Abstract
Objective
Genetic factors confer risk for neuropsychiatric phenotypes, but the polygenic etiology of these phenotypes makes identification of genetic culprits challenging. An approach to this challenge is to examine the effects of genetic variation on relevant endophenotypes, such as hippocampal volume loss. Smaller hippocampus is associated with gene variants of the renin-angiotensin system (RAS), a system implicated in vascular disease. However, no studies have investigated longitudinally the effects of genetic variation of RAS on the hippocampus.
Method
We examined the effects of polymorphisms of AGTR1, the gene encoding angiotensin-II type 1 receptor of RAS, on longitudinal hippocampal volumes of older adults. 138 older adults (age ≥ 60 years) were followed for an average of about four years. Subjects underwent repeated structural MRI and comprehensive neurocognitive testing, and were genotyped for four AGTR1 SNPs with low pairwise linkage disequilibrium values and apolipoprotein E (APOE) genotype.
Results
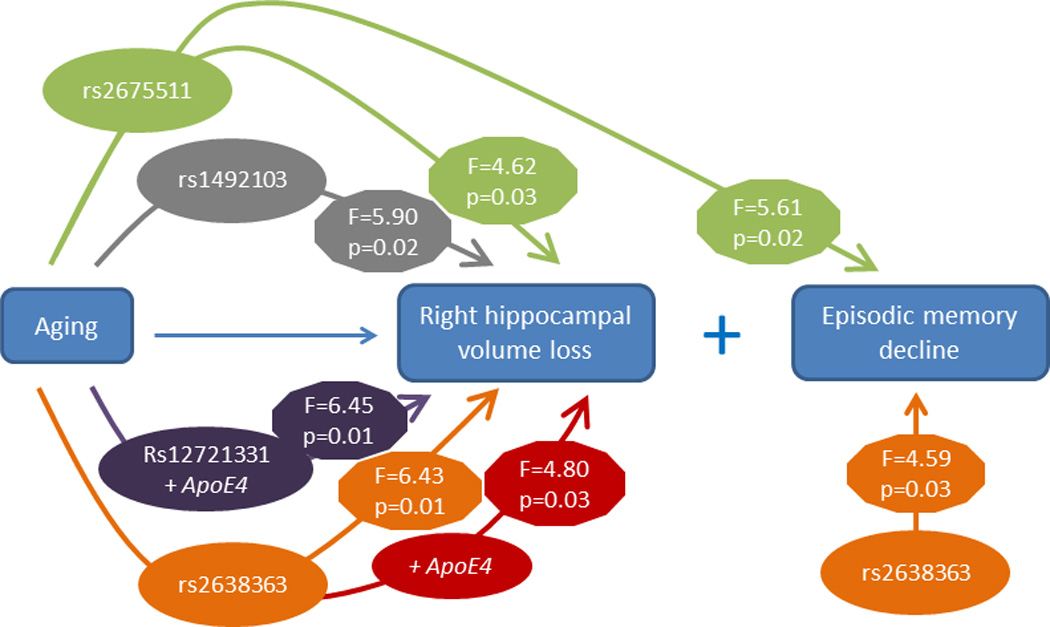
Genetic variants at three AGTR1 SNPs (rs2638363, rs1492103, rs2675511) were independently associated with accelerated hippocampal volume loss over the four-year follow-up in the right but not left hemisphere. Intriguingly, these AGTR1 risk alleles also predicted worse episodic memory performance but were not related to other cognitive measures. Two risk variants (rs2638363 and rs12721331) interacted with the APOE4 allele to accelerate right hippocampal volume loss.
Conclusions
Risk genetic variants of RAS may accelerate memory decline in older adults, an effect that may be conferred by accelerated hippocampal volume loss. Molecules involved in this system may hold promise as early therapeutic targets for late-life neuropsychiatric disorders.
Introduction
Genetic factors have been long hypothesized to confer risk for neuropsychiatric phenotypes, but efforts to identify genetic culprits have been challenging. The failure to identify risk genes may be in large part due to the polygenic etiology and phenotypic heterogeneity of neuropsychiatric disorders (1). These disorders result from complex interactions among multiple genes, tissue-specific epigenetic regulation, and environmental influences, which make identification of relationships between single genes and distal phenotypes challenging.
An alternative approach is to examine endophenotypes, measurable constructs that confer risk for complex disorders and are thought to lie in greater etiologic proximity to genetic factors (2). One promising endophenotype for late-life neuropsychiatric disorders is small hippocampal volume (3). Smaller hippocampus has been associated with treatment resistance in late-life depression (4) and predicts progressive cognitive decline in elderly subjects (5). Therefore, efforts to link hippocampal volume reduction with risk genes may be particularly relevant for late-life neuropsychiatric syndromes.
A novel and biologically plausible gene to investigate in these relationships is AGTR1, the gene encoding angiotensin-II type 1 (AT1) receptor in humans. Angiotensin II and AT1 receptors are the primary effector of the renin-angiotensin system (RAS) in several organs, including the brain. RAS is an important regulator of the stress response and AT1 receptors are expressed in brain regions that modulate stress and emotion, including the hypothalamus, amygdala and hippocampus (6, 7). In animal studies, RAS activation leads to hyperactivity of the stress system and heightened anxious behavior, whereas blockade of AT1 receptors dampens stress responses and ameliorates anxious and depressive behavior (6, 8). As RAS also plays central role in blood pressure regulation and has been implicated in vascular disease (9), examining this system may be particularly relevant for older adults with depression, since late-life depression is often characterized by vascular comorbidity (10).
