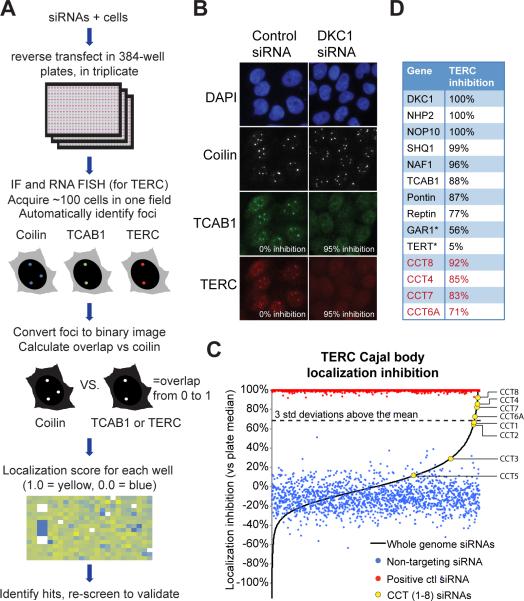
Using a human siRNA library containing 21,121 siRNA pools (4 siRNAs per pool) each targeting a single gene, we reverse transfected TERC-expressing HeLa S3 in 384-well optical plates, in triplicate (Figure 1A). Cells were fixed and immunofluorescence was performed for coilin, a Cajal body marker, and TCAB1 using antibodies against each endogenous protein, followed by RNA FISH for TERC. Images of a single field in each well (approximately 100 cells) were captured robotically and images were loaded into an analysis pipeline, which automatically identified nuclei, coilin foci, TCAB1 foci, and TERC foci. For each image we calculated an localization score – the fraction of foci pixels in the TERC or TCAB1 image that were also foci in the coilin image, i.e. foci overlap using coilin as the baseline and normalized to negative controls to calculate “localization inhibition”, with 0% indicating no TERC or TCAB1 disruption and 100% representing complete lack of TERC or TCAB1 in Cajal bodies (Figure 1B for example images). For TERC and TCAB1, the average Z’-factors across all plates were 0.68 and 0.81 respectively, well above the conventional 0.5 value that designates an assay amenable to large-scale screening (Figure S1D) (Zhang et al., 1999). Whereas positive control dyskerin siRNA wells (eight per plate, n=1608) yielded a median TERC localization inhibition of 99.6%, the median TERC inhibition of all the experimental siRNAs was -0.3% with a standard deviation of 22.4%, demonstrating that the vast majority of experimental siRNAs had no effect on TERC localization to Cajal bodies (Figure S1C). After removing toxic siRNAs and coilin-eliminating siRNAs, and applying a hit cutoff of three standard deviations above the mean (corresponding to 67.3% inhibition), we found that 119 siRNA pools qualified as TERC inhibition hits (Figure 1C). Hits included virtually all known components of the H/ACA box snoRNP complex, which is required for TERC biogenesis: DKC1, NHP2, NOP10, NAF1, and SHQ1 (Figure 1D). Hits also included TCAB1 and the telomerase assembly factors Pontin and Reptin. TERT siRNAs had no effect on TERC localization, which may be a consequence of poor knockdown. In addition to these known telomerase biogenesis and assembly factors, which validate the screening approach, hits included four members of the TCP-1 Ring Complex (TRiC): CCT8, CCT4, CCT7 and CCT6A (Figure 1D). Two other TRiC members fell just below the hit cutoff: CCT1 and CCT2 (Figure 1C). CCT6B siRNAs had no effect, which was expected as its expression is restricted to testicular tissues (Kubota et al., 1997). In the primary screen, CCT3 and CCT5 also showed no effect, which we subsequently determined were false negatives (see Figure S2A). Analyzing the TCAB1 channel in a similar manner, we found that TRiC subunits were also required for TCAB1 localization to Cajal bodies (Figure S1F), demonstrating that multiple telomerase components were perturbed due to TRiC depletion.
Figure 1. TRiC is a recurrent hit in a genetic screen for regulators of telomerase trafficking.
A) High content screening workflow. Reverse transfection of HeLa cells into 384-well plates with siRNA pools targeting individual genes, stained for Coilin, TCAB1, and TERC and imaged at 200x to determine overlap between the Coilin and TERC or TCAB1 channels. Heat map of data from one plate; well color indicates localization score. Yellow (localization), blue (no localization).
B) Representative screen images for negative control and positive control (Dyskerin siRNA) images, shown with percent localization inhibition.
C) Graphical representation of screen data. Y-axis, percent inhibition of TERC localization to Cajal bodies. X-axis, dimensionless. Red dots, positive control wells (siDKC1; n=1,608). Blue dots, negative control wells (siNon-targeting; n=1,608). Black line represents the median value (of triplicates) from each siRNA pool, in ascending order (n=21,119). Yellow dots show siRNA pools targeting CCT/TRiC genes. Dashed line, three standard deviation cutoff.
D) Localization inhibition for known telomerase components and biogenesis factors (black) and TRiC subunits called as hits (red). All called as hits except those marked by asterisk.