Despite these theoretical implications, few studies have examined the relationship between genetic variation in AGTR1 and either neuropsychiatric phenotypes or hippocampal morphology. Studies on the common rs5186 AGTR1 (A-to-C) polymorphism have reported antidepressant response differences between genotypes in elderly depressed subjects (11–13). In a broader analysis of single nucleotide polymorphisms (SNPs) in AGTR1, our group reported that allele frequency differences in two AGTR1 SNPs increased the odds of late-life depression (14). We also found cross-sectional associations between right hippocampal volume and four AGTR1 intronic SNPs, namely rs2638363, rs1492103, rs2675511, and rs12721331 (14). To our knowledge, no studies have examined the effects of AGTR1 polymorphisms on longitudinal changes in hippocampal morphology in older adults. This is particularly important as reduction in hippocampal volume is associated with subsequent cognitive decline (5).
To extend our previous findings demonstrating cross-sectional relationships between AGTR1 and hippocampal morphology (14), we examined the effects of AGTR1 genotype on longitudinal change in hippocampus volume. Our a priori hypothesis was that the gene variants we previously associated with smaller cross-sectional hippocampus volume would also be associated with greater hippocampal volume loss over time. We also sought to determine if these gene variants were associated with differences in cognitive function over time, particularly in domains involving the hippocampus, such as episodic memory. To control for the effects of depression that has been shown to lead via chronic hyperactivity of the stress system to smaller hippocampal volumes (15), we examined two cohorts consisting of elderly depressed and non-depressed subjects. In secondary analyses, we examined whether depression diagnosis or apolipoprotein E (APOE) genotype had synergistic effects with AGTR1 genotypes on hippocampal volume change.
Methods
Study subjects and clinical care
All subjects were age 60 years or older and participated in the Conte Center for the Neuroscience of Depression in Late Life and the Neurocognitive Outcomes of Depression in the Elderly (NCODE) studies conducted at Duke University Medical Center (DUMC). The DUMC Institutional Review Board approved these studies and written informed consent was provided by eligible subjects.
Participants consisted of two cohorts, depressed patients and non-depressed comparison subjects. Depressed subjects were recruited primarily by clinical referral and secondarily by advertisements and met criteria for MDD as outlined in the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition. Diagnosis was based on the Diagnostic Interview Schedule (DIS, 16) and was confirmed by a geriatric psychiatrist during baseline clinical evaluation. Non-depressed subjects were community dwelling older adults recruited either from the Duke University Aging Center Subject Registry or through advertisements. All subjects underwent cognitive screening with the mini-mental state exam (MMSE, 17). Medical comorbidity was assessed with a previously used self-report questionnaire (18).
Exclusion criteria for all subjects were as follows: 1) presence of other major psychiatric disorder, such as schizophrenia or bipolar disorder; 2) history of substance abuse or dependence; 3) presence of neurologic disease; 4) metal in the body or other contraindication for magnetic resonance imaging (MRI); and 5) screening MMSE score lower than 25. Furthermore, non-depressed subjects were excluded if they had evidence of past psychiatric disorder based on the DIS.
Antidepressant treatment followed the Duke Somatic Treatment Algorithm for Geriatric Depression (19), which allows step-wise use of commercially available antidepressant modalities. The majority of depressed subjects were prescribed sertraline on study entry, but treatment differed among subjects and was guided by previous medication trials and depression severity. Following failed trials, switches to other antidepressant agents and augmentation strategies were allowed as clinically indicated. Treatment alternatives included psychotherapy and electroconvulsive therapy.
The current longitudinal study extends our past work (14), where in a separate cohort we found a relationship between four AGTR1 SNPs and right hippocampal volumes. Although there is some overlap in subjects, the current sample is much larger (138 vs. 70 subjects) and imaging data differ from that prior study. The prior study used 3T MRI data; the current study uses 1.5T MRI data as longitudinal 3T data were not available for the majority of participants. Similarly to our previous report and based on work showing racial differences in AGTR1 allele frequencies (14, 20), we limited the current analyses to Caucasian subjects with genotype data for all four AGTR1 SNPs and at least two hippocampal volume measurements (a baseline and at least one follow-up assessment).
Neuropsychological assessments
Neuropsychological testing was administered to study participants at baseline and then annually, regardless of the presence or absence of depressive symptoms. The battery is described fully elsewhere (21) and has been successfully employed in a number of clinical and epidemiological settings (22). Testing was administered by a trained psychometric technician supervised by a licensed clinical psychologist.
We created composite variables from the broader neuropsychological test battery that represented cognitive domains that may be adversely affected by aging. This was achieved by grouping neuropsychological tests into rational constructs similarly to previously published studies (23). We created Z-scores for each measure based on the performance of all participants and summed the Z-scores for all tests within each domain. Internal consistency for each domain was assured using Cronbach's coefficient alpha (CoA). Following this approach, we created four composite neurocognitive measures: a) episodic memory (Logical Memory, Benton Visual Retention Test, Word Learning Immediate, and Word List Recall, CoA 0.88); b) executive speed (Trails A time, Trails B time, Symbol-Digit Modality Test, CoA 0.86); c) verbal fluency (Verbal fluency test, Controlled Oral Word Association test, CoA 0.74); and d) working memory (Digits forward, digits backward, and digits ascending, CoA 0.74).
Genotyping and genetic analyses
All genotyping was performed on DNA from whole peripheral blood using the Gentra PureGene system (Qiagen, Valencia, CA, USA). Assays employed quality control procedures, which included serial genotyping of blinded duplicate samples. Quality requirements for each assay were met only if duplicate samples matched 100%. Efficiency of 95% was further required for each assay before statistical analyses. Deviations from Hardy–Weinberg Equilibrium (HWE) have been previously tested for all SNPs separately in the depressed and non-depressed cohorts (14), using exact tests per the Genetic Data Analysis program (24). Genotyped SNPs were identified using Linkage Disequilibrium (LD) Select (25). Selected SNPs had LD r2<0.64 and minor allele frequency of at least 0.10 in the HapMap project (www.hapmap.org), which was based on European ancestry Utah residents and provided genetic coverage of the entire AGTR1. In this study, we focused on the four SNPs that showed significant cross-sectional relationships with hippocampal volumes among the ten SNPs tested in our previous report (14). To examine whether these four SNPs can be treated as independent signals in our analyses, we estimated the pairwise LDs between all SNPs in our study population. All pairwise r2 values were found to be below 0.5. APOE genotype was determined using previously published methods (26).
MRI acquisition and analysis
Each subject was screened for contraindications and was scanned with a 1.5 Tesla, whole-body MRI system (Signa, GE Medical Systems, Milwaukee, WI) approximately every two years. All acquisitions were performed with the standard head (volumetric) radiofrequency coil. Using a previously described MRI acquisition protocol (27, 28), we first confirmed alignment by a rapid sagittal localizer scan and then obtained two dual-echo, fast spin-echo acquisitions: one in the axial plane for cerebral morphometry and one in a coronal oblique plane for hippocampal morphometry.
Images were then analyzed at the Duke Neuropsychiatric Imaging Research Laboratory (NIRL). Segmentation of tissue and measurement of total cerebral volume, which included the total white and gray matter and CSF volumes in both hemispheres, was performed as previously described (27). Image analysts received extensive training and reliability was established before any data processing by repeated measurements on multiple MRIs separated by at least a week. Intraclass correlation coefficients were as follows: left hippocampus=0.8; right hippocampus=0.7; and total cerebral volume=0.997.
Delineation of the hippocampus was based on previously described methods (28). Analysts began with the most posterior coronal slice and moved anteriorly, measuring the hippocampus where the pulvinar nucleus of the thalamus obscured the crura fornicis on each side. The fimbria and the thin strip of gray matter along the medial border of the hippocampus were transected at their narrowest points. Tracing continued around the hippocampal body to the starting point. The anterior border of the hippocampus was defined as the slice on which the inferolateral ventricle appeared horizontally without any body of gray matter visible below it. The amygdala-hippocampal transition zone, which was transected at its narrowest point, appeared as a diffuse area of gray matter between the anterior portion of the hippocampus and the posterior portion of the amygdala.
Statistical analyses
All statistical tests were performed with SAS version 9.2 (Cary, NC, USA). The level of statistical significance was set a priori at α=0.05 and all P values were two-tailed. To maximize power in our analyses, we dichotomized each SNP into two genotype groups, the major allele homozygotes and the minor allele carriers. This further increased comparability with our previous report (14), where the same genotype groups were used. The two diagnostic cohorts (depressed and non-depressed) were compared at baseline for differences in demographic variables and baseline measures. Categorical variables were compared with χ2 tests, equal-variance continuous variables with pooled two-sample t tests, and unequal-variance continuous variables with Satterthwaite t tests.
We next examined the longitudinal effects of AGTR1 polymorphisms on hippocampal volumes and composite cognitive measures. To analyze these longitudinal data, we performed linear mixed effects models (29) using the PROC MIXED command in SAS 9.3. Analyses included the maximum number of subjects with data for longitudinal time points and for all variables included in the models. In these models, each subject was the independent sampling unit and measurement of each subject at a particular point in time was the observation unit. Separate models were created for the right and left hippocampus and for each composite cognitive measure, with each AGTR1 SNP genotype group, time (as a continuous variable), and genotype by time interaction as the main independent variables. In order to account for between-subject variability in brain volume, we included cerebral volume as covariate in models examining hippocampal volumes as dependent variable. Other covariates included in all models were sex, age, diagnostic group (depressed/non-depressed), and the respective baseline hippocampal volume or cognitive measure. The primary effects of interest were the genotype main effect, which examines the genotype effect irrespective of time, and the genotype by time interaction, which examines the rate of change in the mean hippocampal volume or cognitive measure over time between the two genotype groups for each AGTR1 SNP. Secondary analyses tested three-way depression by genotype by time and AGTR1 genotype by APOE genotype by time interactions. All models were initially run with the interaction terms, and these terms remained in the model if significant but were excluded if non-significant.