TRiC is required for TERC and scaRNA trafficking
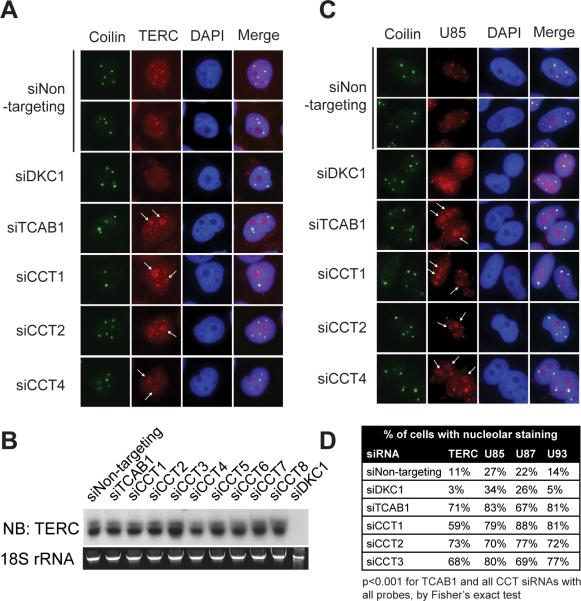
To validate and expand these results at a conventional scale, we depleted TRiC components in HeLa cells using RNA interference and assessed TERC localization at high magnification. Depletion of TRiC subunits led to mislocalization of endogenous TERC to nucleoli, results indistinguishable from depletion of TCAB1 itself (Figure 2A,D)(p<0.001 by Fisher's exact test). In contrast, depletion of dyskerin (DKC1) resulted in a global reduction in TERC signal, consistent with the function of dyskerin in TERC stability and assembly. Unlike depletion of dyskerin, neither TCAB1 depletion nor depletion of any TRiC subunit reduced TERC level measured by northern blot (Figure 2B). Localization of TERC stably overexpressed in HeLa cells was similarly dependent on TRiC, as depletion of any TRiC subunit led to mislocalization to nucleoli, demonstrating that overexpression of TERC cannot compensate for TRiC loss (Figure S2A). To test whether TRiC was required more generally for localization of scaRNAs in Cajal bodies, we used FISH to examine trafficking of scaRNAs containing either an H/ACA box only (U93) or an H/ACA box and a C/D box (U85 and U87). In all cases, we found that TRiC knockdown resulted in partial mislocalization of these scaRNAs to nucleoli, to a degree that phenocopied TCAB1 knockdown (Figure 2C,D, S2B, S2C) (p<0.001 by Fisher's exact test). These data demonstrate that TRiC is required for the proper localization of TERC, as well as multiple scaRNAs, to Cajal bodies.
Figure 2. TRiC is required for TERC and scaRNA trafficking.
A) RNA FISH for TERC (red) and IF for coilin (green) in HeLaS3 cells transfected with indicated siRNAs. DAPI, blue.
B) Northern blot for TERC in HeLa S3 cells transfected with indicated siRNAs; 18S rRNA, loading control.
C) RNA FISH for U85 (red) and IF for coilin (green) in HeLaS3 cells transfected with indicated siRNAs. DAPI, blue.
D) Quantification of nucleolar staining for TERC, U85, U87, and U93 scaRNAs from (A, C, S2B, S2C). 150 cells analyzed per condition, p<0.001 for either TCAB1 siRNAs or each CCT siRNAs vs. siNon-targeting controls, Fisher's exact test.
Telomerase recruitment and telomere addition depend upon TRiC
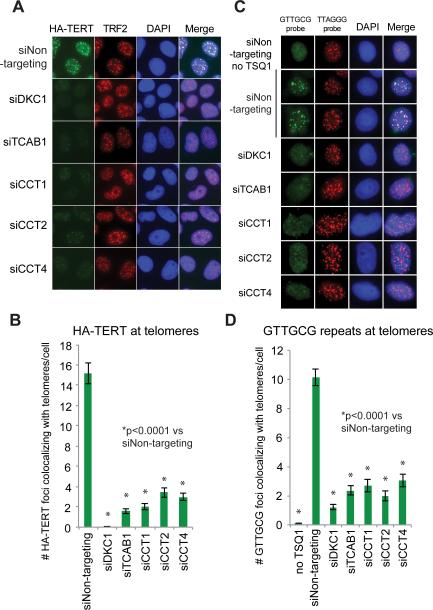
Association between TERC and TCAB1 is required for telomerase to localize to Cajal bodies and, subsequently, for telomerase to localize to telomeres (Cristofari et al., 2007; Stern et al., 2012; Venteicher et al., 2009; Zhong et al., 2011, 2012). To assess telomerase at telomeres, we overexpressed both TERC and HA-tagged TERT in HeLa cells, which causes ectopic telomerase to localize to telomeres in a high proportion of cells (Cristofari and Lingner, 2006; Zhong et al., 2012). We visualized the colocalization of HA-TERT and telomeres by immunofluorescence using anti-HA antibodies and antibodies against the telomere binding protein TRF2 (Figure 3A). Telomerase was efficiently recruited to telomeres in control cells, whereas siRNA-mediated depletion of DKC1 or TCAB1 strongly decreased that recruitment. Depletion of TRiC subunits significantly reduced the average frequency of telomerase foci at telomeres from 15 HA-TERT foci at telomeres in control cells to 2-4 in TRiC-depleted cells (p<0.0001, Student's t-test) (Figure 3B). Thus, recruitment of telomerase to telomeres – a rate-limiting step in telomere synthesis – is exquisitely dependent on the TRiC complex.
Figure 3. Telomerase recruitment and telomere addition depend upon TRiC.
A) Colocalization of HA-TERT and TRF2 by IF in cells transfected with indicated siRNAs and HA-TERT and TERC plasmids.
B) Quantification of HA-TERT at telomeres from (A). 150 cells analyzed per condition. Mean +/− standard error of the mean (SEM), p<0.0001 for each siRNA vs siNon-targeting control by two-tailed Student's t-test.
C) FISH for incorporation of mutant telomeres (GTTGCG) and FISH for wild-type telomere sequences (TTAGGG) in HeLa S3 cells transfected with indicated siRNAs.
D) Quantification of GTTGCG foci at telomeres (TTAGGG foci) from (C). 150 cells analyzed per condition. Mean +/− SEM, p<0.0001 for each siRNA vs siNon-targeting control by two-tailed Student's t-test.
To determine whether telomere addition requires TRiC, we employed an assay that allows direct visualization of newly added telomere repeats, programmed by a TERC variant containing a mutant template region: GTTGCG (Diolaiti et al., 2013). When expressed in HeLa cells for four days, this mutant TERC (TSQ1), together with endogenous TERT and other telomerase cofactors, efficiently mediated addition of variant GTTGCG repeats that were visualized by combined DNA FISH for the mutant sequences and for natural telomere repeats (TTAGGG)(Figure 3C). Control cells contained, an average of ~10 telomeres per cell with detectable GTTGCG signal, demonstrating that telomere elongation occurred at these telomeres over the four-day period (Figure 3D). Depletion of TRiC subunits, DKC1 or TCAB1 using siRNA strongly decreased addition of new repeats to fewer than three telomeres per cell, indicating that TRiC is required for telomere synthesis. Together, these data show that loss of TRiC leads to mislocalization of telomerase to nucleoli, inhibits telomerase recruitment to telomeres, and prevents telomere addition by telomerase.