Results
Sample demographics and baseline measures
The primary sample included 138 elderly subjects (79 depressed and 59 non-depressed) with genotype data for AGTR1 SNPs and at least two MRI scans (a baseline and one follow-up assessment). The demographics and clinical data are shown on Table 1. Age ranged between 60 and 84 for the depressed cohort and between 60 and 82 for the non-depressed cohort. The two groups were similar in genotype frequencies, age, MMSE scores and baseline left and right hippocampal volumes. However, the percentage of female subjects and the educational level were higher in the non-depressed group. The percentage of patients who reported hypertension was higher in the depressed group. Finally, we found that the number of patients who reported a history of either cardiac complaints or hypertension did not significantly differ between the four AGTR1 or APOE genotype groups (data not shown). The maximum number of MRI measures per subject was 5 and there was no significant difference in length of study follow-up between diagnostic cohorts (depressed: 1435.3 (502.5) days, non-depressed: 1495.9 (785.8) days; Satterthwaite t-test = 130, 2 df, t = 0.55, P = 0.5855).
Table 1.
Demographic variables and genotype frequencies
| Demographic variables (N=138) |
Depressed (N = 79) | Non-depressed (N = 59) | Test statistic | P value | ||
|---|---|---|---|---|---|---|
| Means or Ns |
SDs | Means or Ns | SDs | |||
| Age, years | 69.6 | 6.7 | 69.7 | 5.8 | T136 = 0.05 | 0.9611 |
| Sex | χ2 = 4.08, 1 df | 0.0433 | ||||
| Female | 52 | - | 48 | - | ||
| Male | 27 | - | 11 | - | ||
| Education, years | 14.5 | 2.1 | 15.7 | 1.6 | T134 = 4.08 | 0.0004 |
| History of cardiac complaint | χ2 = 0.2, 1 df | 0.6517 | ||||
| Yes | 14 | - | 9 | - | ||
| No | 63 | - | 50 | - | ||
| History of hypertension | χ2 = 5.82, 1 df | 0.0158 | ||||
| Yes | 29 | 11 | ||||
| No | 48 | 48 | ||||
| Mini-mental state exam | 28.8 | 1.3 | 29.1 | 1.1 | T132 = 1.35 | 0.1808 |
| Baseline right hippocampal volume, ml | 3.08 | 0.39 | 3.13 | 0.42 | T136= 0.85 | 0.3968 |
| Baseline left hippocampal volume, ml | 2.97 | 0.4 | 3.00 | 0.45 | T136 = 0.47 | 0.6395 |
| Cerebrum, ml | 1164.8 | 128.3 | 1140.6 | 117.2 | T136 = 1.12 | 0.2645 |
| rs2638363 (A/G) | χ2 = 2.39, 1 df | 0.1220 | ||||
| GG | 58 | - | 36 | - | ||
| AA/AG | 21 | - | 23 | - | ||
| rs1492103 (C/T) | χ2 = 0.87, 1 df | 0.3522 | ||||
| TT | 58 | - | 39 | - | ||
| CC/CT | 21 | - | 20 | - | ||
| rs12721331 (C/T) | χ2 = 1.43, 1 df | 0.2312 | ||||
| TT | 70 | - | 48 | - | ||
| CC/CT | 9 | - | 11 | - | ||
| rs2675511 (A/G) | χ2 = 1.03, 1 df | 0.3107 | ||||
| AA | 40 | - | 35 | - | ||
| GG/GA | 39 | - | 24 | - | ||
| APOE | χ2 = 0.46, 1 df | 0.4994 | ||||
| APOE4 carriers | 20 | - | 18 | - | ||
| No APOE4 | 59 | - | 41 | - | ||
Continuous variables are presented as means and standard deviations (SDs). Categorical variables are presented as Ns. All continuous variables had equal variances and were tested with pooled T-test, except for education which exhibited unequal variances between the two diagnostic groups and were thus tested using Satterthwaite T-test. Reported P values are two-tailed.
Abbreviations: SD, standard deviation; MMSE, mini-mental state exam
Longitudinal effects of AGTR1 SNPs on hippocampal volumes
We created mixed models examining hippocampus volume in the left or right hemisphere as repeatedly measured dependent variables. Independent variables included diagnosis (depressed/non-depressed), age, sex, cerebral volume, baseline hippocampal volume, time, and AGTR1 SNP genotype. To test the hypothesis that SNPs would differentially affect hippocampal volume over time, we included a SNP by time interaction term. Table 2 presents the effects of the four AGTR1 SNPs examined and their interactions with time on left and right hippocampal volumes. When examining models of right hippocampal volume, three of the four SNPs (rs2638363, rs1492103, rs2675511) showed both a statistically significant primary effect as well as an interactive effect with time (Table 2). The primary effect of AGTR1 SNPs replicated our previous findings (14), despite the nearly double sample size and different MRI field strength. Again, homozygous subjects for the vascular risk alleles (rs2638363 GG, rs1492103 TT, and rs2675511 AA) exhibited smaller hippocampal volumes when compared to individuals who were heterozygous or homozygous for the alternate allele. The SNP by time interaction tested for differences between SNP alleles on change in hippocampal volume over time. When examining these interactions, individuals homozygous for the risk alleles exhibited accelerated decrease in right hippocampal volume over time as compared with individuals with alternate genotypes. However, the four SNPs exhibited neither a significant primary effect nor a significant gene by time interaction on left hippocampal volume. In other words, the effect of all three SNPs was selective and lateralized to the right hemisphere.