TRiC interacts with the TCAB1 WD40 domain and is required for TCAB1 accumulation
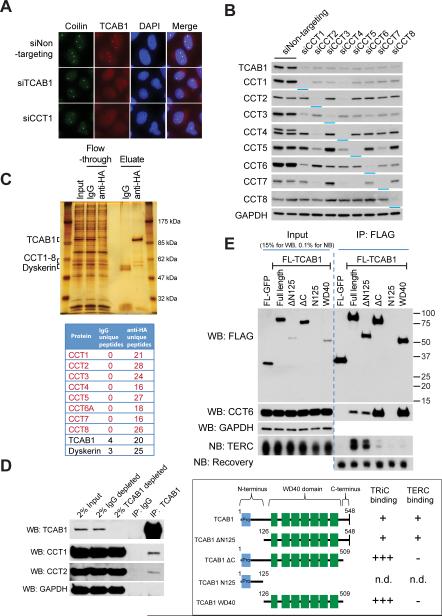
The similar requirements for TCAB1 and TRiC in telomerase localization, recruitment to telomeres and telomere synthesis, together with the observation that TRiC knockdown led to reduced TCAB1 foci in our genetic screen (Figure S1F), led us to hypothesize that the requirement for TRiC in telomerase function was based upon a role for TRiC in TCAB1 biogenesis. To test this idea, we depleted TRiC subunits in HeLa S3 cells and monitored TCAB1 foci by immunofluorescence and overall TCAB1 levels by western blot. Depletion of any of the eight TRiC subunits with siRNA strongly diminished TCAB1 foci (Figure 4A and S3A) and markedly reduced TCAB1 protein levels (Figure 4B) without altering TCAB1 mRNA levels (Figure S3B). Each CCT siRNA led to co-depletion of specific subsets of CCT proteins, likely reflecting patterns of codependence of CCT components within the TRiC complex (Figure 4B). To determine the selectivity of this TRiC-TCAB1 relationship, we depleted two Hsp70-family molecular chaperones: HSPA8 (Hsc70) and HSPA5 (GRP-78), both of which are required for the proper folding of a diverse set of cytoplasmic proteins (Liu et al., 2012). Depletion of these Hsp70-family chaperones had no effect on TCAB1 protein accumulation (Figure S3C), showing that disruption of TCAB1 is a TRiC-specific response rather than a result of general protein folding disruption.
Figure 4. TRiC interacts with the TCAB1 WD40 domain and is required for TCAB1 accumulation.
A) IF for TCAB1 (red) and coilin (green) in HeLa S3 cells transfected with siRNAs against TCAB1 and CCT1. DAPI, blue.
B) Western blot for TCAB1 and TRiC subunits in HeLa S3 cells transfected with siRNAs against eight TRiC subunits; GAPDH, loading control. Blue bars highlight targeted knockdown in each lane.
C) Top: dual-affinity chromatography purified AH3-TCAB1 from HeLa S3 cells, silver stain. Bottom: unique peptides for each protein identified by mass spectrometry. IgG in eluate, pre-clear negative control step before anti-HA pulldown.
D) Western blot for TCAB1 and TRiC subunits from co-IP of endogenous TCAB1 from HeLa S3 cells. IgG, negative control.
E) Co-IP of in vitro translated FLAG constructs in RRL incubated with in vitro transcribed TERC. Top, Western blot (FLAG, CCT6, GAPDH) and Northern blots (TERC, recovery control). Bottom, schematic of each construct and degree of TRiC and TERC interaction. n.d., not determined because N125 did not accumulate to detectable levels.
To determine whether TCAB1 and TRiC interact, we purified TCAB1 complexes through dual affinity chromatography coupled with mass spectrometry (MS). TCAB1 tagged at its N-terminus with Staph Protein A, a TEV cleavage site and three HA epitopes (AH3-TCAB1) was stably expressed in HeLa S3 cells by retroviral transduction, purified on rabbit IgG, eluted with TEV protease, captured again using anti-HA resin, then eluted, fractionated by SDS-PAGE and analyzed using nano-liquid chromatography-tandem MS (nanoLC-MS/MS). High peptide coverage was obtained for TCAB1, for the TCAB1-associated protein dyskerin, and for all eight subunits of the TRiC complex (Figure 4C). Immunoprecipitation of endogenous TCAB1 using TCAB1 polyclonal antibodies effectively depleted HeLa cell extracts of TCAB1 protein and revealed endogenous CCT1 and CCT2 in association with TCAB1 (Figure 4D). Pulldown of TCAB1 did not deplete CCT1 or CCT2, indicating that only a small fraction of TRiC is associated with TCAB1 (Figure 4D). These findings show that TCAB1 and TRiC associate at the endogenous level and suggest a substoichiometric or transient association.
The predicted structure of TCAB1 includes an unstructured N-terminal region, a WD40 domain with seven WD40 repeats forming a β-propeller – a protein structure with complex topology and multiple regions of hydrophobicity – followed by a short C-terminal extension (Figure 4E). Analysis of a series of N-terminal deletion mutants of TCAB1 revealed that the WD40 domain and the C-terminal extension were sufficient for TRiC association and for Cajal body-localization in HeLa cells (Fig. S3D, S3E). Further impingement on the WD40 domain, or deletion of the short C-terminal extension, prevented accumulation in human cells (data not shown), therefore we expressed additional mutant proteins in rabbit reticulocyte lysate (RRL), an extract that enables translation and folding, but lacks active protein degradation machinery. Deletion of the C-terminal region (ΔC, deletion of residues 510-548) resulted in efficient translation and an enhanced association with rabbit TRiC in the extract by co-IP (Figure 4E). Expression of the WD40 domain alone (residues 126-509, WD40) resulted in low levels of this isolated domain, but in significantly greater binding to TRiC compared to full-length TCAB1. These data show that the WD40 domain of TCAB1 mediates association with TRiC, but that stabilization of TCAB1 and efficient release from TRiC also require the C-terminal region of TCAB1.