Table 2.
Longitudinal effects of AGTR1 SNPs on hippocampal volumes
| Right hippocampal volume | Left hippocampal volume | |||
|---|---|---|---|---|
| F value | P value | F value | P value | |
| rs2638363 | 5.96 | 0.0160 | 0.81 | 0.3693 |
| rs2638363 by time | 6.43 | 0.0124 | 0.11 | 0.7400 |
| rs2638363 by APOE by time | 4.80 | 0.0303 | 0.14 | 0.7115 |
| rs1492103 | 5.01 | 0.0269 | 0.72 | 0.3967 |
| rs1492103 by time | 5.90 | 0.0166 | 0.10 | 0.7558 |
| rs1492103 by APOE by time | 3.78 | 0.0541 | 0.01 | 0.9100 |
| rs12721331 | 0.20 | 0.6564 | 0.96 | 0.3297 |
| rs12721331 by time | 1.29 | 0.2574 | 0.09 | 0.7709 |
| rs12721331 by APOE by time | 6.45 | 0.0123 | 0.07 | 0.7982 |
| rs2675511 | 4.06 | 0.0459 | 0.30 | 0.5842 |
| rs2675511 by time | 4.62 | 0.0335 | 0.23 | 0.6352 |
| rs2675511 by APOE by time | 0.75 | 0.3890 | 0.3 | 0.5876 |
Analyses conducted using linear mixed effects models. These models tested for the effect of AGTR1 SNP genotype on hippocampal volume and also examined AGTR1 by time and AGTR1 by APOE by time interactions. All models included 138 subjects and controlled for sex, age, diagnostic group (depressed/non-depressed), baseline hippocampal volume, and total cerebral volume. Values in bold denote statistically significant parameters, defined as P < 0.05. Reported P values are two-tailed.
In secondary analyses we tested whether AGTR1 genetic variation has synergistic effects with depression or APOE genotype on the rate of hippocampal volume change. There were no significant three-way depression by AGTR1 by time interactions for any of the SNPs for either the left or right hippocampus. However, we did observe significant gene-gene interactions. APOE genotype had statistically significant epistatic effects with rs2638363 and rs12721331 for the right hippocampus volume over time (Table 2). In both cases, presence of both the risk AGTR1 allele and APOE4 allele was associated with accelerated hippocampal volume loss. We observed a similar trend that did not achieve statistical significance for the interaction between rs1492103, APOE, and time (Table 2).
Longitudinal effects of AGTR1 SNPs on composite cognitive measures
We next tested whether AGTR1 variants confer risk for cognitive decline. A total of 138 elderly subjects (63 depressed and 75 nondepressed) had both genotype data and neurocognitive measures and were included in these analyses. This sample partially overlaps with the sample examining hippocampal volumes, with 81 subjects (36 depressed and 45 nondepressed) included in both analyses. As expected depressed subjects performed significantly worse than nondepressed subjects on univariate comparisons of the four composite cognitive measures of episodic memory (T106=5.14, P<0.0001), executive speed (T67=4.36, P<0.0001), verbal fluency (T136=5.18, P<0.0001), and working memory (T114=2.97, P=0.0037).
We used mixed models to examine the effect of AGTR1 genotypes and their interaction with time on each of the composite cognitive measures. All models controlled for diagnosis (depressed/non-depressed), age, sex, education, time, and baseline cognitive measure score. In these models, only rs2638363 showed a significant main effect of genotype on composite episodic memory performance (F1,126=4.59, P=0.0340), but did not exhibit a statistically significant genotype by time interaction. Supportingly, rs1492103 exhibited a trend for a direct effect on episodic memory (F1,126=3.85, P=0.0519), but this did not achieve statistical significance. Finally, rs2675511 exhibited a gene by time interaction (F1,125=5.61, P=0.0194) predicting worsening episodic memory performance. In accordance with SNP effects on hippocampal volumes, homozygous subjects for the vascular risk alleles (rs2638363 GG, rs1492103 TT, and rs2675511 AA) exhibited lower episodic memory scores. None of the four genetic variants exhibited either a statistically significant direct effect or interaction with time effect on executive speed, verbal fluency, or working memory.
Discussion
Replicating and extending our previous findings of AGTR1 effects on right hippocampal morphology (14), three of the same SNPs predicted longitudinal hippocampal volume change in the right but not the left hemisphere in elderly subjects. The direction of the observed relationships was similar to the cross-sectional study, that is, each of the previously identified “risk” genotypes (rs2638363 GG, rs1492103 TT, and rs2675511 AA) predicted longitudinally greater shrinkage of the right hippocampus when compared to the alternate genotypes. Intriguingly, these AGTR1 risk alleles also predicted poorer performance in episodic memory, a neurocognitive measure mediated by the hippocampus, but had no effects on other cognitive measures. Additionally, two risk variants (rs2638363 and rs12721331) showed epistatic effects with the APOE4 allele, a known risk factor for dementia (26), on right hippocampal volume loss over time. Importantly, the SNPs examined are in low pairwise LD (r2 < 0.5) and thus represent independent signals. The presence of several signals within the AGTR1 locus that are independently associated with hippocampal volume changes and memory decline lends further support to the reliability of these associations. Taken together, our findings strongly support that AGTR1 gene variants via both direct and epistatic effects may confer vulnerability for progressive memory decline by accelerating hippocampal volume loss.