To determine which TCAB1 sequences are involved in binding TERC, we performed IP-northern assays in RRL with FLAG-tagged TCAB1 mutant proteins after adding in vitro transcribed TERC. In validation studies, the association between wild-type TCAB1 and TERC in this assay was dependent upon an intact CAB box, as either of two inactivating mutations in the CAB box, m1-UGAc or m2-UcAG (Cristofari et al., 2007; Jady et al., 2004), markedly diminished binding to TCAB1 (Figure S3F). Full-length TCAB1 and ΔN125 bound wild-type TERC similarly well, but association between ΔC and WD40 fragments of TCAB1 with TERC was greatly reduced (Figure 4E). These data suggest that the WD40 domain, together with the short C-terminal extension, may comprise a single functional domain that both binds TERC and depends upon TRiC for folding.
TRiC is required to fold TCAB1, enabling TCAB1 to bind TERC
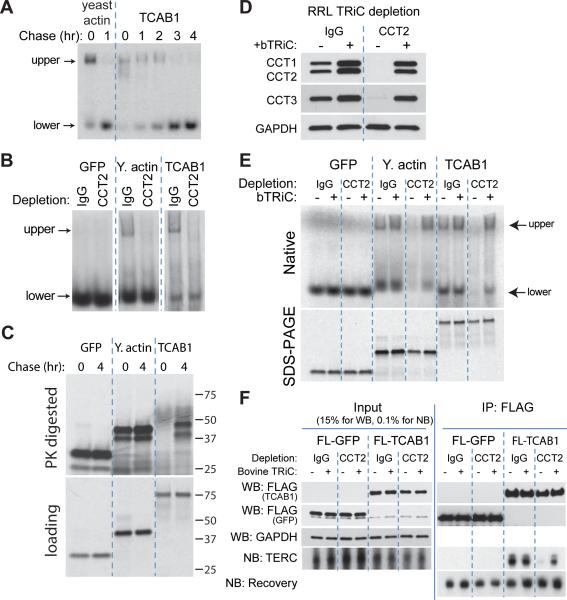
Although the kinetics of dissociation vary by substrate, TRiC clients are only transiently associated with the complex, and are released after folding is complete. To determine whether TCAB1 is a TRiC substrate, we in vitro translated TCAB1, or yeast actin as a positive control, in the presence of 35S-methionine in RRL. After the initial labeling, reactions were chased with cold methionine for the indicated number of hours and proteins were fractionated by native PAGE. Actin is a known substrate of TRiC and is folded in a TRiC-dependent manner in RRL (Gao et al., 1992). Migration of newly synthesized yeast actin was retarded in the gel, whereas after a one hour chase, actin migrated more rapidly, consistent with being released from TRiC (Vainberg et al., 1998) (Figure 5A). In vitro translated TCAB1 similarly migrated in two forms. Newly translated TCAB1 protein was detected almost exclusively in a slow migrating species that co-migrated with newly synthesized actin. Over a four hour chase period, TCAB1 became rapidly migrating and formed a discrete lower band. We verified that both radiolabeled species were in fact TCAB1 by translating FLAG-TCAB1 and performing a western blot after native PAGE (Figure S4A). TRiC, visualized by CCT2 western blotting, co-migrated with the retarded, newly synthesized form of TCAB1 in RRL (Figure S4A). We showed that the upper TCAB1 band was TRiC-bound by immunodepleting CCT2 after TCAB1 translation (Figure S4B), leading to a significant decrease in the upper bands of both yeast actin and TCAB1, whiles preserving the lower bands for both proteins (Figure 5B). These results demonstrate that TCAB1 polypeptides are bound by TRiC during or soon after translation and are released from association with TRiC over time.
Figure 5. TRiC is required to fold TCAB1, enabling TCAB1 to bind TERC.
A) Time course of [35S]-Met-labeled actin or TCAB1 in RRL, chased with cold Met for indicated time, native PAGE. Upper, slowing migrating species. Lower, rapidly migrating species.
B) Immunodepletion of CCT2 from RRL extracts in which [35S]-Met-labeled proteins (GFP, actin or TCAB1) were translated, native PAGE. IgG, negative control.
C) Partial PK digestion of [35S]-Met labeled proteins (GFP, actin or TCAB1) in RRL chased with cold Met for indicated times, top. Total, undigested protein, bottom. SDS-PAGE.
D) Immunodepletion of CCT2 from RRL extracts. IgG, negative control. +bTRiC, add-back of purified bovine TRiC. Western blot for CCT1, CCT2, CCT3 and GAPDH.
E) Immunodepletion of CCT2 from RRL extracts (as in D) prior to translation of [35S]-Met-labeled proteins (GFP, actin or TCAB1). Native PAGE, top. SDS-PAGE for loading, bottom. +bTRiC, add-back of purified bovine TRiC.
F) Co-IP of TCAB1 and TERC from RRL. Western (FLAG, GAPDH) and Northern (TERC, recovery) blots from co-IP of FLAG constructs in vitro translated in previously IgG or CCT2-immunodepleted RRL, as in (D), incubated with in vitro transcribed TERC.
We hypothesized that TRiC released TCAB1 once folding was complete. Partial resistance to protease digestion is a characteristic of many correctly folded, compact proteins (Frydman et al., 1994). To optimize this approach for natively folded TCAB1, we performed a dose response proteinase K (PK) digestion on HeLa lysate and analyzed protease-resistant TCAB1 fragments by western blotting (Figure S4C). Partial PK digestion at 1.2 μg/mL generated two protease-resistance TCAB1 bands between 37 and 50 kDa, while higher concentrations degraded the protein completely and lower concentrations had no effect. We then examined the protease resistance of in vitro translated TCAB1 before and after TRiC-release, to assess whether TRiC binding caused TCAB1 to adopt a similarly protease-resistant structure. PK degraded all protein at zero hours, but resulted in a protected species at four hours, characterized by two discrete protease-resistant TCAB1 bands (Figure 5C) similar in size to those generated by partial digestion of cellular TCAB1 (Figure S4C). Importantly, the input of total TCAB1 protein treated with PK was comparable at both time points (Figure 5C, bottom). These data indicate that TCAB1 is partially resistant to protease treatment only after being released from TRiC, suggesting that this TRiC-released TCAB1 species is compact and correctly folded.
To understand if TRiC is required for folding TCAB1, we immunodepleted TRiC from RRL using antibodies to CCT2, and then reconstituted the complex to endogenous levels using purified bovine TRiC, or used mock depletion with IgG as a negative control (Figure 5D). Proteins were then synthesized under TRiC-depleted and TRiC-add back conditions. Translation of either yeast actin or TCAB1 in TRiC-depleted RRL resulted in a marked reduction of both the TRiC-associated form (>50% reduction vs. mock depleted controls) and the folded species for each protein (>90% reduction vs. mock depleted controls) by native PAGE (Figure 5E, S4D). Add-back of bovine TRiC to the depleted RRL restored both the TRiC-bound (>100% vs. mock depleted controls) and folded species (>50% vs. mock depleted controls) of actin and TCAB1 (Figure 5E, S4D). Depletion of TRiC, or TRiC add-back, did not significantly change the overall amount of translated proteins by SDS-PAGE (Figure 5E, lower panel). These data show that TRiC is required for TCAB1 to be folded to a compact, rapidly migrating conformation in RRL.
TCAB1 associates with TERC and with scaRNAs by binding the CAB box sequence common to these RNAs (Tycowski et al., 2009; Venteicher et al., 2009). To understand whether TRiC-mediated folding is required for TCAB1 to associate with TERC, we tested the effects of TRiC depletion in RRL on TCAB1-TERC binding by IP-northern (Figure 4). TCAB1 translated in TRiC-depleted RRL bound <10% of the TERC bound by TCAB1 translated in mock-depleted RRL (Figure 5F, S4E). Add-back of bovine TRiC to TRiC-depleted extracts restored the ability of TCAB1 to associate with TERC to >60% of mock-depleted level (Figure 5F, S4E). Taken together, these data demonstrate that TCAB1 is an obligate TRiC substrate, requiring TRiC for folding and TERC binding, and that loss of TRiC cannot be compensated by the presence of other chaperones.
Disruption of TRiC-mediated folding underlies the defects in the TCAB1-mutant form of DC
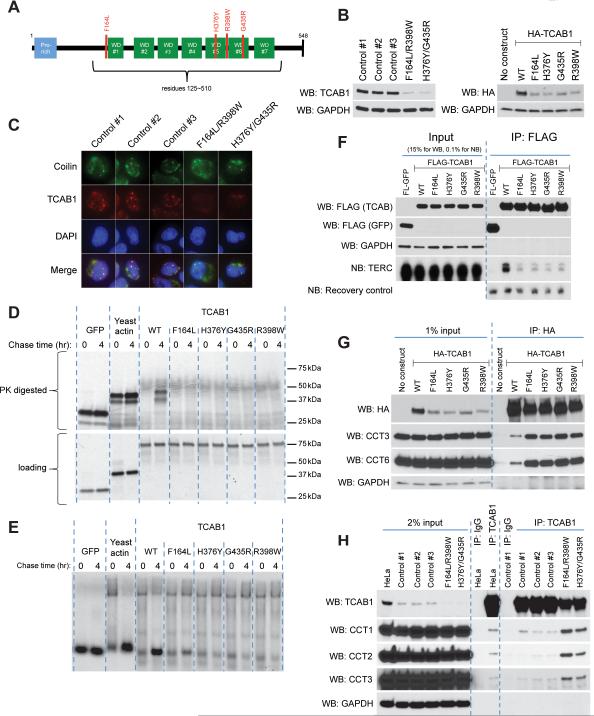
The effects of TRiC depletion on the telomerase pathway – diminished TCAB1 protein, loss of TERC and TCAB1 from Cajal bodies, and a failure of telomere synthesis – closely resembled the underlying features of a form of DC caused by compound heterozygous mutations in TCAB1 (Zhong et al., 2011). For each of four identified disease-causing alleles, missense mutations in TCAB1 were located in or near loop regions within the WD40 domain, the principal substrate for TRiC-mediated folding (Figure 6A). We therefore hypothesized that the single amino acid mutations in TCAB1 specifically disrupted folding of TCAB1 by TRiC. TCAB1 protein levels were markedly reduced in Epstein-Barr Virus (EBV)-transformed lymphoblasts derived from two DC patients with compound heterozygous TCAB1 mutations, compared to levels in unrelated individuals with wild-type TCAB1 (Figure 6B). Furthermore, each of the mutant TCAB1 alleles also showed impaired protein accumulation when stably overexpressed in HeLa cells, indicating that diminished expression is an intrinsic property of the mutant proteins and not related to mRNA level (Figure 6B, S5A). Consistent with diminished expression, TCAB1 protein failed to localize properly to Cajal bodies, both in patient lymphoblasts (Figure 6C) and in stably transduced HeLa cells where TCAB1 was detected in the cytoplasm (Figure S5B), where TRiC resides almost exclusively (Figure S5C) (Zhong et al., 2011).
Figure 6. Disruption of TRiC-mediated folding underlies the defects in the TCAB1-mutant form of DC.
A) Schematic of TCAB1 domains and dyskeratosis congenita (DC)-associated mutations.
B) Western blots of endogenous TCAB1 from control or DC patient-derived lymphoblasts (left) or stably overexpressed HA-tagged TCAB1 (wild-type or individual mutants) from HeLa S3 cells (right).
C) IF for Coilin and TCAB1 in DC patient-derived lymphoblasts.
D) Partial PK digestion [35S]-Met labeled TCAB1 proteins in RRL chased with cold Met for indicated times, SDS-PAGE, top. Undigested loading, bottom.