Our study offers novel insights into the role of RAS in hippocampal atrophy and cognitive decline. Hippocampal atrophy has been shown to predict cognitive decline in late life and time to conversion to Alzheimer disease (5, 30). Prior animal and human studies support involvement of the RAS in cognitive syndromes (6, 14, 31). RAS activation modulates cerebral blood flow, increases brain vulnerability to ischemia, and promotes brain inflammation (6, 32). These effects render this system particularly relevant in late life, where vascular and inflammatory processes are hypothesized to contribute to depressive and cognitive syndromes (10, 33). On the other hand, AT1 receptor blockade dampens stress responses, ameliorates anxious and depressive behaviors, and reduces brain inflammation and vulnerability to ischemia (6, 8, 32). Thus, it is plausible that perturbations in RAS could heighten the risk for development of dementia by accelerating age-related changes in brain morphology, particularly in brain regions most vulnerable to the effects of aging, such as the hippocampus. Although RAS activity may be modulated by functional variants at the AGTR1 locus, the molecular effects of the risk AGTR1 SNPs are currently not clear. Despite their intronic location, these SNPs could have functional effects. For instance, intronic SNPs may be located in enhancer regions that influence the three-dimensional changes in chromatin conformation necessary for transcription regulation (34). Alternately, these SNPs may be tagging other functional variants in nearby regions of the AGTR1 locus. Finally, as supported by the epistatic effects with APOE observed in our study, these SNPs may have functional effects via interaction with other genes that lie in common biological pathways central for the development of brain pathology and dementia. Future studies on the functional effects of these genetic variants are warranted and may shed light on the mechanisms linking AGTR1 and cognitive syndromes.
Intriguingly, the effects of AGTR1 variation were lateralized to the right hippocampus. This is in accordance with our previous cross-sectional study (14). Although the significance of this lateralization is unclear, hippocampal asymmetry may be an important endophenotype that has been underemphasized by previous studies. Hippocampal asymmetry has been observed in patients with severe depression and in non-depressed relatives of depressed subjects (35, 36). Furthermore, unilateral hippocampal volume changes may be associated with decline in specific cognitive outcomes (5, 37) and with decreased likelihood to achieve antidepressant remission (4). Thus, hippocampal asymmetry may be an early step in the pathogenesis of some late-life mood and cognitive syndromes. This asymmetry could be induced by differential hemispheric effects of the RAS on the hippocampus. Notably, activity of angiotensinase, an enzyme that metabolizes angiotensin, has been found to be distributed asymmetrically between the left and right hippocampi of the rat brain (38, 39). This may also reflect differential activity of angiotensin and potentially asymmetric effects of AGTR1 genetic variants between the two hemispheres. Whether this mechanism holds true and the clinical significance of these asymmetries remain to be determined.
Several limitations should be considered when interpreting our findings. Although we controlled for the effects of depression diagnosis and basic demographic factors on hippocampal morphology, we did not control for antidepressant treatments that may have neurotrophic effects on the hippocampus (40) and even reverse hippocampal volume loss in some cases of depression (37). Depressed participants were treated based on an algorithm rather than a rigid clinical trial. Although this makes our approach comparable to clinical practice, it makes it challenging to elucidate the effects of antidepressants. Data for each subject on antidepressant use prior to enrollment, as well as specific treatment modalities, duration and doses used over the study period were not known. Thus, it is possible that variable treatments between genotype groups might have influenced our results. However, the assignment of treatment modalities occurred randomly, treating clinicians were blinded regarding genotype groups, and thus systematic errors cannot account for our findings. Another limitation was our focus on variation at a single genetic locus and a single brain region. This was based on our sample size and the a-priori plausible involvement of AGTR1 and hippocampal pathology in late-life neuropsychiatric syndromes. Larger longitudinal studies are warranted for a more systematic examination of the multiple brain regions, genes, and biological pathways involved in the pathogenesis of these syndromes. Our study shows that RAS is one such pathway that should be included in future pathogenetic models.
In summary, this is the first study to explore the longitudinal effects of AGTR1 genetic variation on hippocampal morphology and cognitive decline. In contrast with cross-sectional approaches that cannot establish temporal relationships, the current study shows that older adults homozygous for AGTR1 risk variants exhibit accelerated hippocampal volume loss and memory decline. Our study exemplifies how examining the longitudinal effects of biologically plausible genotypes on relevant endophenotypes may provide novel insights into the pathogenesis of complex phenotypes. It further suggests that molecules involved in RAS may serve as early therapeutic targets for late-life neuropsychiatric syndromes.
Figure 1.