E) Time course of [35S]-Met-labeled TCAB1 proteins in RRL, chased with cold Met for indicated time, native PAGE. See (D) for loading.
F) Co-IP of TCAB1 mutants with TERC. Western (FLAG, GAPDH) and Northern (TERC, recovery) blots from co-IP of FLAG-tagged TCAB1 proteins in vitro translated in RRL and incubated with in vitro transcribed TERC.
G) Co-IP of TCAB1 mutants stably expressed in HeLa S3 cells with endogenous TRiC. Western blot.
H) Co-IP of endogenous TCAB1 and endogenous TRiC from either HeLa S3 cells or DC patient-derived lymphoblasts. Control 1, 2 and 3 are unrelated healthy donor lymphoblasts. F146L/R398 and H376Y/G435R are TCAB1-mutant patient lymphoblasts. IgG, negative control.
To determine whether DC-derived TCAB1 mutants fail to accumulate because of a defect in folding by TRiC, we assessed the resistance to protease digestion of mutant versus wild-type TCAB1 in patient-derived lymphoblasts. As in HeLa cells and RRL, wild-type TCAB1 in lymphoblasts displayed two protease-resistant bands between 50 and 37 kDa, however mutant TCAB1 in DC-patient derived lymphoblasts showed no resistance to protease treatment under the same conditions (Figure S5D). Mutant TCAB1 accumulated to lower levels than wild-type TCAB1, raising the possibility that the difference in protease resistance was due to a difference in total protein level. We therefore examined DC-derived TCAB1 mutants versus wild-type TCAB1 expressed in RRL, where the mutant and wild-type TCAB1 proteins accumulate at comparable levels (Figure 6D, bottom). Unlike wild-type TCAB1, which was partially protected from PK digestion at 4 hours, each DC-derived TCAB1 mutant failed to show PK-resistance at 4 hours (Figure 6D, top). To determine whether this inability to fold properly corresponded to differences in TRiC binding or release, TCAB1 proteins were assayed after pulse-chase in RRL by native PAGE. In vitro translated TCAB1 mutants were initially bound by TRiC to the same degree as wild-type TCAB1, but whereas wild-type TCAB1 was released after several hours, the TCAB1 mutants remained TRiC-associated and were impaired in progression to the compact, rapidly migrating form (Figure 6E, see 6D for loading). We assessed whether this impaired folding of the DC-derived TCAB1 mutants affected their ability to bind TERC. Compared with wild-type TCAB1, in vitro translated TCAB1 mutants showed a marked decrease in TERC association by IP-northern (Figure 6F). These data show that DC-associated TCAB1 mutants cannot be properly folded by TRiC, show enhanced TRiC association and are impaired in their ability to bind TERC.
The increased association of TCAB1 mutants with TRiC was recapitulated in human cells, as measured by IP-western of HA-tagged TCAB1. Despite accumulating at a lower level, each TCAB1 mutant showed a steady-state association with TRiC substantially greater than that of wild-type TCAB1 (Figure 6G). We used extracts from DC patient lymphoblasts to determine whether this increased association with TRiC was evident when TCAB1 mutants were expressed at endogenous levels. Although TCAB1-mutant proteins were expressed at reduced levels in DC lymphopblasts, immunoprecipitation of endogenous mutant TCAB1 protein pulled down significantly more TRiC compared with immunoprecipitation of wild-type TCAB1 from healthy donor control lymphoblasts (Figure 6H). Taken together, these data show that the TCAB1-mutant form of DC is caused by amino acid mutations that disrupt TCAB1 folding by TRiC, providing important evidence that telomere maintenance by telomerase is exquisitely dependent on TRiC and that interfering with this process results in severe disease in humans.
DISCUSSION
A model for TRiC-mediated folding of TCAB1 in telomere maintenance
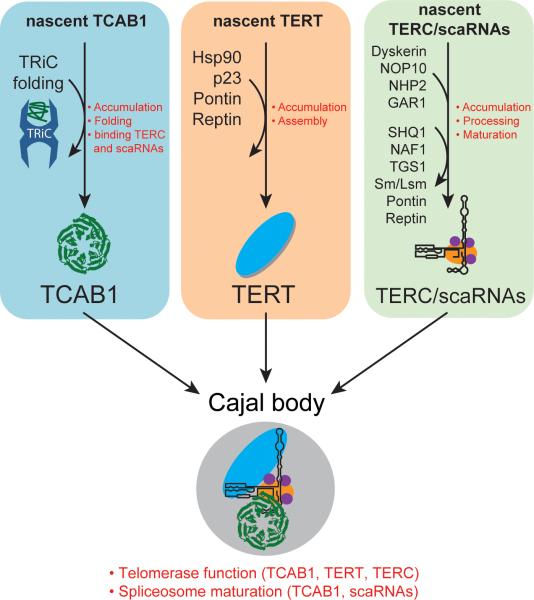
In searching for new regulators of telomerase trafficking using a genome-wide loss-of-function high content screen, we identified the TRiC chaperonin as a critical node in the telomerase pathway and one whose actions are subverted in disease. Our data support a model in which newly translated TCAB1 protein associates with TRiC, through an interaction that requires the WD40 domain of TCAB1. TRiC folds TCAB1 in the cytoplasm, and once correctly folded, TCAB1 translocates to the nucleus where it binds the TERC CAB box, and likely associates with dyskerin through protein-protein contacts, to facilitate localization of telomerase within Cajal bodies. This localization step is essential for telomere maintenance, as TCAB1 is also required for recruitment of telomerase to telomeres, a step governed by the interaction between TERT and TPP1 (Nandakumar et al., 2012; Stern et al., 2012; Venteicher et al., 2009; Zhong et al., 2011, 2012). Consistent with a requirement in folding TCAB1, depletion of any TRiC subunit resulted in reduced TCAB1 protein in Cajal bodies, mislocalization of TERC to nucleoli and a marked impairment of telomerase recruitment to telomeres. TRiC is also needed for correct trafficking of scaRNAs to Cajal bodies, as loss of TRiC resulted in inappropriate trafficking of scaRNAs to nucleoli. Because scaRNAs serve as guides for the pseudouridylation of splicing RNAs, our data suggest that, through folding TCAB1, TRiC may be required for the assembly of splicing RNPs and perhaps for efficient RNA splicing.