Acknowledgments
Funding
This work was funded by research grants R01 MH077745, R01 MH054846, and K24 MH070027.
Footnotes
Disclosure
The authors declare no conflicts of interest.
References
- 1.Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 3.Hasler G, Drevets WC, Manji HK, Charney DS. Discovering endophenotypes for major depression. Neuropsychopharmacology. 2004;29:1765–1781. doi: 10.1038/sj.npp.1300506. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh MH, McQuoid DR, Levy RM, Payne ME, MacFall JR, Steffens DC. Hippocampal volume and antidepressant response in geriatric depression. Int J Geriatr Psychiatry. 2002;17:519–525. doi: 10.1002/gps.611. [DOI] [PubMed] [Google Scholar]
- 5.Steffens DC, McQuoid DR, Payne ME, Potter GG. Change in hippocampal volume on magnetic resonance imaging and cognitive decline among older depressed and nondepressed subjects in the neurocognitive outcomes of depression in the elderly study. Am J Geriatr Psychiatry. 2011;19:4–12. doi: 10.1097/JGP.0b013e3181d6c245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saavedra JM, Sánchez-Lemus E, Benicky J. Blockade of brain angiotensin II AT1 receptors ameliorates stress, anxiety, brain inflammation and ischemia: Therapeutic implications. Psychoneuroendocrinology. 2011;36:1–18. doi: 10.1016/j.psyneuen.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsutsumi K, Saavedra JM. Characterization and development of angiotensin II receptor subtypes (AT1 and AT2) in rat brain. Am J Physiol. 1991;261(1 Pt 2):R209–R216. doi: 10.1152/ajpregu.1991.261.1.R209. [DOI] [PubMed] [Google Scholar]
- 8.Nayak V, Patil PA. Antidepressant activity of fosinopril, ramipril and losartan, but not of lisinopril in depressive paradigms of albino rats and mice. Indian J Exp Biol. 2008;46:180–184. [PubMed] [Google Scholar]
- 9.Duprez DA. Role of the renin-angiotensin-aldosterone system in vascular remodeling and inflammation: a clinical review. J Hypertens. 2006;24:983–991. doi: 10.1097/01.hjh.0000226182.60321.69. [DOI] [PubMed] [Google Scholar]
- 10.Taylor WD, Aizenstein HJ, Alexopoulos GS. The vascular depression hypothesis: mechanisms linking vascular disease with depression. Mol Psychiatry. 2013;18:963–974. doi: 10.1038/mp.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bondy B, Baghai TC, Zill P, Schule C, Eser D, Deiml T, Zwanzger P, Ella R, Rupprecht R. Genetic variants in the angiotensin I-converting-enzyme (ACE) and angiotensin II receptor (AT1) gene and clinical outcome in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1094–1099. doi: 10.1016/j.pnpbp.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Kondo DG, Speer MC, Krishnan KR, McQuoid DR, Slifer SH, Pieper CF, Billups AV, Steffens DC. Association of AGTR1 with 18-month treatment outcome in late-life depression. The Am J Geriatr Psychiatry. 2007;15:564–572. doi: 10.1097/JGP.0b013e31805470a4. [DOI] [PubMed] [Google Scholar]
- 13.Saab YB, Gard PR, Yeoman MS, Mfarrej B, El-Moalem H, Ingram MJ. Renin–angiotensin-system gene polymorphisms and depression. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:1113–1118. doi: 10.1016/j.pnpbp.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Taylor WD, Benjamin S, McQuoid DR, Payne ME, Krishnan RR, MacFall JR, Ashley Koch A. AGTR1 gene variation: Association with depression and frontotemporal morphology. Psychiatry Res. 2012;202:104–109. doi: 10.1016/j.pscychresns.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheline YI, Gado MH, Kraemer HC. Untreated depression and hippocampal volume loss. Am J Psychiatry. 2003;160:1516–1518. doi: 10.1176/appi.ajp.160.8.1516. [DOI] [PubMed] [Google Scholar]
- 16.Robins LN, Helzer JE, Croughan J, Ratcliff KS. National Institute of Mental Health Diagnostic Interview Schedule. Its history, characteristics, and validity. Arch Gen Psychiatry. 1981;38:381–389. doi: 10.1001/archpsyc.1981.01780290015001. [DOI] [PubMed] [Google Scholar]
- 17.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 18.Taylor WD, McQuoid DR, Krishnan KR. Medical comorbidity in late-life depression. Int J Geriatr Psychiatry. 2004;19:935–943. doi: 10.1002/gps.1186. [DOI] [PubMed] [Google Scholar]
- 19.Steffens DC, McQuoid DR, Krishnan KRR. The Duke Somatic Treatment Algorithm for Geriatric Depression (STAGED) approach. Psychopharmacology Bulletin. 2002;36:58–68. [PubMed] [Google Scholar]
- 20.Hindorff LA, Heckbert SR, Tracy R, Tang Z, Psaty BM, Edwards KL, Siscovick DS, Kronmal RA, Nazar-Stewart V. Angiotensin II type 1 receptor polymorphisms in the cardiovascular health study: relation to blood pressure, ethnicity, and cardiovascular events. Am J Hypertens. 2002;15:1050–1056. doi: 10.1016/s0895-7061(02)03063-7. [DOI] [PubMed] [Google Scholar]