Our data indicate that TRiC-dependent folding of TCAB1 represents a third biogenesis pathway required for telomerase function. Most well understood are the RNA-dependent steps needed for stability, processing and assembly of TERC. TERC biogenesis relies on the machinery devoted to related snoRNAs, including the dyskerin core complex, assembly factors Shq1 and Naf1, as well as factors needed for RNA end processing and capping (Figure 7, green box). Factors have also been identified that are required for TERT accumulation and assembly into the telomerase complex. Pontin and Reptin are AAA+ ATPases that are required for both TERC accumulation and TERT assembly into the telomerase holoenzyme (Venteicher et al., 2008). Additionally, the molecular chaperones p23 and Hsp90 both bind TERT, and inhibition of this interaction reduces accumulation of TERT protein and blocks assembly of the telomerase catalytic core (Holt et al., 1999) (Figure 7, orange box). In contrast to these two pathways focused on the catalytic core of the enzyme, our findings reveal a proteostasis pathway that is not essential for the biogenesis of TERT or TERC, but is critical for the folding of TCAB1 and therefore for localization of the telomerase complex in Cajal bodies (Figure 7, blue box).
Figure 7. Model of telomerase biogenesis and trafficking.
A) Three arms of telomerase holoenzyme biogenesis: TCAB1, TERT and TERC undergo independent and essential biogenesis steps before being assembled into active telomerase. Curved arrows, factors transiently associated. Curved lines, factors permanently associated. Red text, processes mediated by the listed factors. Factors not necessarily listed in chronological order and not all-inclusive.
TRiC-mediated folding of TCAB1 is required for prevention of dyskeratosis congenita
TRiC depletion using siRNA caused biochemical and cell biological phenotypes similar to those seen in cells from dyskeratosis congenita (DC) patients with missense mutations in TCAB1, including reduced TCAB1 protein level and mislocalization of TERC to nucleoli. Based on these similarities, we investigated the hypothesis that the missense mutations in TCAB1, located in the WD40 repeat domain, represent folding mutations – sequence changes that disrupt the ability of TCAB1 to be correctly folded by TRiC. Our data indicated that each patient-derived TCAB1 mutant protein failed to be released from TRiC, could not achieve a protease-resistant form, exhibited marked reduction in binding TERC and showed a significant increase in association with TRiC. All these findings are consistent with a disruption of TCAB1 folding by the amino acid mutations in DC patients. Because mutant TCAB1 readily binds TRiC, we conclude that the missense mutations disrupt the folding process itself, and that misfolded TCAB1 is eventually released and destroyed, after failing an important quality control step. Although misfolded TCAB1 is reduced in level and is functionally impaired, it is important to note that reduced levels of mutant TCAB1 protein remain detectable in the nucleus and even in Cajal bodies. These findings suggest that the folding mutations are hypomorphic in nature, severely impairing, but not eliminating TCAB1 function.
Folding mutations yield hypomorphic disease variants in highly conserved proteins
Thus far, no complete loss-of-function mutations in any telomerase component have been identified in DC patients, suggesting that complete inactivation of telomerase function may be incompatible with viability in humans (Zaug et al., 2013). Homozygous null mutations in TCAB1 would be even less likely, given the potential importance of TCAB1 in the pseudouridylation of splicing RNAs. The challenge in generating a telomere-disease phenotype lies in reducing telomere maintenance levels below a certain threshold, while preserving sufficient reserves in telomere maintenance to avoid embryonic lethality. For multifunctional proteins, their functions in other pathways need to be preserved as well. The best example is dyskerin, which is required for pseudouridylation of ribosomal RNAs and splicing RNAs. To avoid the embryonic lethality of dyskerin inactivation, DC mutations in dyskerin are missense mutations that cluster at a particular surface of the protein, sufficiently preserving its pseudouridylation function to allow viability, while at the same time compromising its ability to support telomerase biogenesis (Mitchell et al., 1999; Rashid et al., 2006; Wong and Collins, 2006). The exquisite dependence of TCAB1 on the TRiC pathway allowed for the emergence of TCAB1 folding mutations that accomplish this titration of TCAB1 activity by a different means – by disrupting TRiC-mediated folding and by globally reducing TCAB1 levels and function. It is interesting to note that the vast majority of disease-associated mutations in TERT and TERC are point mutations, which commonly preserve some residual activity. It is likely that a significant subset of these mutations disrupt folding of TERT protein or TERC RNA, and that such mutations would operate in similar fashion to the TCAB1 mutations in reducing the level and activity of TERT or TERC.
The mechanisms by which mutations in other TRiC substrates cause disease differ. Mutant huntingtin (Htt) protein generates large aggregates, eventually overwhelming the proteostasis network and causing proteotoxicity (Behrends et al., 2006; Kitamura et al., 2006; Tam et al., 2006). Instead of aggregating, VHL mutants either prevent TRiC interaction or prevent TRiC release (Feldman et al., 2003), the latter mechanism being similar to the effects of the TCAB1 mutations. The identification of TCAB1 as a TRiC substrate is surprising because TCAB1 is both nuclear and an RNA-binding protein, which are rare among TRiC substrates (Yam et al., 2008). Thus, TRiC may be responsible for folding a larger variety of protein classes than previously thought, expanding its potential contribution to the proteostasis network.
Proteostasis, aging, telomerase function and disease
A dramatic decline in proteostasis has been linked to aging in diverse organisms, and this decline drives age-related pathologies (Hartl et al., 2011; Taylor and Dillin, 2011). TRiC, specifically, has been implicated in human age-related degeneration via its interaction with the htt protein (Kitamura et al., 2006; Tam et al., 2006). The evidence that chaperones (TRiC, Hsp90, p23) impinge on multiple steps of telomerase biogenesis leads us to speculate that reduced proteostasis with aging may lead to a decline in telomerase function, particularly because proteostatic networks tend to lack significant excess capacity (Morimoto, 2008). Impaired telomerase function, in turn, could lead to age-related telomere dysfunction, particularly in those tissues that depend on telomerase during aging, such as bone marrow, gastrointestinal epithelium, skin, liver and lung.
Our data identify an exquisite dependence of telomerase on TRiC and reveal that TRiC-mediated folding of TCAB1 is essential for telomere maintenance in human tissues and that disruption of this process leads to dyskeratosis congenita. These findings have important implications in understanding telomere dysfunction in aging and in human disease.
EXPERIMENTAL PROCEDURES
Cell culture, transfections, and viral transductions
HeLa S3 and 293T cells were cultured in DMEM supplemented with 10% FBS. Lymphoblasts were cultured in RPMI 1640 supplemented with 10% FBS. Lipofectamine 2000 (Life Technologies, 11668019) was used for all cDNA transfection experiments. Dharmafect 4 (Thermo, T-2004-01) was used for all siRNA transfections. All siRNAs were siGENOME pools purchased from Dharmacon/Thermo and used at 25 or 50 nM. Cells were assayed 72 hr after transfection. For transduction, 293T cells were transfected with the appropriate target plasmid and packaging constructs (lentiviral or retroviral) overnight; 48 hr later, viral supernatant was collected. Cells were transduced in the presence of 5 μg/mL Polybrene and selected for 3-7 days depending on the selection marker.
Western and Northern blotting
Cells were lysed in NP40 buffer (25 mM HEPES-KOH, 150 mM KCl, 1.5 mM MgCl2, 0.5% NP40, 10% Glycerol, pH 7.5) supplemented with protease inhibitors (Roche, 11836153001) for 20 min on ice. Lysate was clarified by centrifugation at 16,000 xg for 10 min and quantified by BCA assay (Pierce, 23225). For Western blotting, 10-50 μg was separated by SDS-PAGE, transferred onto PVDF membrane (GE Healthcare, RPN303F), and blotted according to standard protocols. 5% milk in TBST (0.1% Tween) was used for all blocking and antibody incubation steps; TBST (0.1% Tween) was used for all washes. For Northern blotting, RNA was extracted with TRIzol (Life Technologies, 15596-026), separated using 5% urea TBE gel electrophoresis, transferred onto Hybond-N membrane (GE Healthcare, RPN303N), and hybridized with ULTRAhyb (Ambion, AM8670). TERC probe was generated by random body labeling with full length TERC cDNA.
TERC in vitro binding assays
TERC or TERC mutant RNA (m1, m2) was in vitro transcribed (IVT) using the MEGAScript T7 kit (Life Technologies, AM1334). Target DNA was PCR amplified with a minimal T7 promoter at the 5’ end and 240 ng of each PCR product was used in the transcription reaction. The reaction was incubated for 6 hrs at 37°C and RNA was purified using TRIzol reagent (Life Technologies, 15596-026). In vitro transcription/translation of TCAB1 or TCAB1 mutants is described in detail in the “In vitro transcription/translation” section; RRL reaction was incubated for 2 hr at 30°C, then 1 μg IVT RNA was added and the reaction was incubated for another 2 hr at 30°C. The reaction was diluted in 500 μL of NP40 buffer (see “western blotting” section) and FLAG immunoprecipitation was performed as described in “Immunoprecipitations and immunodepletions”. RNA was analyzed by Northern blotting.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Artandi and Frydman labs for helpful discussions and advice. We thank the Stanford High-Throughput Bioscience Center, with support from NIH NCRR Instrumentation grant S10RR026338. We are grateful to Anthony Tomlinson for generously providing purified bovine TRiC, and Sharon Savage for supplying DC patient lymphoblasts. AF was supported by Stanford Cancer Biology Training Program (T32 CA09302) and a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. FZ was supported by a fellowship from the Agency for Science, Technology, and Research (A*STAR), Singapore. This work was supported by NIH grants AG033747, CA125453, CA111691 and AG036695, and by the Glenn Foundation for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.F., F.Z., J.F. and S.A. designed the experiments. A.F. and F.Z. performed the experiments. A.V., Z.M., and T.V. performed TCAB1 IP-MS studies. A.F. and S.A. wrote the manuscript with input from all other co-authors.
REFERENCES
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanèse V, Yam AY-W, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. The telomere syndromes. Nat. Rev. Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista LF, Artandi SE. Understanding telomere diseases through analysis of patient-derived iPS cells. Curr. Opin. Genet. Dev. 2013;23:526–533. doi: 10.1016/j.gde.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Böttcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolaiti ME, Cimini BA, Kageyama R, Charles FA, Stohr BA. In situ visualization of telomere elongation patterns in human cells. Nucleic Acids Res. 2013;41:e176. doi: 10.1093/nar/gkt689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA New York N. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol. Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack C-G, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kubota H, Hynes GM, Kerr SM, Willison KR. Tissue-specific subunit of the mouse cytosolic chaperonin-containing TCP-1. FEBS Lett. 1997;402:53–56. doi: 10.1016/s0014-5793(96)01501-3. [DOI] [PubMed] [Google Scholar]
- Liu T, Daniels CK, Cao S. Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol. Ther. 2012;136:354–374. doi: 10.1016/j.pharmthera.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Lingner J. Replication of telomeres and the regulation of telomerase. Cold Spring Harb. Perspect. Biol. 2013;5:a010405. doi: 10.1101/cshperspect.a010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Sexton AN, Youmans DT, Collins K. Specificity Requirements for Human Telomere Protein Interaction with Telomerase Holoenzyme. J. Biol. Chem. 2012;287:34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol. Cell. Biol. 2012;32:2384–2395. doi: 10.1128/MCB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011;3:a004440. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Weissman JS, Dillin A. Differential Scales of Protein Quality Control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Wong JMY, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K, Cech TR. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013;41:8969–8978. doi: 10.1093/nar/gkt653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Chung, Oldenburg A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LFZ, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.