- 21.Steffens DC, Welsh-Bohmer KA, Burke JR, Plassman BL, Beyer JL, Gersing KR, Potter GG. Methodology and preliminary results from the neurocognitive outcomes of depression in the elderly study. J Geriatr Psychiatry Neurol. 2004;17:202–211. doi: 10.1177/0891988704269819. [DOI] [PubMed] [Google Scholar]
- 22.Tschanz JT, Welsh-Bohmer KA, Skoog I, West N, Norton MC, Wyse BW, Nickles R, Breitner JC. Dementia diagnoses from clinical and neuropsychological data compared: the Cache County study. Neurology. 2000;54:1290–1296. doi: 10.1212/wnl.54.6.1290. [DOI] [PubMed] [Google Scholar]
- 23.Sheline YI, Pieper CF, Barch DM, Welsh-Bohmer K, McKinstry RC, MacFall JR, D'Angelo G, Garcia KS, Gersing K, Wilkins C, Taylor W, Steffens DC, Krishnan RR, Doraiswamy PM. Support for the vascular depression hypothesis in late-life depression: results of a 2-site, prospective, antidepressant treatment trial. Arch Gen Psychiatry. 2010;67:277–285. doi: 10.1001/archgenpsychiatry.2009.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaykin D, Zhivotovsky L, Weir BS. Exact tests for association between alleles at arbitrary numbers of loci. Genetica. 1995;96:169–178. doi: 10.1007/BF01441162. [DOI] [PubMed] [Google Scholar]
- 25.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;48:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 27.Payne ME, Fetzer DL, MacFall JR, Provenzale JM, Byrum CE, Krishnan KRR. Development of a semi-automated method for quantification of MRI gray and white matter lesions in geriatric subjects. Psychiatry Res. 2002;115:63–77. doi: 10.1016/s0925-4927(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Steffens DC, Byrum CE, McQuoid DR, Greenberg DL, Payne ME, Blitchington TF, MacFall JR, Krishnan KR. Hippocampal volume in geriatric depression. Biol Psychiatry. 2000;48:301–309. doi: 10.1016/s0006-3223(00)00829-5. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J, Edwards LJ, Maldonado-Molina MM, Komro KA, Muller KE. Real longitudinal data analysis for real people: building a good enough mixed model. Stat Med. 2010;29:504–520. doi: 10.1002/sim.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Honig LS, Mayeux R, Stern Y, Tabert MH, de Leon MJ. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- 31.Wright JW, Harding JW. The brain RAS and Alzheimer's disease. Exp Neurol. 2010;223:326–333. doi: 10.1016/j.expneurol.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Saavedra JM, Nishimura Y. Angiotensin and cerebral blood flow. Cell Mol Neurobiol. 1999;19:553–573. doi: 10.1023/a:1006995016403. [DOI] [PubMed] [Google Scholar]
- 33.Alexopoulos GS, Morimoto SS. The inflammation hypothesis in geriatric depression. Int J Geriatr Psychiatry. 2011;26:1109–1118. doi: 10.1002/gps.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zannas AS, Binder EB. Gene-environment interactions at the FKBP5 locus: sensitive periods, mechanisms and pleiomorphism. Genes Brain and Behavior. 2014;13:25–37. doi: 10.1111/gbb.12104. [DOI] [PubMed] [Google Scholar]
- 35.Boccardi M, Almici M, Bresciani L, Caroli A, Bonetti M, Monchieri S, Gennarelli M, Frisoni GB. Clinical and medial temporal features in a family with mood disorders. Neurosci Lett. 2010;468:93–97. doi: 10.1016/j.neulet.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 36.Mervaala E, Föhr J, Könönen M, Valkonen-Korhonen M, Vainio P, Partanen K, Partanen J, Tiihonen J, Viinamäki H, Karjalainen AK, Lehtonen J. Quantitative MRI of the hippocampus and amygdala in severe depression. Psychol Med. 2000;30:117–125. doi: 10.1017/s0033291799001567. [DOI] [PubMed] [Google Scholar]
- 37.Hou Z, Yuan Y, Zhang Z, Bai F, Hou G, You J. Longitudinal changes in hippocampal volumes and cognition in remitted geriatric depressive disorder. Behav Brain Res. 2012;227:30–35. doi: 10.1016/j.bbr.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 38.Banegas I, Prieto I, Alba F, Vives F, Araque A, Segarra AB, Durán R, de Gasparo M, Ramírez M. Angiotensinase activity is asymmetrically distributed in the amygdala, hippocampus and prefrontal cortex of the rat. Behav Brain Res. 2005;156:321–326. doi: 10.1016/j.bbr.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Wu HM, Wang C, Wang XL, Wang L, Chang CW, Wang P, Gao GD. Correlations between angiotensinase activity asymmetries in the brain and paw preference in rats. Neuropeptides. 2010;44:253–259. doi: 10.1016/j.npep.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Castrén E. Neurotrophic effects of antidepressant drugs. Curr Opin Pharmacol. 2004;4:58–64. doi: 10.1016/j.coph.2003.10.004. [DOI] [PubMed] [Google Scholar]


